Engineering a stable future for DNA-origami as a biomaterial
Abstract
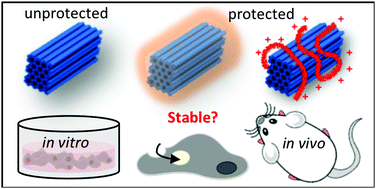
DNA as a biomaterial has evoked great interest as a potential platform for therapeutics and diagnostics and as hydrogel scaffolds due to the relative ease of programming its robust and uniform shape, site-specific functionality and controlled responsive behavior. However, for a stable self-assembled product, a relatively high cation concentration is required to prevent denaturation. Physiological and cell-culture conditions do not match these concentrations and present additional nucleases that cause a serious threat to the integrity of DNA-based materials. For the translation of this promising technology towards bioengineering challenges, stability needs to be guaranteed. Over the past years, various methods have been developed addressing the stability-related weaknesses of DNA-origami. This mini-review explains the common stability issues and compares the stabilization strategies recently developed. We present a detailed overview of each method in order to ease the selection process on which method to use for future users of DNA-origami as a biomaterial.
- This article is part of the themed collection: Biomaterials Science Emerging Investigators


 Please wait while we load your content...
Please wait while we load your content...