Open Access Article
Open Access ArticleBiological activity of human mesenchymal stromal cells on polymeric electrospun scaffolds†
Febriyani F. R.
Damanik
ab,
Gabriele
Spadolini
ac,
Joris
Rotmans
d,
Silvia
Farè
 c and
Lorenzo
Moroni
c and
Lorenzo
Moroni
 *ab
*ab
aUniversity of Twente, Drienerlolaan 5, Zuidhorst 145, 7522 NB Enschede, the Netherlands
bMaastricht University, MERLN Institute for Technology-Inspired Regenerative Medicine, Universiteitsingel 40, 6229 ER Maastricht, the Netherlands. E-mail: l.moroni@maastrichtuniversity.nl
cPolitecnico di Milano, Department of Chemistry, Materials and Chemical Engineering “G. Natta”, Piazza L. Da Vinci 32, Milan, Italy
dDepartment of Internal Medicine, Leiden University Medical Center, Albinusdreef 2, 2333ZA Leiden, the Netherlands
First published on 21st December 2018
Abstract
Electrospinning provides a simple robust method to manufacture scaffolds for tissue engineering applications. Though varieties of materials can be used, optimization and biocompatibility tests are required to provide functional tissue regeneration. Moreover, many studies are limited to 2D electrospun constructs rather than 3D templates due to the production of high density packed fibres, which result in poor cell infiltration. Here, we optimised electrospinning parameters for three different polymers: poly(ε-caprolactone) (PCL), polylactic acid (PLA) and poly(ethylene oxide terephthalate)/poly(butylene terephthalate) (PA) copolymers. Human mesenchymal stromal cells (hMSCs) were cultured on scaffolds for 14 days to study the scaffolds’ biocompatibility and their multi-lineage differentiation potential or maintenance of stemness in the absence of chemical stimuli. For all scaffolds, a high and stable metabolic activity was measured throughout the culture time with a high proliferation rate compared to day 1 (PCL 5.8-, PLA 4-, PA 4.9-fold). The metabolism of hMSCs was also measured through glucose and lactate concentrations, showing no cytotoxic levels up to 14 days. Total glycosaminoglycan (GAG) production was the highest in PA electrospun scaffolds. When normalized to DNA, GAG production was the highest in PLA and PA scaffolds. All scaffolds were prone to differentiate to an osteogenic lineage, with PCL providing the highest alkaline phosphatase and collagen type Ia gene upregulation. As PA had the most stable fibre formation, it was chosen as a template to further incorporate stromal cell-derived factor-1 (SDF-1) and granulocyte colony-stimulating factor (G-CSF), and stimulate higher hMSC infiltration. These scaffolds provided significantly higher hMSC infiltration than normal PA scaffolds. In conclusion, our optimized biocompatible electrospun scaffolds have shown promising regulation of hMSC fate. When combined with migratory stimulating cytokines, these scaffolds may overcome the known challenges of poor cellular infiltration typical of micro- and nano-fibrillary random meshes.
1. Introduction
Electrospinning is a robust and simple technique to produce nanofibres from a wide variety of polymers.1 By controlling different parameters, electrospinning offers several advantages. These include extremely high surface-to-volume ratio, tuneable nanofibre composition, porosity, and flexibility to conform to a wide variety of shapes and sizes.2,3 Hence, electrospun nanofibres have been extensively investigated in the past decade for their use in various applications, such as filtration, optical and chemical sensors, electrode materials and scaffolds.4Human mesenchymal stromal cells (hMSCs) are found throughout the body and play a crucial role in tissue regeneration by migrating to sites of injury and replacing dysfunctional cells via their multipotent differentiation capacity.5 Studies showing short life spans of hMSCs have administered them without supporting tissues or biocompatible substrates.6 Electrospun scaffolds have been well known to provide a template for cells to grow and develop into functional engineered tissues. This is done by mimicking the natural tissue environment as the diameter of the electrospun fibres can be tuned to match the fibrils of the extracellular matrix (ECM).7,8 Cell behaviour can be quite diverse in response to a scaffold, and features such as surface topography, chemistry and mechanical properties are known to have an influence on cell activity.9 The mechanical and physical properties of copolymer families such as poly(ethylene oxide terephthalate)/poly(butylene terephthalate) (PA) can be easily tailored by varying the PEOT and PBT weight ratio and the length of the PEG segments,10 and hence can be a suitable material platform for electrospinning applications. Moreover, many other biodegradable polymers such as poly(ε-caprolactone) (PCL) and polylactic acid (PLA) have been used to obtain electrospun scaffolds that showed in vitro and in vivo biocompatibility.11,12 However, widespread success in clinical applications remains dependent on understanding the basic cell–scaffold interactions in these nano- and micro-fibrillar meshes.
