Conjugation of carboxymethyl cellulose and dopamine for cell sheet harvesting†
Soyoung
Hong
 a,
Jeong Hey
Sunwoo
ab,
Ji Seon
Kim
a,
Jeong Hey
Sunwoo
ab,
Ji Seon
Kim
 a,
Hungwon
Tchah
*b and
Changmo
Hwang
a,
Hungwon
Tchah
*b and
Changmo
Hwang
 *ac
*ac
aBiomedical Engineering Research Center, Asan Institute for Life Sciences, Asan Medical Center, Seoul 05505, Republic of Korea. E-mail: hwang.changmo1@gmail.com
bDepartment of Convergence Medicine, University of Ulsan College of Medicine & Asan Institute for Life Sciences, Asan Medical Center, Seoul 05505, Republic of Korea. E-mail: hwtchah@amc.seoul.kr
cDepartment of Convergence Medicine, University of Ulsan College of Medicine & Asan Institute for Life Sciences, Asan Medical Center, Seoul 05505, Republic of Korea
First published on 29th October 2018
Abstract
In this study, we investigated the feasibility of enzymatic digestion of polysaccharides for cell sheet harvesting. Cellulose was digested using cellulase; in brief, cellulose was pre-coated under a confluent cell layer, and then enzymatic digestion of cellulose under the confluent cell layer enabled cell detachment with minimal cell damage, yielding cell sheets. For the surface adhesion of the cellulose, carboxymethyl cellulose (CMC) molecules were conjugated with dopamine (DA), and the synthesized CMC–DA was pre-treated onto the surface of the culture plates. Then, human mesenchymal stem cells (hMSCs) or corneal limbal epithelial cells (hCLEs) were cultured on the pre-coated CMC–DA and harvested using cellulase containing cell culture medium. Single hMSCs treated with cellulase showed higher proliferative activity, showing an aggregated morphology compared with trypsin-treated hMSCs. Additionally, hMSC sheets were detached from the pre-coated CMC–DA surface 10 min after cellulase treatment. Also, hCLE sheets were generated with a well-preserved morphology and transparency after cellulase-assisted cell sheet generation. These results demonstrate that the strategy of CMC–DA coating combined with cellulase enzymatic harvesting is an effective option for harvesting cell sheets.
Introduction
In the fields of tissue engineering and regenerative medicine, cell sheet engineering has multiple applications, including myocardial,1 esophageal,2 and corneal regeneration,3 in cell-based therapies.When forming cell sheets, the autocrine extracellular matrix (ECM) of the cells could play an important role in the adhesion and coordination of interactions among the cells,4 and maintain stable cell–cell interactions among confluent cells.5In vitro, cell adhesion plays an integral role in cell communication and regulation of the ability of a single cell to stick to another cell or an ECM. Most mammalian cells are anchorage-dependent and attach firmly to the substrate or the surface of tissue culture polystyrene (TCPS)6 with the use of integrins, cadherins, and focal adhesion kinase (FAK). The detachment of cells cultured on TCPS usually requires harsh enzymatic methods, such as the trypsin/ethylenediaminetetraacetic acid (EDTA)-based detaching procedure (trypsinization), which have deleterious effects on the morphology and secretory function of the cells being harvested. Removal by trypsinization tends to yield disaggregated cells with a rounded appearance and damage to the ECM underlying the cells. Conventionally, cell sheets are harvested using temperature-responsive culture dishes and proteolytic enzymes, such as dispase. Temperature-responsive culture dishes treated with poly(N-isopropylacrylamide) (pNIPAAM) are generally used to generate cell sheets. The cells adhered to and cultured on the pNIPAAM-treated surface are released from pNIPAAM at temperatures below 32 °C.7 Although this temperature-responsive cell sheet harvesting technique has proven to be an effective in vivo strategy,8 it has some limitations. The UpCell™ dishes for temperature-sensitive cell sheet generation are expensive and difficult to apply to numerous experiments for clinical applications. Under laboratory conditions, expensive equipment and procedures are required, such as electron beam (EB) and plasma polymerization for coating pNIPAAM to the culture plate.9 Also, temperature changes during cell detachment may affect gene expression in the detached cells.10
In this article, we suggest a simple method of cell sheet generation that does not use expensive facilities or materials or cause harmful reactions. We used cellulose and cellulase for cell sheet generation without using a trypsin-based enzyme. Cellulose is a polysaccharide that consists of glucose units and depolymerizes to yield glucose after digestion by cellulase, an enzyme that lyses cellulose. Cellulose is applied to tissue engineering11 due to its biocompatibility,12 biodegradability,13 and mechanical strength.14 Additionally, methylcellulose has been used as one of the constituents of the ophthalmic drops for dry eye.15 Cellulase is an enzyme that hydrolyzes cellulose into the simple sugar beta-glucose.16 Cellulase showed biocompatibility in in vitro and in vivo studies including cytotoxicity and animal experiments.17,18 In a previous trial investigating cell sheet engineering with cellulose and cellulase, Sakai et al. demonstrated a fibroblast cell sheet harvested with crosslinked cellulose–tyramine conjugate hydrogel.19 Also, Ko et al. generated an endothelial cell sheet using a cellulose coating with the addition of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and digestion of the coated cellulose with cellulase.20
Here, we also investigated the feasibility of harvesting the cell sheet cultured on the adhered cellulose confluently by degrading cellulose using cellulase. To pre-coat cellulose on to the cell culture surface, the surface coating process is a key strategy because the cellulose detachment reaction must be prohibited during the cell culture period under wet conditions until cell sheet harvesting with cellulase. Dopamine (DA) is present in mussel adhesive proteins, Mefp-3,5,21 which contain catecholic amino acids and have attracted substantial interest because of the biomedical applications of their adhesive properties.22,23 Self-polymerization of dopamine formed an adherent polydopamine in a typical buffer (10 mM Tris, pH 8.5).23 The polydopamine can serve as a bridge for secondary surface-mediated reactions for grafted polymer coatings. Hyaluronic acid,24 alginate,25 poly(ethylenimine) (PEI),26 dextran,27 and chitosan28,29 have been reported as biopolymers that can be conjugated with dopamine or catecholamines to form adhesive polymers. In this study, we also conjugated dopamine (DA) with carboxymethyl cellulose (CMC) to conduct a simple coating with cellulose by dopamine bonding on the surface of the culture plate for cell sheet engineering.