A major challenge encountered in electrospinning for tissue engineering is the heterogeneity of cellular distribution and migration in the scaffold with increasing depth under passive, static seeding conditions.13 This can potentially stagnate further applications of electrospun scaffolds in various tissue engineering applications due to the high packing density of the fibrous network.14 Hence, it is of great importance to formulate a method to fabricate cell-permeable electrospun scaffolds with enhanced cell distribution and migration throughout the scaffolds’ volume. Several methods have reported to address these issues. Decreasing the fibre diameter provided an adverse outcome due to the increase of fibre-to-fibre contact per unit length, and eventually a decrease in the mean pore radius.15 Ekaputra et al.16 showed no significant improvement upon blending the desired polymer with a water soluble polymer compared to conventionally electrospun fibres. The method of leaching out a sacrificial polymer from an electrospun scaffold is a multiple step process and might change the chemical and mechanical properties of the remaining electrospun polymer, ultimately affecting the cell behaviour.13,17 Another approach consisted of electrospraying a cellular solution simultaneously with electrospinning or coaxial electrospinning to encapsulate cells. However, the complexity of the setup, and additional exposure of a high electric field and shear stress to the cells, may limit this approach.18–20
Numerous studies have shown that the chemokine stromal cell-derived factor-1 (SDF-1) is crucial for stem/progenitor and mesenchymal cell chemotaxis.21,22 Similarly, granulocyte colony-stimulating factor (G-CSF) significantly enhances the homing capacity of these stem cells. In this study, we aimed to make use of electrospinning to preselect three polymer types and fine tune their fibre diameter to the same magnitude as that of the fibrils in the ECM and compare their efficacy as a substrate for cell growth and distribution. Comparative studies of PCL, PLA and PA on electrospinning parameters, stem cell activity and fate have not been explored to this extent. This is important as many individual studies on PCL, PLA or PA alone have shown different results. Moreover, gene analysis was performed to display the 3D scaffold potential for hMSC differentiation capacity. Finally, we also combine this cellular analysis with efforts to solve the well-known cell infiltration issues of electrospun scaffolds in a straightforward manner. We used a one-step simple process to improve cell filtration throughout the scaffold by incorporating chemokines, G-SCF and SDF-1, to promote hMSC migration and evaluate its functionality through direct and indirect seeding of loaded scaffolds.
2. Experimental
2.1. Scaffold preparation
Three different polymers were used to electrospun polymeric fibrous scaffolds: poly(ε-caprolactone) (PCL, Mw = 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Dalton, Sigma Aldrich), polylactic acid (PLA, Mw = 200
000 Dalton, Sigma Aldrich), polylactic acid (PLA, Mw = 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Dalton, 2002D, Folienwerk Wolfen GmbH) and the copolymer poly(ethylene oxide terephthalate)/poly(butylene terephthalate) (PA) (PEG Mw = 300 Dalton, weight ratio 55
000 Dalton, 2002D, Folienwerk Wolfen GmbH) and the copolymer poly(ethylene oxide terephthalate)/poly(butylene terephthalate) (PA) (PEG Mw = 300 Dalton, weight ratio 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 of PEOT
45 of PEOT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBT). Polymer solutions were prepared by dissolution in chloroform (Sigma Aldrich) overnight at different concentrations within the range of 10–20% (w/v) (ESI Table 1†). A custom electrospinning chamber with environmental control (20 °C, 30% humidity) was used to fabricate the scaffolds. The prepared solutions were loaded into a syringe and placed in a syringe pump (KDS 100, KD Scientific). A metallic needle was connected to a syringe tip that acted as the spinneret. Aluminium foil was used as a collector. Different flow rates (0.5–20 ml h−1) and voltages (12–25 kV) were used and examined. Chemokine incorporation was done with PA scaffolds showing the best results in terms of flow rate versus control of fibre diameter. Incorporation of granulocyte colony-stimulating factor and stromal cell-derived factor (G-CSF, SDF-1; PeproTech) in PA fibres was performed by mixing the chemokines with 20% (w/v) polymer solution and electrospinning at 15 kV, 1 ml h−1 flow rate and 20 cm working distance. We used 6 μg ml−1 SDF-1 or G-SCF as the final concentration for electrospinning. A list of parameter specification for different polymer types can be seen in ESI Table 1.†
PBT). Polymer solutions were prepared by dissolution in chloroform (Sigma Aldrich) overnight at different concentrations within the range of 10–20% (w/v) (ESI Table 1†). A custom electrospinning chamber with environmental control (20 °C, 30% humidity) was used to fabricate the scaffolds. The prepared solutions were loaded into a syringe and placed in a syringe pump (KDS 100, KD Scientific). A metallic needle was connected to a syringe tip that acted as the spinneret. Aluminium foil was used as a collector. Different flow rates (0.5–20 ml h−1) and voltages (12–25 kV) were used and examined. Chemokine incorporation was done with PA scaffolds showing the best results in terms of flow rate versus control of fibre diameter. Incorporation of granulocyte colony-stimulating factor and stromal cell-derived factor (G-CSF, SDF-1; PeproTech) in PA fibres was performed by mixing the chemokines with 20% (w/v) polymer solution and electrospinning at 15 kV, 1 ml h−1 flow rate and 20 cm working distance. We used 6 μg ml−1 SDF-1 or G-SCF as the final concentration for electrospinning. A list of parameter specification for different polymer types can be seen in ESI Table 1.†
2.2. Scaffold characterisation
Electrospun fibres were gold sputtered (Cressington 108 auto) at 40 mA and 100 mTorr for 30 seconds. Fibre morphology was observed using a scanning electron microscope (SEM) (XL 30 ESEM-FEG, Philips/FEI). Images at 50–10000× magnification were captured and examined for homogeneity and fibre surface topography. A magnification of 500× was used to measure the fibre's diameter. A minimum of 5 images per sample were randomly acquired with a minimum of 20 fibres measured to determine fibre diameters using ImageJ (National Institutes of Health, Bethesda, MD, USA). Selected electrospinning parameters were chosen from the highest reproducibility and similar fibre diameters.2.3. Cell isolation and expansion
Bone marrow aspirates were obtained through written informed consent, in conformity with national laws and after a local ethical committee approval. Ethical approval for the use of bone marrow samples was obtained from the ethical advisory board of the Medisch Spectrum Twente, Enschede. All methods were carried out in accordance with local and relevant guidelines and regulations. Bone marrow derived mesenchymal stromal cells (hMSCs) were isolated and expanded as previously described.23 hMSCs were cultured in a culture medium comprising α-MEM (Gibco), fetal bovine serum (10%, Lonza), ascorbic acid (0.2 mM, Gibco), L-glutamine (2 mM, Gibco), penicillin (100 U ml−1) and streptomycin (100 mg ml−1, Gibco). hMSCs were expanded at an initial seeding density of 3000 cells per cm2 in a basic culture medium and refreshed every 2–3 days. Cells were harvested at 80–90% confluency, before trypsinisation for cell seeding on electrospun scaffolds. All cell experiments were performed under a 5% CO2 humid atmosphere at 37 °C.2.4. In vitro study
In vitro experiments were performed with cells from a 53-year-old male and seeded at passage 4. PCL, PLA and PA electrospun scaffolds 13 mm in diameter and ∼500 μm in thickness were disinfected in 70% ethanol for 10 minutes 5 times and the ethanol was allowed to evaporate upon the last incubation. The scaffolds were then washed twice with sterile PBS (Gibco), transferred to a non-treated 24 well plate (Nunc) and incubated in a culture medium overnight. After the removal of the media, the scaffolds were seeded with 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells in 250 μl culture medium. An additional 750 μl culture medium was added after 4 hours at 37 °C to allow cell attachment. The culture was observed at day 1, 7 and 14, if not mentioned otherwise.
000 cells in 250 μl culture medium. An additional 750 μl culture medium was added after 4 hours at 37 °C to allow cell attachment. The culture was observed at day 1, 7 and 14, if not mentioned otherwise.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per scaffold were used. In transwells, 5000 cells were seeded on top of the transwell membrane. The culture was observed at day 1 and day 3. The experiments were performed in triplicate.
000 cells per scaffold were used. In transwells, 5000 cells were seeded on top of the transwell membrane. The culture was observed at day 1 and day 3. The experiments were performed in triplicate.