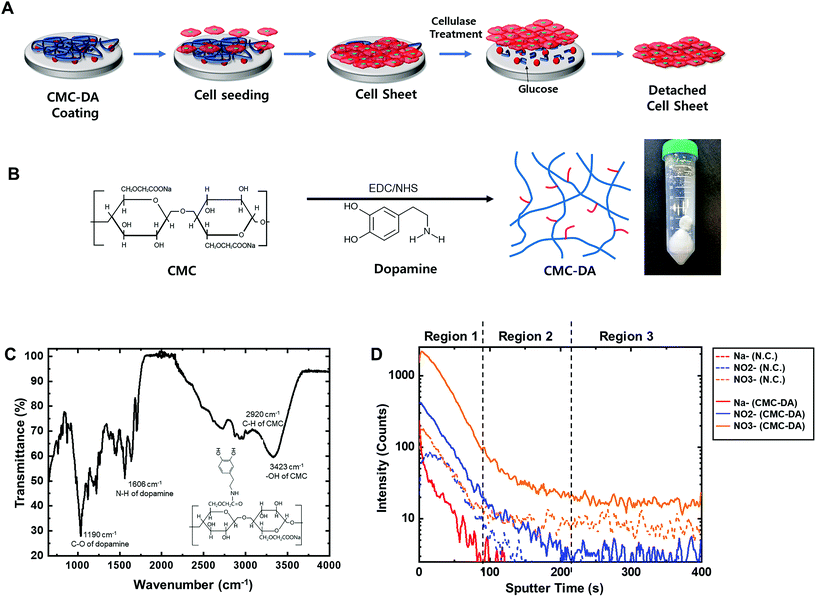
In brief, CMC and DA were conjugated and the synthesized CMC–DA was pre-coated under the confluent cell layer; then the enzymatic digestion of cellulose via the addition of cellulase below the confluent cell layer enabled cell detachment with minimal cell damage, yielding cell sheets (Fig. 1A). Human mesenchymal stem cells (hMSCs) and human limbal epithelial cells (hCLEs) were examined under the conditions of CMC–DA coating/cellulase liberation for cell sheets.
Materials and methods
Synthesis of CMC–DA conjugates
CMC–DA was synthesized based on a previously reported method (Fig. 1B).19 Briefly, carboxymethyl cellulose sodium salt (CMC, medium viscosity, Sigma, Korea) was dissolved in 50 mM 4-morpholineethanesulfonic acid (MES hydrate, Sigma) buffer (pH 5.5) with 1% (w/v) concentration for 12 h at room temperature (RT). Both N-hydroxysuccinimide (NHS, 50 mM, Sigma) and N-(3-dimethylaminopropyl)-N′-ethyl carbodiimide hydrochloride (EDC, 100 mM, Sigma) were added to the CMC solution, and gently stirred for 2 h. Dopamine hydrochloride (DA, 50 mM, Sigma) was dissolved in the CMC solution and stirred for 1 day at RT. The CMC–DA solution was dialyzed using a dialysis membrane (MWCO 3.5 kDa, Spectrum Labs, CA, USA) to remove unreacted molecules against 5 mM hydrochloric acid (HCl, Duksan, Korea) in distilled water, preventing the dopamine from self-polymerization. The dialyzed CMC–DA solution was lyophilized for 2 days. The resultant powder was stored at −20 °C before use. The chemical structure of the synthesized CMC–DA was characterized via attenuated total reflectance Fourier transform infrared spectroscopy (ATR-FTIR, Nicolet Is50, Thermo Fisher Scientific Co.) at Hanyang University in South Korea.Coating of CMC–DA on surfaces
1 to 5 mg ml−1 CMC–DA was dissolved in 10 mM Tris buffer solution (Sigma Aldrich) (pH 8.5). Glass coverslips (Paul Marienfeld, Germany) were cleaned with 99.9% ethyl alcohol (EtOH, Duksan) and sonicated for 20 min. The Transwell membranes or coverslips were coated with the prepared CMC–DA solution for 2 h at RT. After the coating step, the Transwell plate membranes or coverslips were washed with distilled water twice and dried in a heating oven for 6 h at 40 °C. Sterilization of the Transwell plate membrane was conducted under an ultraviolet (UV) lamp (λ = 254 nm, 15 W) for 30 min.Characterization of the CMC–DA coated surfaces
The CMC–DA coated surface was characterized by using various analytical tools. Measurements were conducted using time of flight secondary ion mass spectrometry (TOF-SIMS, TOF.SIMS-5, ION TOF GmbH, Germany) at the Seoul National University, and ATR-FTIR (Nicolet Is50, Thermo Fisher Scientific Co.) at Hanyang University in South Korea. Secondary ion mass spectrometry (SIMS) can analyze a cation or an anion that is released while striking a surface with a primary ion to obtain the surface structure and chemical composition.30 The CMC–DA-coated polyester samples were collected on a TOF-SIMS instrument equipped with a liquid ion source. A Bi3+ ion beam (30 kV, 0.6 pA) was used to generate the spectra in both high spatial and high spectral resolution modes. The SIMS negative ion spectra were calibrated to the H, C, O, N, and Na peaks.Human mesenchymal stem cell culture
hMSCs (donated by the Stem Cell Center, Asan Medical Center), originating from human umbilical cord Wharton's jelly, were cultured in 1 g l−1 glucose Dulbecco's modified Eagle's medium (DMEM, Thermo Fisher Scientific) with 10% (v/v) fetal bovine serum (FBS, Corning, NY, USA), and 1% (v/v) antibiotic–antimycotic (AA, Gibco, Korea) at a temperature of 37 °C under a humidified atmosphere of 5% CO2. The cells were cultured to 80% confluence in T-75 flasks (Thermo Fisher Scientific).Primary culture of human corneal limbal epithelial cell sheets
Human corneal limbal epithelial cells (hCLEs) were obtained from the Institutional Review Board of the Asan Medical Center (S2014-0955-0005) after the central cornea was used for transplantation. The iris, ciliary body, and Descemet's membrane with corneal endothelium, conjunctiva, and excess sclera were surgically removed from the corneosclera. Limbal epithelium was isolated by treatment with 0.2% (w/v) dispase II (Roche, Korea) at 4 °C overnight. Subsequently, the epithelium was treated with 0.25% (w/v) trypsin/0.01% (w/v) EDTA (Gibco) at 37 °C for 1 h to generate isolated cells. Twelve 2 mm explants were obtained from each limbal rim using a blade. HCLEs were cultured with the superficial limbal rims on cell culture plates (3450, Corning). The components of the epithelial medium were as follows: 96% (v/v) DMEM/nutrient mixture F-12 (F12, Gibco), 4% (v/v) FBS (Gibco), 10 ng ml−1 recombinant human fibroblast growth factor 7 (KGF, 100-19, Peprotech, Korea), Y27632 (10 μM, Enzo Biochem, NY, USA), 10 μg ml−1 insulin (I0516, Sigma), 0.5 μg ml−1 hydrocortisone (H0888, Sigma), 2 nM triiodothyronine (T6397, Sigma), 250 ng ml−1 isoproterenol (I6379, Sigma), and 1% AA (Gibco). The medium was changed at day 3, day 5, and every day after day 7.Cell sheet harvest on the CMC–DA coating dish