2.5. Biochemical assay
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 Alamar Blue (Sigma Aldrich) in the culture medium for 4 hours at 37 °C in the dark. 100 μl of the solution from each sample were measured in triplicate with a spectrofluorometer at a fluorescence excitation wavelength of 570 nm and emission at 585 nm (Viktor3, PerkinElmer). Fluorescence levels were normalized by cell number to achieve a per cell measurement.
10 Alamar Blue (Sigma Aldrich) in the culture medium for 4 hours at 37 °C in the dark. 100 μl of the solution from each sample were measured in triplicate with a spectrofluorometer at a fluorescence excitation wavelength of 570 nm and emission at 585 nm (Viktor3, PerkinElmer). Fluorescence levels were normalized by cell number to achieve a per cell measurement.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4° C. The aqueous phase containing RNA was collected, precipitated with 70% ethanol, transferred to Nucleospin RNA columns (Macherey-Nagel), and processed following the manufacturer's instructions. The quantity and quality of RNA were analysed using an ND100 spectrophotometer at 260 nm (Nanodrop Technologies, USA). cDNA was synthesized from 500 ng of RNA, using a SensiFast cDNA synthesis kit (BioRad) according to the manufacturer's protocol. Quantitative PCR was performed on an iQ5 detection system (Bio-Rad) and fold induction was calculated using the Pfaffl method. The primer sequences are provided in ESI Table S2.†
000 rpm at 4° C. The aqueous phase containing RNA was collected, precipitated with 70% ethanol, transferred to Nucleospin RNA columns (Macherey-Nagel), and processed following the manufacturer's instructions. The quantity and quality of RNA were analysed using an ND100 spectrophotometer at 260 nm (Nanodrop Technologies, USA). cDNA was synthesized from 500 ng of RNA, using a SensiFast cDNA synthesis kit (BioRad) according to the manufacturer's protocol. Quantitative PCR was performed on an iQ5 detection system (Bio-Rad) and fold induction was calculated using the Pfaffl method. The primer sequences are provided in ESI Table S2.†
2.6. Imaging
2.7. Statistical analysis
All data are expressed as mean ± s.d. (n = 3), if not stated otherwise. Biochemical assays were performed in triplicate, if not stated otherwise. Statistical analysis was performed with two-way analysis of variance (ANOVA) with Bonferroni's multiple comparison test (p < 0.05), unless otherwise indicated in the figure legends. For all figures the following applies: * = p < 0.05, ** = p < 0.01, *** = p < 0.001.3. Results
3.1. Electrospun fibre fabrication from different polymer solutions and parameters
PCL, PLA and PA at different concentrations were electrospun with different voltages, working distances and flow rates to fabricate different fibre diameters and surface topographies on the fibres. Different surface topographies can be obtained on the scaffolds by tuning the electrospinning parameters. As seen in Fig. 1a–c, tuning different parameters generated pores of 10–50 μm size, rough porous surface topographies of fibres, and bead–string fibres. By controlling the different parameters, and allowing only one parameter to change, in this case the flow rate (Fig. 1g–n), different fibre diameters can be obtained. By analysing the fibre diameter reproducibility and similarity, selected electrospinning process parameters (Table 1) for the different considered polymers were chosen to prepare the scaffolds for in vitro studies. | ||
| Fig. 1 Scanning electron microscopy images of electrospun fibres and fibre diameter analysis. (a–f) Images of PCL, PLA and PA. (a–c) Different surface topographies created by electrospinning: (a) semi-rough circular pores, (b) rough porous fibres, and (c) beads and strings. (d–f) Fibres represented in Table 1, providing different parameters to fabricate similar diameter fibres of different polymer compositions. (g–l) Images of PA at different flow rates: (g) 1, (h) 2.5, (i) 5, (j) 10, (k) 15, and (l) 20 ml h−1. Scale bar: 50 μm. (m) The correlation between fibre diameter and flow rate upon electrospinning PCL, PLA and PA solutions. All data are shown as mean ± s.d. (n = 20). | ||
| Polymer | Concentration (w/v) | Voltage | Working distance | Flow rate | Fibre diameter |
|---|---|---|---|---|---|
| PCL | 20% | 12 kV | 20 cm | 0.8 ml h−1 | 4.387 ± 0.567 μm |
| PLA | 15% | 12 kV | 20 cm | 7.5 ml h−1 | 4.504 ± 1.064 μm |
| PA | 20% | 15 kV | 20 cm | 1.0 ml h−1 | 6.313 ± 1.004 μm |
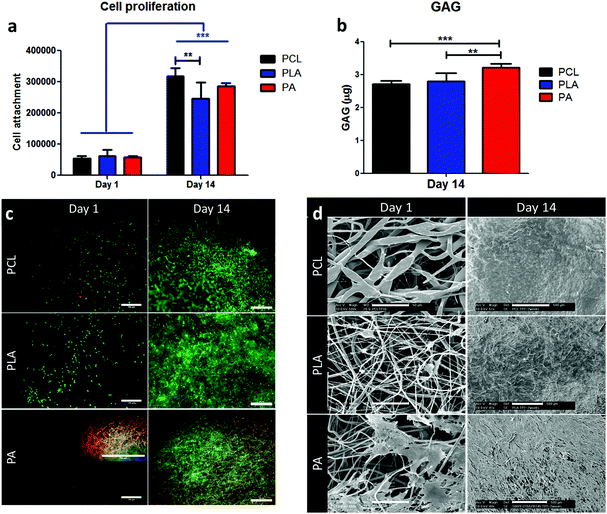
3.2. Biocompatibility of electrospun scaffolds through cell proliferation, viability and morphology
After two weeks of culture, the cells proliferated significantly (Fig. 2a), resulting in a 5.8-fold increase for PCL, 4-fold for PLA and 4.9-fold for PA. PCL provided significantly higher proliferation in comparison with PLA scaffolds. At day 14, the amount of GAG per scaffold was quantified and showed significantly higher GAG in PA scaffolds compared to PCL and PLA. When normalized to the amount of DNA, PA and PLA showed significantly higher GAG compared to PCL scaffolds (ESI Fig. 1†). Cell viability was qualitatively measured by imaging cells using live/dead staining. As shown in Fig. 2c, the cells observed in both PCL and PLA scaffolds showed a majority of live cells, with very limited or no dead cells. Despite the high autofluorescence of PA scaffolds, the day 1 inset showed viable live cells. At day 14, cell viability was confirmed, showing no presence of dead cells. For all polymer types, cells were homogenously distributed on the scaffolds. SEM images provided a highly detailed cell morphology (Fig. 2d). In particular, PCL and PLA scaffolds showed more singular cells and single cell–fibre contact, while PA substrates displayed more cells, cell–fibre contact, and cell–cell contact. At day 14, cells formed a confluent monolayer on the scaffolds with possible ECM production.3.3. Metabolic activity analysis on cells seeded on electrospun scaffolds
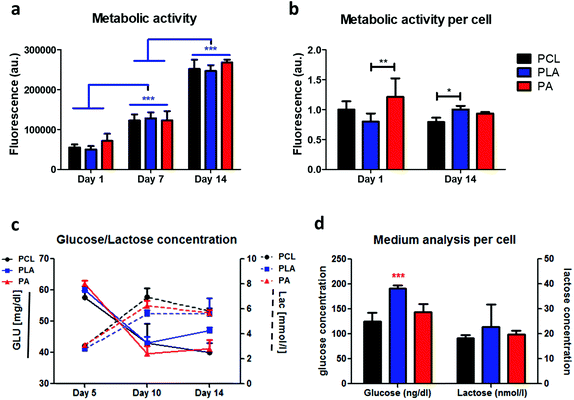
The total metabolic activity was observed to increase with the days of culture (Fig. 3a). A significantly higher metabolic activity was observed for the PA scaffold in comparison with PLA at day 1 (Fig. 3b). Moreover, with respect to the amount of initial glucose, 100 mg dl−1 (11 mmol l−1) in the medium upon refreshing, around 40% of glucose was consumed from day 1 to day 5, and significantly more from day 5 to day 10 in all the considered scaffolds, with cells on PA consuming the most at 60.33%. There was no significant difference in terms of glucose consumption at day 14 in comparison with day 10. In regard to lactate concentration, an average build-up of 27 mg dl−1 (3 mmol l−1) was exhibited at day 5, and a significant increase on all scaffold types at day 10 was measured, with PCL producing the most quantity at 62.6 mg dl−1 (6.95 mmol l−1). No significant difference was observed when comparing day 10 and 14 lactate production (Fig. 3c). The amounts of consumed glucose and lactate were normalized by the amount of cells. A significantly higher glucose consumption was found for PLA with respect to PCL (Fig. 3d).3.4. Gene expression level on electrospun scaffolds versus unseeded and tissue culture plates
Considering hMSCs, which were not seeded on electrospun scaffolds (day 0), as a reference, seeded hMSCs after 14 days of culture in electrospun scaffolds and tissue culture plates (TCPs) were downregulated in stemness markers (Fig. 4a). For both stemness markers, activated leukocyte cell adhesion molecule (ALCAM) and octamer-binding transcription factor 4 (OCT4), hMSCs on electrospun scaffolds displayed on average 10- and 16-fold downregulation. Both types of downregulation were statistically significant to TCP of 5- and 7-fold. CD63 was also downregulated by an average of 4-fold, with a slightly higher expression compared to TCP, although not statistically significant (Fig. 4a). Comparing differentiation markers, at day 14 using TCP as a reference (Fig. 4b–g, ESI Fig. 2†) it is possible to detect the influence of the substrate in hMSC differentiation. For the osteogenic markers OPN and OCN (Fig. 4b and c), upregulation was observed for all electrospun scaffolds with an average of 2- and 7-fold (Fig. 