Cell culture inserts (Transwell, 3450, Corning) were coated with 1 mg ml−1 CMC–DA (2 h, RT) and cultured for 3 to 12 days. hMSCs or hCLEs were seeded in a density of 1.8 × 105 cells per cm2 or 4–6 × 105 cells per cm2. After the hMSCs or hCLEs reached the confluence point, the medium was changed daily until hyperconfluence. Each cell sheet was treated with a medium containing cellulase (50 U ml−1, Sigma) at the bottom of the insert at 37 °C for 10 min to 1 h. After detaching hCLE cell sheets from the insert, the sheets were aspirated using a pipette and washed using phosphate-buffered saline (PBS, Gibco) and transferred to a fresh cell culture dish. The harvested sheets were used for hematoxylin and eosin (H&E) staining and immunohistochemistry. Both hMSC and hCLE sheets were fixed in 10% (v/v) neutral buffered formalin (Sigma) and a 3% (w/v) agar/paraffin block.Cellulase cytotoxicity
Cell proliferation was analyzed using CCK-8 (Dojindo Molecular Technologies, Japan) according to the manufacturer's instructions. Briefly, 10% (v/v) CCK-8 solution was mixed in a cell culture medium, placed in cell culture wells, and incubated at 37 °C under 5% CO2 for 2 h. The supernatant was collected and placed in a 96-well plate, and the absorption at 450 nm was determined using a plate reader (Sunrise™, TECAN, Switzerland). The sheets were evaluated using a live/dead assay (Life Technologies, Carlsbad, CA, USA). Briefly, the samples were incubated at 37 °C under 5% CO2 for 30 min in the live/dead solution (Invitrogen, Korea) and observed using a fluorescence microscope (EVOS, Life-Technologies, Korea). Digital images were captured and processed using the ImageJ (NIH, USA) image processing software (see the ESI†).31Morphology of detached single hMSC cells
To immobilize the cells on the cover glass, 2–3 drops of the cell suspension solutions were incubated on poly-L-lysine-coated cover glass for 20 min. After 20 min, the cells were fixed with 1% glutaraldehyde (Sigma) and 1% paraformaldehyde (Sigma) in 0.1 M sodium cacodylate buffer (Sigma), pH 7.4, for 30 min at 4 °C. The samples were rinsed 3 times with 0.1 M sodium cacodylate buffer, pH 7.4, for 10 min. After washing, the samples were then fixed in 1% osmium tetraoxide (OsO4, Wako, Japan) for 30 min at RT. Then the samples were again rinsed 3 times with 0.1 M sodium cacodylate buffer, pH 7.4, for 10 min. Dehydration of the fixed cells was performed using an ethanol series (50%, 60%, 70%, 80%, 90%, 100%) for 20 min in each, and the cells were immersed in ethanol and isoamyl acetate. Finally, the cells were dried at the critical point using CO2 and coated with platinum (Pt) by sputtering. The sample was observed with a field-emission scanning electron microscope (FE-SEM, S-4700, Hitachi, Japan) at an accelerating voltage of 10 kV.Relative gene expression: single hMSCs
After the treatment of cellulase or trypsin, hMSCs were harvested using the TRIzol reagent (Thermo Fisher Scientific), according to the manufacturer's instructions for total RNA extraction. Quantitative real-time polymerase chain reactions (qPCRs) were performed with an Applied Biosystems 7500 Real-Time PCR system (Applied Biosystems, Korea). All experiments were carried out in triplicate. Quantitative polymerase chain reaction was performed using cDNA as a template for the different groups of hMSCs to reveal the expression levels of the following genes: human FAK, RhoA, RhoB, and P53 (Table S1†). The primer sequences for each gene used in this study are shown in Table S1.† The comparative Ct method (2−ΔΔCT) was used to calculate the relative changes in gene expression determined from real-time quantitative PCR.32Immunofluorescence staining of cell sheets
Paraffin-embedded cell sheets were blocked, and the sections with the cells were permeabilized with 0.2% Triton X-100 (Sigma) in PBS. The sections were incubated for 1 h at 37 °C with the following primary antibodies: anti-human collagen I and IV, laminin, phalloidin, ZO-1, and p64 (Table S2†). The sections were then incubated for 1 h at RT with either Alexa Fluor 488 goat-anti-mouse or goat-anti-rabbit secondary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100). Cell nuclei were counterstained using 4′6-diamidino-2-phenylindole (DAPI) for nucleic acid (Invitrogen) staining. The sections were viewed using an LSM780 confocal microscope (Carl Zeiss, Germany).
100). Cell nuclei were counterstained using 4′6-diamidino-2-phenylindole (DAPI) for nucleic acid (Invitrogen) staining. The sections were viewed using an LSM780 confocal microscope (Carl Zeiss, Germany).