4d). In alkaline phosphatase (ALP) gene expression, PCL scaffolds provided a significant upregulation of 4-fold compared to PLA and PA scaffolds of 2.5- and 3-fold. Collagen type 1a (Col1a) gene expression also obtained a significantly higher upregulation in PCL scaffolds of 32-fold, compared to PA (17-fold upregulation) and PLA (24-fold upregulation) scaffolds (Fig. 4e). For other osteogenic markers, such as runt-related transcription factor 2 (Runx2) and bone morphogenetic protein 2 (BMP2), PLA scaffolds provided a similar gene regulation as TCP in Runx2 and significantly higher gene regulation than PCL and PA scaffolds (Fig. 4f and g). For chondrogenic markers, collagen type 2a (Col2a) and sox9 downregulation was observed when compared to TCP. PLA scaffolds exhibited a lower downregulation of 2-fold, significantly lower than PA scaffolds in Col2a gene expression (ESI Fig. 2†).3.5. Stimulating active migration of hMSCs to the depth of the electrospun scaffold
As PA showed more homogeneous retention of fibre morphology with a fibre diameter increasing with increasing flow rate up to 20 ml h−1, we chose these conditions to electrospin scaffolds incorporating G-CSF and SDF-1 to induce the migration of hMSCs to the inner part of the electrospun scaffolds. As reported in Fig. 5a–c, normal PA scaffolds did not provide a high amount of cell filtration despite the high cell density on the top surface. By incorporating G-SCF and SDF-1 chemokines, cell filtration increased significantly (Fig. 5d–i). Dual layer scaffolds, where fibres loaded with G-SCF or SDF-1 chemokines are found on the bottom layer of the scaffolds, actively attracted hMSCs to infiltrate to the bottom layer of the scaffolds (Fig. 5j–o). Fig. 5p shows that PA with G-SCF and SDF-1 chemokines tended to have a higher metabolic activity compared to PA only. Direct contact of hMSCs with PA loaded with chemokines resulted in a higher significant difference compared to PA only upon initial seeding. At day 3, PA + SDF-1 provided a significant higher metabolic activity. Direct contact with PA + G-CSF showed a significantly higher metabolic activity in comparison with PA/G-SCF at day 1. This was also observed for SDF-1 at day 3, with a slightly higher metabolic activity for PA + G-CSF in comparison with PA/G-SCF. Cell migration studies (Fig. 5q) using transwell membranes showed a significant increase of migratory cells when PA scaffolds were loaded with G-SCF and SDF-1 chemokines, with PA mixed with chemokines being more statistically significant than PA with an additional layer of chemokines. Furthermore, the stained images of migratory cells supported our results demonstrating that loaded scaffolds induced more hMSC migration (ESI Fig. 3†).4. Discussion
Here, we used three different polymers to electrospin scaffolds mimicking the physical structure of the natural tissue environment by fine tuning different processing parameters. By controlling the different parameters of electrospinning, such as solution concentration and composition, voltage, working distance, flow rate, humidity and temperature, one can tune the resulting fibre diameter and topography.2 This versatility can provide many solutions for various applications.4,17 A standard control to check if electrospinning produces high-quality fibres would be to evaluate if the fibres are distinguishable, smooth and without beads. However, non-standard fibres (Fig. 1a–c) can eventually provide alternative applications. The creation of circular pores has been shown to provide higher cell attachment and increase ECM protein secretion.24 Porous fibre surfaces increased focal adhesion strength and have been shown to contribute to hMSC differentiation to osteogenic lineages through the activation of ERK 1/2 by the Grb2-Sos-Ras pathway.25,26 Moreover, beads can be used as a drug reservoir and have diverse applications in controlled drug delivery.27 Fibre diameters of about 5 μm were chosen to mimic a similar magnitude of native fibrils and provide an optimal template for cells to attach and distribute well around the fibres with diameters smaller than those of the cells.28 Increasing flow rate resulted in an increase of the fibre diameter, which is supported by many other studies with other polymers.2 When spinning different polymer compositions, tuning the different parameters might be necessary to provide similar fibre diameters. Different polymers have different molecular weights and chemical compositions, which reflect the number of entanglement and the type of polymer chain in the solution. This has a significant effect on rheological and electrical properties such as viscosity, surface tension, conductivity and dielectric strength,3 which are important for the morphology of the electrospun fibres, hence resulting in differences in the fibre diameter despite similar flow rates.In tissue engineering applications, fibrous electrospun scaffolds require an optimal template for cells to seed, migrate, and grow. Successful regeneration of biological tissues and organs calls for the development of fibrous structures with fibre architectures beneficial for cell deposition and proliferation. Here, we demonstrated how our scaffolds produced a high proliferation rate up to 6-fold higher after two weeks of culture, and provided good distribution of the cells and high cell confluency. PCL scaffolds displayed the highest increase in proliferation with significantly higher cell attachment at day 14 compared to PLA. However, PLA showed higher initial cell attachment. This could be due to the relatively higher hydrophilicity of the PLA used here compared to PCL, in addition to its lower molecular weight.29,30 PLA has been known to degrade through bulk hydrolysis of ester bonds releasing lactic acid. This may cause a drop in pH levels and increase ionic strength by releasing lactate ions that might hamper cell proliferation.31 Hence, despite being more hydrophilic than PCL, PLA provided lower proliferation at day 14. Upon encountering hydrophobic surfaces such as PA, the cells were prone to aggregate and trigger the production of ECM proteins such as GAG and collagen.32 Moreover, PA has been shown to be more hydrophobic compared to the other considered polymer types.24 This could explain the necessity of cell–cell contact, and higher secretion of total GAG, as the cells seem to be more grouped and less spread compared to PLA and PCL.
One of the key factors of biocompatibility of an implanted biomaterial is cell viability and metabolic activity. Here, we showed that cells are viable in our electrospun scaffolds, with a majority of live cells and limited or no dead cells. Alamar Blue assay33 supported these findings showing a significantly high increase of metabolic activity throughout the culture time, and a stable activity when normalised to the number of cells. Initially, significantly higher metabolic activity was seen in PA in comparison with PLA scaffolds, perhaps due to the higher hydrophobicity of the scaffold providing the need to secrete proper proteins for optimal adherence. In recent studies, a higher amount of the ECM, which can be correlated to higher metabolic activity, was produced in less hydrophilic surfaces to maintain a more optimal environment for the cells to grow.24 Another factor to evaluate cell/scaffold interaction would be the concentration of glucose and lactate. Cells utilize glucose available in the culture medium (100 mg dl−1) for energy and produce lactate as a metabolic by-product. High levels of lactate (≥20 mmol L−1) can significantly inhibit cell growth.31 Hence, monitoring glucose and lactate concentrations is a crucial assessment for culture performance. As the medium was refreshed at day 1, 5, 7, and 10, we show a high metabolism of glucose with a significant increase at day 10 and 14 compared to day 5. The metabolic waste lactate was at a level not considered to be cytotoxic.34 However, increased levels of lactate likely due to the degradation of PLA in combination with a reduced pH may hamper cell growth. A possible explanation to the medium analysis at day 14 normalized by cell amount, with PLA providing a significantly higher glucose consumption, reflects and supports the analysed cell metabolic activity.