Statistical analysis
All the data are reported as means ± standard deviations. Differences between the experimental groups were analyzed by two-sample t-tests, and a p-value less than 0.05 was considered statistically significant.Results
Synthesis of CMC–DA
To easily attach CMC to the surface of the culture plates, dopamine was incorporated into the CMC carboxyl group. From infrared spectroscopy (IR) spectra (Fig. 1C), it was estimated that the amino groups in dopamine and the carboxyl groups in CMC were conjugated: C–O (1190 cm−1) and N–H (1606 cm−1) of dopamine, C–H (2920 cm−1) and –OH (3423 cm−1) of CMC. These results demonstrated that CMC–DA was successfully conjugated.CMC–DA coated surface
A thin layer of enzymatically digestible CMC–DA was pre-coated on the surface of the cell culture dish to prepare a surface that allowed the cells to adhere and grow to form a cell sheet. Fig. 1D and S1† show the negative depth profile and ion spectra of uncoated and CMC–DA-coated surfaces obtained by TOF-SIMS. Although the spectra of CMC–DA were very similar to the spectra of pure coating (Fig. S1†), there were differences in the fragmentation pattern (relative intensities of peaks at different positions) that were calibrated using C–, CH–, O–, OH–, C2–, and C2H–. The peaks at m/z 23 (Na−), 46 (NO2−), and 62 (NO3−) can be attributed to the CMC–DA-coated surface (Fig. 1D). The changes in the ion intensity with the sputter time reflect in-depth variations, but they are also strongly dependent on the CMC–DA from which the ions were emitted. The depth profile presents negative ions assigned to Na−, NO2−, and NO3−, indicating the presence of CMC–DA on the surface. Note that the intensity is reported using a logarithmic scale, which emphasizes the low intensity signals. In the depth profile, the intensity of Na− showed higher values for the CMC–DA-coated surface compared to the low intensity of the negative control. Na− is a possible ion assignment for negative ion fragments originating from CMC as well as from CMC–DA. NO2− and NO3− are possible ion assignments for negative ion fragments originating from dopamine as well as from CMC–DA. Three regions can be identified in Fig. 1D. The first one, corresponding to the first 80 s of sputtering, is characterized by high intensity and relatively constant Na−, NO2−, and NO3− signals.Optimization of CMC–DA and cellulase concentration in hMSC culture
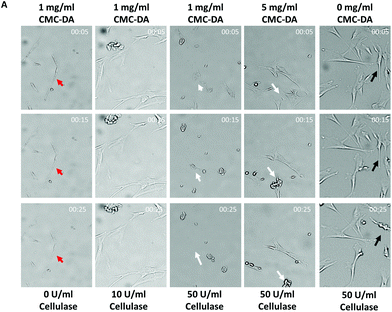
hMSCs were proliferated on the surface of the CMC–DA-coated culture plates compared to the uncoated plates (data not shown).33To investigate the concentration of CMC–DA pre-coating and cellulase for fast detachment, cell dynamic morphological changes were observed without any mechanical forces during the treatment of various concentrations of cellulase (Fig. 2). After pre-coating the culture plates with 0, 1, and 5 mg ml−1 of CMC–DA, hMSCs (5 × 103 cm−2) were cultured for 3 days until enzymatic detachment with 0, 10, 20, 50, and 100 U ml−1 cellulase in the medium. In general, the hMSCs had an elongated and spindle-shaped morphology. Fig. 2 shows that the hMSC morphology dramatically changed 5 min after 50 U ml−1 cellulase treatment under a concentration of 1 mg ml−1 CMC–DA pre-coated surface.34 All the hMSCs on the 1 mg ml−1 CMC–DA pre-coated surface detached under a concentration of 50 U ml−1 cellulase 25 min after treatment. hMSCs that were pre-coated with 1 mg ml−1 CMC–DA did not detach in 0 and 10 U ml−1 cellulase until after 25 min. Most hMSCs pre-coated with 5 mg ml−1 CMC–DA contracted; some cells were attached to the surface at 25 min in 50 U ml−1 cellulase. hMSCs on the non-coated surface attached to the surface; some cells contracted at 25 min in 50 U ml−1 cellulase. The data summarized in Table 1 reveal that the fastest detachment occurred with 1 mg ml−1 CMC–DA coating and 50 U ml−1 cellulase treatment.
| CMC–DA coated concentration (mg ml−1) | 1 | 1 | 1 | 5 | 0 |
|---|---|---|---|---|---|
| Cellulase concentration (U ml−1) | 0 | 10 | 50 | 50 | 50 |
| Detachment starting time (min) | n/a | n/a | 15 | 15 | 60 |
Cytotoxicity studies were conducted on confluent hMSCs. Fig. S3† shows the normalized cell viability of cellulase with various concentrations (0, 10, 20, 50, and 100 U ml−1 in the medium) after 1 h in the cultured cells. Fig. S3† reveals over 80% viability of cellulase-treated hMSCs compared to hMSCs not treated with cellulase.
Human mesenchymal stem cell detachment with cellulase
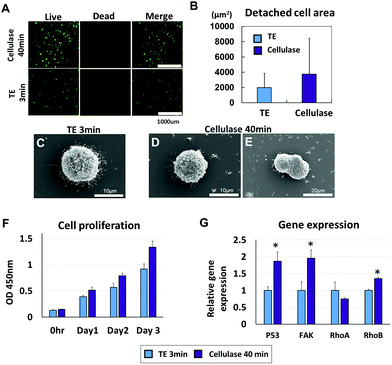
The effects of cellulase on cell viability were investigated and compared to the conventional detachment method using trypsin/EDTA (TE) treatment. The hMSC viability after detachment with cellulase for 40 min was compared to the viability after trypsinization for 3 min with a live/dead assay; both groups showed over 90% viability (Fig. 3A). In terms of an average detached cell area after the treatment of trypsin/EDTA vs. cellulase (Fig. 3B), the cellulase-treated hMSCs had higher values based on the live/dead assay results (TE 3 min: 2003.57 ± 1875.04 μm2, cellulase 40 min: 3747.77 ± 4685.51 μm2). Compared to the trypsinized hMSCs, the cellulase-treated hMSCs were characterized by a wider and higher cell area distribution frequency (Fig. S2†). Scanning electron microscopy (SEM) images showed numerous microvilli from hMSCs in single cells after detachment treatment, exhibiting the characteristics of a migratory/invasive phenotype (Fig. 3C),35–38 whereas hMSC aggregates revealed a smooth spheroid-like appearance in the cellulase-treated group (Fig. 3D and E). Additionally, it was observed that a significant aggregation of hMSCs into cellular clumps occurred instantly upon treatment with cellulase (Fig. 3E). The proliferation after cellulase or trypsin/EDTA treatment was monitored at 0 h, and then at 1, 2, and 3 days using CCK-8 analysis (Fig. 3F). Cellulase-treated hMSCs were proliferated for 40 min and had significantly more viability after detachment compared to hMSCs treated with trypsin/EDTA. At day 3, hMSCs treated with cellulase showed more proliferation (TE 3 min: 0.9195 ± 0.07, cellulase 40 min: 1.3342 ± 0.052). Gene expression after 3 min of trypsin treatment or 40 min of cellulase treatment is shown in Fig. 3H. hMSCs treated with cellulase for 40 min expressed more FAK, p53, and RhoB compared to trypsin/EDTA treatment for 3 min (P53 TE 3 min: 1 ± 0.11, cellulase 40 min: 1.86 ± 0.27; FAK TE 3 min: 1 ± 0.26, cellulase 40 min: 1.96 ± 0.24; RhoA TE 3 min: 1 ± 0.25, cellulase 40 min: 0.75 ± 0.03; and RhoB TE 3 min: 1 ± 0.03, cellulase 40 min: 1.35 ± 0.03).Human mesenchymal stem cell sheets using enzymatic digestion
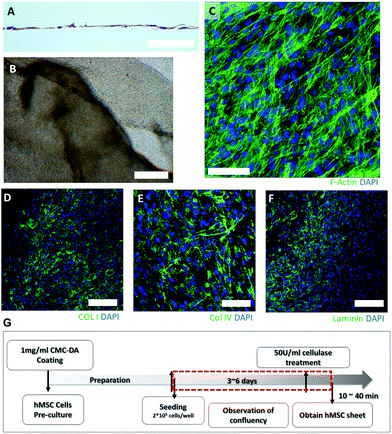
To generate the hMSC sheets, hMSCs on the CMC–DA pre-coated culture dishes were cultured until confluent cell sheets formed. These sheets were then detached from the tissue culture plastic using enzymatic digestion with cellulase. Fig. 4A and B show that a single hMSC layer was formed at day 6. Movie S1† shows the detachment process, under a microscope, of the hMSC sheets after cellulase treatment. The hMSC sheets formed quickly and finally they aggregated into spherical shapes when suspended in the media (data not shown). The ECM markers of hMSCs, collagen type I and IV and laminin, were localized in the cytosol of the hMSCs on day 6 (Fig. 4C–F).Application of enzymatic digestion for the limbal cell sheet
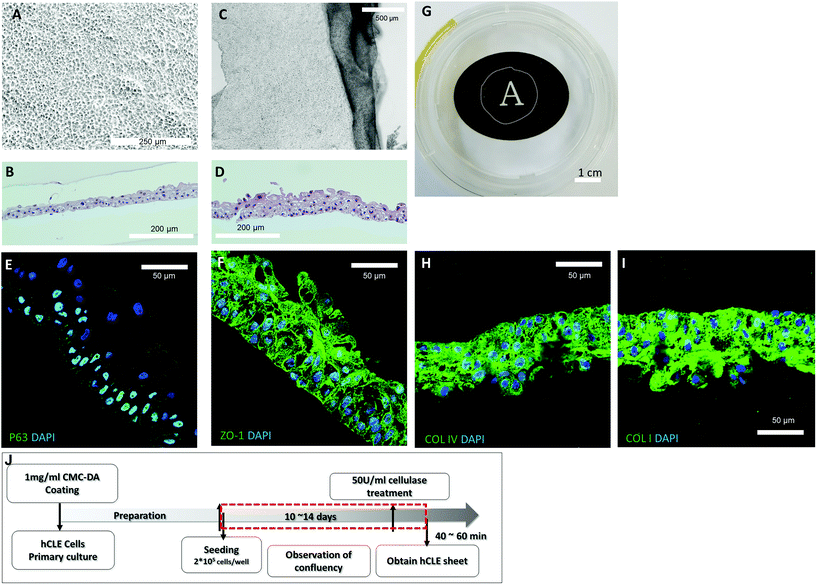
To confirm the feasibility of the developed method, hCLE sheets were generated using the enzymatic digestion methodology. Cell proliferation was observed for 10 to 14 days until confluency was achieved on the Transwell membrane (Fig. 5A). hCLEs grown on pre-coated CMC–DA were highly compact and polygonal, and formed stratified epithelial sheets with two to three cell layers consisting of small cuboidal basal cells (Fig. 5A and B). After digestion using cellulase for more than 40 min, hCLE sheets detached and contracted into four to five cellular layers, and were half of their original size (Fig. 5C and D). The putative epithelial stem cell marker p63 was expressed in the basal cell layers, and the cell junction marker ZO-1 was expressed in the generated hCLE sheets, representing cell-to-cell interactions among the hCLE sheets (Fig. 5E and F). The hCLE sheets maintain the extracellular matrix (ECM), collagen type I and IV, after detachment from the culture surface (Fig. 5H and I). Additionally, the generated hCLE sheets were transparent (Fig. 5G) and of various sizes (Fig. S4†).Discussion
CMC–DA for cell sheet engineering
This study confirmed the hypothesis that a pre-coated CMC–DA surface is a substrate for cell sheet engineering with enzymatic lysis by cellulase using hMSCs and hCLEs. Cellulose conjugated with dopamine was used to coat the surface of a Transwell membrane by simple coating under pH 8.5 conditions; hMSCs or hCLEs were seeded and cultured to confluency. Each sheet was released from the membrane by immersion and permeation with cellulase from underneath the Transwell membrane. The cellulase was allowed to pass through the Transwell membrane, then slide underneath the coated CMC–DA and degrade CMC into glucose, releasing the cell sheet from the CMC–DA.Cellulose and its derivatives are useful materials for numerous pharmaceutical, medical, and tissue-engineering applications due to their biocompatibility and biodegradability.39–42 In this study, we attempted to synthesize CMC with catechol moieties using aqueous-phase carbodiimide activation chemistry for obtaining CMC–DA. This was achieved through the generation of covalent cross-linking between CMC molecules via an enzyme-catalyzed reaction that resulted in the conjugation of carboxyl groups to amines in dopamine. In cell engineering, dopamine has also been used for mouse embryonic stem cell culture on polydimethylsiloxane (PDMS) by gelatin–dopamine grafting for cellular attachment.43 To our knowledge, CMC–DA is the first conjugation polymer, and dopamine was conjugated and coated for the first time for cell sheet engineering in this study. CMC–DA was coated onto the culture plate by chemical immobilization between oxidized catechol groups and the culture plate.21,23 We confirmed that CMC was successfully immobilized on the surface of the plate with the TOF-SIMS data (Fig. 1D and S1†).
Cell sheet research has existed for over 10 years,44 and fast cell detachment in cell sheet engineering remains an important subtopic.45 To optimize the process, fast hMSC detachment was examined with various concentrations of CMC–DA and cellulase. All hMSCs detached most rapidly from a 1 mg ml−1 CMC–DA coating surface with 50 U ml−1 cellulase. The higher cellulase concentration, 100 U ml−1, also effected the same detachment time with 50 U ml−1 cellulase treatment (data not shown). Also, the permeability of a CMC–DA-coated Transwell membrane with DMEM was examined with various concentrations of CMC–DA coating without cell culture (Fig. S5†). The 5 mg ml−1 CMC–DA coated membrane was not permeated until about 24 h compared to 5 min permeation time under 1 mg ml−1 CMC–DA conditions. However, after cellulase treatment on the CMC–DA coated surface, the permeation time was dramatically reduced. This means that the pre-coated CMC–DA covered the membrane's pores, and cellulase treatment removed the pre-coated CMC–DA on the pore of the membrane. Also, the glucose contents were increased with the CMC–DA-coated surface after 100 U ml−1 cellulase treatment for 3 hours (Fig. S6†).
Cell behavior after treatment with cellulase and detachment
After cellulase treatment, hMSC viability, proliferation, morphology, and gene expression were examined and compared to the single trypsinized hMSCs. Compared to the trypsinized cells, the surface morphology of the cellulase-treated hMSCs showed loss of microvilli and were observed to be in aggregated cell clumps under SEM (Fig. 3C–E). This is consistent with the detached cell size, showing a higher detached cell size (Fig. 3B).A more migratory invasive phenotype resulted, with increased cell surface lamellipodia and filopodia46,47 transiently pointing in all directions. On flat surfaces, the transient filopodia of individual cell migration are known to quickly disappear during spreading.48 Kurashina et al. reported that, similar to our findings, under SEM, trypsinized cells showed more lamellipodia and filopodia compared to collagenase-treated cells.49 These results suggest that cellulase degrades the pre-coated cellulose, leaving the cell membrane intact, but trypsin hydrolyses cell membrane proteins. Additionally, hMSC proliferation was higher in the cultured hMSCs treated with cellulase for 40 min and then trypsinized for 3 min after detachment.
In cell sheet generation, the deposited ECM and the cell–cell junction play important roles in contiguous cell sheets.50 FAK plays a role in the regulation of the cytoskeleton,51 cell adhesion and detachment, and creating combinational signaling complexes.52,53 There was more FAK expression in cellulase-treated hMSCs compared to the detached hMSCs after trypsinization; these observations showed that the digestion of cellulose occurred without breaking molecular bonds in the cellular membrane or ECM.
The RhoA and RhoB genes are Rho family GTPase proteins and they play important roles in the morphological cellular changes triggered by extracellular stimuli;54 they also regulate the cell cytoskeleton by mechanical signaling.55 RhoA stimulates actin polymerization and regulates cell adhesion molecules.56 RhoB inhibits migration and invasion in several cancers.57 Cell adhesion mediated by cadherin receptors has been shown to modulate the activity of Rho proteins.58 From these points of view, it was hypothesized that both RhoA and RhoB change with cell adhesion strength after trypsinization or cellulase treatment. p53 is a tumor suppressor protein known to be deleted or mutated in numerous human cancers.59 Honoki et al. reported that disruption of p53 may lead to cell cycle acceleration through the downregulation of p21 and the upregulation of CDK and cyclins of MSCs.60 Also, Golubovskaya et al. reported that FAK binds directly to p53.61 Our results showed that p53 has a role as a cell cycle and cell adhesion regulator in cell sheet generation. In summary, enzymatic cell detachment is most probably caused by the digestion of the pre-coated CMC–DA induced by cellulase, based on the results.
Human mesenchymal stem cell/human corneal limbal epithelial cell sheet engineering
Production of hMSC/hCLE sheets showed various separation times depending on cell type, cell passage number, seeding density, and culture period (Table 2). In general, the hMSC sheets were separated within 10 min in our environment, while the hCLE sheets were acquired more than 40 min after cellulase treatment. Fig. 2A shows that detached hMSCs contracted dramatically in response to cellulase treatment. Although all the pre-coated CMC on the surface was not degraded, some hMSCs on the degraded CMC detached, and then the entire cell sheet was separated from the CMC as a result of the contracting force of confluent hMSC sheets. The cell sheet detachment time depends on the thermo-responsive surface fabrication method, cell types, and culture dishes.9| Cell type | Seeding density (cells per cm2) | Culture duration (day) | Time of detachment (min) |
|---|---|---|---|
| hMSC | 1.8 × 105 | 3–4 | 10–40 |
| hCLE | 2.1 × 105 | 5 | 40–60 |
| 4–6 × 104 | 10–15 |
hCLEs have been shown to be effective for corneal regeneration. However, hCLE sheets without substrates have been rarely reported to our knowledge. For corneal regeneration, the cell source used was the oral mucosal epithelial single cells62–64 or limbal epithelial cells. Li et al. reported a study in which human limbal cells were seeded and cultured on 3D Matrigel.65 Utheim et al. cultured hCLEs on human amniotic membranes for 2 weeks.66 In experiments using temperature-responsive culture dishes, oral mucosal epithelial cells67,68 and human limbal epithelial cells were used for cell sheets, with implications for corneal disease treatment. Human limbal cells have been cultured on feeder cells for cell sheet generation.69–71 Also, human corneal limbal epithelial single cells have been cultured with keratinocyte growth factor (KGF) and Rho kinase inhibitor for cultivation.72 The animal experiments have also been carried out with rabbit limbal cell sheets without any feeder or support materials for cell sheet engineering.73
We made hCLE sheets without a substrate (Fig. 5C) for in vivo applications. Additionally, hCLE sheets with cellular junctions (Fig. 5E) created the autocrine ECM with the intact cell sheet (Fig. 5H and I). We are also testing the hCLE sheets in in vivo experiments for corneal regeneration. The hCLE sheets were transplanted on the removed epithelium of the rabbit cornea, and corneal regeneration was observed for 17 weeks post-operation. The transplanted hCLE sheets have confirmed the adherence on the impaired epithelium (data not shown). However, the detachment time of the cell sheet needs to be reduced within minutes when testing with hCLE sheets.
Conclusions
In this paper, we propose a novel method using cellulase treatment for collecting cell sheets from a CMC–DA-coated surface. The efficacy of the method was experimentally verified using hMSC and hCLE sheets. However, the safety of the cell sheets after cellulase treatment should be investigated at the pre-clinical stage before clinical use. As an extension of this research, we are studying the efficacy of hCLE sheets for corneal epithelial regeneration in a rabbit model.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Electron Microscopy Core Facility and the CLSM (Confocal Laser Scanning Microscopy) Core Facility at the ConveRgence mEDicine research cenTer (CREDIT), Asan Medical Center, for support and instrumentation.This study was supported by grants from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number HI17C0193), and the ASAN Institute for Life Sciences, ASAN Medical Center, Seoul, Korea (grant number 2017-648).
References
- T. Shimizu, M. Yamato, A. Kikuchi and T. Okano, Biomaterials, 2003, 24, 2309–2316 CrossRef CAS PubMed
.
- A. K. Saxena, H. Ainoedhofer and M. E. Höllwarth, J. Pediatr. Surg., 2009, 44, 896–901 CrossRef PubMed
.
- R. Nakajima, T. Kobayashi, T. Kikuchi, Y. Kitano, H. Watanabe, M. Mizutani, T. Nozaki, N. Senda, K. Saitoh, R. Takagi, M. Yamato, T. Okano and S. Takeda, J. Tissue Eng. Regener. Med., 2015, 9, 637–640 CrossRef CAS PubMed
.
- H. Zhang, S. Liu, B. Zhu, Q. Xu, Y. Ding and Y. Jin, Stem Cell Res. Ther., 2016, 7, 168 CrossRef PubMed
.
- Q. Wei, D. Reidler, M. Y. Shen and H. Huang, BMC Biotechnol., 2013, 13, 17 CrossRef CAS PubMed
.
- G. Sagvolden, I. Giaever, E. O. Pettersen and J. Feder, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 471–476 CrossRef CAS
.
- N. Yamada, T. Okano, H. Sakai, F. Karikusa, Y. Sawasaki and Y. Sakurai, Macromol. Rapid Commun., 1990, 11, 571–576 CrossRef CAS
.
- O. H. Kwon, A. Kikuchi, M. Yamato, Y. Sakurai and T. Okano, J. Biomed. Mater. Res., 2000, 50, 82–89 CrossRef CAS PubMed
.
- N. G. Patel and G. Zhang, Organogenesis, 2013, 9, 93–100 CrossRef PubMed
.
- L. A. Sonna, J. Fujita, S. L. Gaffin and C. M. Lilly, J. Appl. Physiol. (1985), 2002, 92, 1725–1742 CrossRef CAS PubMed
.
- N. Petersen and P. Gatenholm, Appl. Microbiol. Biotechnol., 2011, 91, 1277–1286 CrossRef CAS PubMed
.
- G. Helenius, H. Bäckdahl, A. Bodin, U. Nannmark, P. Gatenholm and B. Risberg, J. Biomed. Mater. Res., Part A, 2006, 76, 431–438 CrossRef PubMed
.
- H. Jung, H. G. Yoon, W. Park, C. Choi, D. B. Wilson, D. H. Shin and Y. J. Kim, Cellulose, 2007, 15, 465–471 CrossRef
.
- H. Bäckdahl, G. Helenius, A. Bodin, U. Nannmark, B. R. Johansson, B. Risberg and P. Gatenholm, Biomaterials, 2006, 27, 2141–2149 CrossRef PubMed
.
- M. Safarzadeh, P. Azizzadeh and P. Akbarshahi, J. Optom., 2017, 10, 258–264 CrossRef PubMed
.
- M. K. Bhat and S. Bhat, Biotechnol. Adv., 1997, 15, 583–620 CrossRef CAS PubMed
.
- B. X. Wang, X. G. Lv, S. Y. Chen, Z. Li, X. X. Sun, C. Feng, H. P. Wang and Y. M. Xu, Cellulose, 2016, 23, 3187–3198 CrossRef CAS
.
- R. J. Greenough, D. J. Everett and M. Stavnsbjerg, Food Chem. Toxicol., 1991, 29, 781–785 CrossRef CAS PubMed
.
- S. Sakai, Y. Ogushi and K. Kawakami, Acta Biomater., 2009, 5, 554–559 CrossRef CAS PubMed
.
- I. K. Ko, K. Kato and H. Iwata, J. Biomater. Sci., Polym. Ed., 2005, 16, 1277–1291 CrossRef CAS PubMed
.
- H. Lee, N. F. Scherer and P. B. Messersmith, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 12999–13003 CrossRef CAS PubMed
.
- N. K. Kaushik, N. Kaushik, S. Pardeshi, J. G. Sharma, S. H. Lee and E. H. Choi, Mar. Drugs, 2015, 13, 6792–6817 CrossRef CAS PubMed
.
- H. Lee, S. M. Dellatore, W. M. Miller and P. B. Messersmith, Science, 2007, 318, 426–430 CrossRef CAS PubMed
.
- Y. Lee, H. Lee, Y. B. Kim, J. Kim, T. Hyeon, H. Park, P. B. Messersmith and T. G. Park, Adv. Mater., 2008, 20, 4154–4157 CAS
.
- S. H. Hong, M. Shin, J. Lee, J. H. Ryu, S. Lee, J. W. Yang, W. D. Kim and H. Lee, Adv. Healthcare Mater., 2016, 5, 75–79 CrossRef CAS PubMed
.
- E. Kim, I. T. Song, S. Lee, J.-S. Kim, H. Lee and J.-H. Jang, Angew. Chem., Int. Ed., 2012, 51, 5598–5601 CrossRef CAS PubMed
.
- J. Y. Park, J. Yeom, J. S. Kim, M. Lee, H. Lee and Y. S. Nam, Macromol. Biosci., 2013, 13, 1511–1519 CrossRef CAS PubMed
.
- Y. Zhao, Y. Wu, L. Wang, M. Zhang, X. Chen, M. Liu, J. Fan, J. Liu, F. Zhou and Z. Wang, Nat. Commun., 2017, 8, 2218–2225 CrossRef PubMed
.
- D. Lee, J. P. Park, M.-Y. Koh, P. Kim, J. Lee, M. Shin and H. Lee, Biomater. Sci., 2018, 6, 1040–1047 RSC
.
- J. C. Vickerman, Analyst, 1994, 119, 513–523 RSC
.
- S. Hong, J. H. Yun, E.-S. Kim, J. S. Kim, H. Tchah and C. Hwang, Invest. Ophthalmol. Visual Sci., 2018, 59, 1475–1485 CrossRef PubMed
.
- T. D. Schmittgen and K. J. Livak, Nat. Protoc., 2008, 3, 1101 CrossRef CAS PubMed
.
- C. Demitri, M. G. Raucci, A. Giuri, V. M. De Benedictis, D. Giugliano, P. Calcagnile, A. Sannino and L. Ambrosio, J. Biomed. Mater. Res., Part A, 2016, 104, 726–733 CrossRef CAS PubMed
.
- H. Motaln, C. Schichor and T. T. Lah, Cancer, 2010, 116, 2519–2530 CrossRef CAS PubMed
.
- A. B. Bohil, B. W. Robertson and R. E. Cheney, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 12411–12416 CrossRef CAS PubMed
.
- J. Albuschies and V. Vogel, Sci. Rep., 2013, 3, 1658 CrossRef CAS PubMed
.
- Y. Klymenko, J. Johnson, B. Bos, R. Lombard, L. Campbell, E. Loughran and M. S. Stack, Neoplasia, 2017, 19, 549–563 CrossRef CAS PubMed
.
- A. Pietuch and A. Janshoff, Open Biol., 2013, 3, 133084 CrossRef PubMed
.
-
K. A. Sindhu, R. Prasanth and V. K. Thakur, Medical Applications of Cellulose and its Derivatives: Present and Future, ch. 16, DOI:10.1002/9781118872246
.
- N. D. Sanandiya and A. K. Siddhanta, RSC Adv., 2016, 6, 92953–92961 RSC
.
- M. M. Pakulska, K. Vulic, R. Y. Tam and M. S. Shoichet, Adv. Mater., 2015, 27, 5002–5008 CrossRef CAS PubMed
.
- S. Ohta, T. Nishiyama, M. Sakoda, K. Machioka, M. Fuke, S. Ichimura, F. Inagaki, A. Shimizu, K. Hasegawa, N. Kokudo, M. Kaneko, Y. Yatomi and T. Ito, J. Biosci. Bioeng., 2015, 119, 718–723 CrossRef CAS PubMed
.
- J. Fu, K. Y. Quek, Y. J. Chuah, C. S. Lim, C. Fan and D.-a. Wang, J. Mater. Chem. B, 2016, 4, 7961–7973 RSC
.
- N. Matsuda, T. Shimizu, M. Yamato and T. Okano, Adv. Mater., 2007, 19, 3089–3099 CrossRef CAS
.
- K. O. Hyeong, K. Akihiko, Y. Masayuki, S. Yasuhisa and O. Teruo, J. Biomed. Mater. Res., 2000, 50, 82–89 CrossRef
.
- J. Gavard, M. Lambert, I. Grosheva, V. Marthiens, T. Irinopoulou, J.-F. Riou, A. Bershadsky and R.-M. Mège, J. Cell Sci., 2004, 117, 257–270 CrossRef CAS PubMed
.
- L. M. Machesky, FEBS Lett., 2008, 582, 2102–2111 CrossRef CAS PubMed
.
- M. A. Partridge, E. E. Marcantonio and R. Assoian, Mol. Biol. Cell, 2006, 17, 4237–4248 CrossRef CAS PubMed
.
- Y. Kurashina, K. Takemura, S. Miyata, J. Komotori and T. Koyama, Biomicrofluidics, 2014, 8, 054118 CrossRef CAS PubMed
.
- M. Yamato and T. Okano, Mater. Today, 2004, 7, 42–47 CrossRef CAS
.
- B. Fabry, A. H. Klemm, S. Kienle, T. E. Schaffer and W. H. Goldmann, Biophys. J., 2011, 101, 2131–2138 CrossRef CAS PubMed
.
- M. A. Wozniak, K. Modzelewska, L. Kwong and P. J. Keely, Biochim. Biophys. Acta, Mol. Cell Res., 2004, 1692, 103–119 CrossRef CAS PubMed
.
- A. Khalili and M. Ahmad, Int. J. Mol. Sci., 2015, 16, 18149 CrossRef CAS PubMed
.
- A. P. Wheeler and A. J. Ridley, Exp. Cell Res., 2004, 301, 43–49 CrossRef CAS PubMed
.
- P. P. Provenzano and P. J. Keely, J. Cell Sci., 2011, 124, 1195–1205 CrossRef CAS PubMed
.
- J. Alblas, L. Ulfman, P. Hordijk and L. Koenderman, Mol. Biol. Cell, 2001, 12, 2137–2145 CrossRef CAS PubMed
.
- K. Jiang, J. Sun, J. Cheng, J. Y. Djeu, S. Wei and S. Sebti, Mol. Cell. Biol., 2004, 24, 5565–5576 CrossRef CAS PubMed
.
- V. M. M. Braga, Curr. Opin. Cell Biol., 2002, 14, 546–556 CrossRef CAS PubMed
.
- M. R. Junttila and G. I. Evan, Nat. Rev. Cancer, 2009, 9, 821–829 CrossRef CAS PubMed
.
- K. Honoki, H. Fujii, Y. Tohma, T. Tsujiuchi, A. Kido, S. Tsukamoto, T. Mori and Y. Tanaka, ISRN Oncol., 2012, 2012, 909453 Search PubMed
.
- V. M. Golubovskaya, R. Finch and W. G. Cance, J. Biol. Chem., 2005, 280, 25008–25021 CrossRef CAS PubMed
.
- T. P. Utheim, O. A. Utheim, Q. E. Khan and A. Sehic, J. Funct. Biomater., 2016, 7, 5 CrossRef PubMed
.
- T. Kobayashi, K. Kan, K. Nishida, M. Yamato and T. Okano, Biomaterials, 2013, 34, 9010–9017 CrossRef CAS PubMed
.
- C. Burillon, L. Huot, V. Justin, S. Nataf, F. Chapuis, E. Decullier and O. Damour, Invest. Ophthalmol. Visual Sci., 2012, 53, 1325–1331 CrossRef PubMed
.
- G. Li, Y. Zhang, S. Cai, M. Sun, J. Wang, S. Li, X. Li, S. Tighe, S. Chen, H. Xie and Y. Zhu, Sci. Rep., 2018, 8, 6566 CrossRef PubMed
.
- O. Utheim, R. Islam, T. Lyberg, B. Roald, J. R. Eidet, M. F. de la Paz, D. A. Dartt, S. Raeder and T. P. Utheim, PLoS One, 2015, 10, e0118517 CrossRef PubMed
.
- T. Soma, R. Hayashi, H. Sugiyama, M. Tsujikawa, S. Kanayama, Y. Oie and K. Nishida, PLoS One, 2014, 9, e110987 CrossRef PubMed
.
- Y. Oie and K. Nishida, BioMed Res. Int., 2013, 2013, 428247 CrossRef PubMed
.
- Y. Li, T. Inoue, F. Takamatsu, T. Kobayashi, A. Shiraishi, N. Maeda, Y. Ohashi and K. Nishida, Invest. Ophthalmol. Visual Sci., 2014, 55, 1453–1462 CrossRef CAS PubMed
.
- K. Watanabe, M. Yamato, Y. Hayashida, J. Yang, A. Kikuchi, T. Okano, Y. Tano and K. Nishida, Biomaterials, 2007, 28, 745–749 CrossRef CAS PubMed
.
- S. Gonzalez, H. Mei, M. N. Nakatsu, E. R. Baclagon and S. X. Deng, Stem Cell Res., 2016, 16, 358–364 CrossRef CAS PubMed
.
- M. Yoshihara, Y. Sasamoto, R. Hayashi, Y. Ishikawa, M. Tsujikawa, Y. Hayashizaki, M. Itoh, H. Kawaji and K. Nishida, Sci. Rep., 2017, 7, 2845 CrossRef PubMed
.
- Y. Hayashida, K. Nishida, M. Yamato, J. Yang, H. Sugiyama, K. Watanabe, Y. Hori, N. Maeda, A. Kikuchi, T. Okano and Y. Tano, Invest. Ophthalmol. Visual Sci., 2006, 47, 552–557 CrossRef PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8bm00971f |
| This journal is © The Royal Society of Chemistry 2019 |