hMSCs are often used in tissue engineering strategies because of their ability to differentiate into multiple cell lineages.35,36 Li et al. found that electrospun PCL scaffolds support the attachment, proliferation and differentiation of hMSCs into adipogenic, chondrogenic, or osteogenic lineages based upon the culture media selected.37 Here, we used a basic medium to examine the potential of MSC differentiation and stemness maintenance of three different chemical composition polymeric scaffolds, without the addition of differentiation supplements. SB-10 antibody, a surface marker of CFU-Fs, reacts with CD166 (also known as ALCAM) found on undifferentiated MSCs, which disappears as soon as the cells initiate osteogenic differentiation and start to secrete alkaline phosphatase on their cell surface.38 This could be the reason why all scaffolds downregulated stemness markers, as all seemed to be prone to differentiate to an osteogenic lineage. High porosity surfaces, as the one obtained in electrospinning, can influence a number of cellular processes.39 A previous study showed that the disordered array of pores resulted in an increase in bone mineral production and osteogenic differentiation of hMSCs even in the absence of chemical supplements.40 Similarly, Kumar et al. showed that the cells are highly sensitive to the scaffold structure with nanofibrous scaffolds, which steered hMSCs down to an osteogenic lineage.41 Moreover, studies comparing 2D and 3D scaffolds showed a higher level of osteoblast proliferation and could be favourable for bone matrix formation.42
The potential of osteogenic differentiation was detected in all our electrospun scaffolds, with the upregulation of ALP, Col1a, OPN and OCN. Osteogenic differentiation of MSCs in vitro can be divided into three stages. The first stage consists of an initial proliferation period, confluency and expression of Runx2 and BMP2.43 Runx2, the earliest transcription factor proven to be essential for bone formation, is activated by DLX3 and a homeodomain transcriptional network through BMP2.44 The second stage is early cell differentiation, which is characterised by the expression of the ALP and Col1 matrix providing a template where the minerals will be deposited.45,46 The final stage is the high expression of OCN and OPN, followed by calcium and phosphate deposition.47 Studies of early markers of osteogenic differentiation are normally performed after three days in culture, between the proliferation stage and the determined stage of differentiation due to their high expression.48,49 Hence, despite the crucial activation of Runx2 to promote ALP, Col1a, OPN and OCN, high expression of these late markers was not seen at day 14. Furthermore, studies have found that short-term BMP-2 treatment is both necessary and sufficient for osteogenic differentiation.50 PCL provided the highest upregulation of ALP and Col1a. Col1a is a general marker of production of the ECM, which could also explain the highest proliferation rate on PCL. Hydrophilic surfaces tend to enhance early osteogenic markers, such as ALP and Col1a, compared to hydrophobic surfaces,51,52 much like that seen when comparing PCL to PA scaffolds. From this theory, PLA scaffolds might have provided a higher upregulation; however one could argue that the degradation of the PLA scaffolds, and the release and chemical actions of lactic ions hampers upregulation,53 as the addition of a decreased pH has been shown to significantly decrease ALP activity and collagen synthesis.54,55
hMSCs can undergo osteoblastic differentiation in response to nanoscale surface features of pits, grooves, symmetry and disorder in absence of media supplements.40,56 Dalby et al. showed that the differentiation mechanism of these cells was different compared to the cells cultured in media supplemented with dexamethasone, despite having similar gene expressions.57 The physical and mechanical properties of the scaffolds did not stimulate chondrogenic differentiation to the same level as shown in the literature.58,59 PLA might provide a potential scaffold for cartilage regeneration, due to its high GAG/DNA and highest gene expression of the Col2a and Sox9 markers, compared to PCL and PA. However, much needs to be modified in the PLA scaffold alone and recent studies have combined PLA with carbon nanotubes, gelatin, collagen, and chitosan to create scaffolds for cartilage regeneration.60,61 For future studies, differentiation media could be used as positive controls, such as the addition of a basic fibroblast growth factor to maintain stemness,62 dexamethasone (DEX) and beta-glycerophosphate to differentiate to osteogenic lineages63,64 and transforming growth factor-beta for a chondrogenic medium.65,66
For the development of electrospun scaffolds for tissue engineering applications, one would need to consider the ability of cells to infiltrate into the pore network. Many electrospun scaffolds provide high cell proliferation, but are somewhat limited to their outside part as the cells are not able to infiltrate their pore network.13 Several studies have been focused to improve cell infiltration. However, they have shown very limited improvement or adverse response in other functionalities of electrospun scaffolds at the interface with cells.16,67,68 Other novel ways to improve cell filtrations would normally require limiting the variety of polymers that can be electrospun, post-processing, and eliminating the simplicity of electrospinning by adding complex setups.13,67,69 Cells can migrate in response to a variety of chemical and physical stimuli. Hence, in this study we used a one-step electrospinning technique by incorporating chemokines, SDF-1 or G-SCF to promote hMSC migration into the depth of the scaffold. PA was the scaffold of choice due to its ability to be electrospun at different flow rates resulting in fine fibres, with increasing fibre diameter. Moreover, the high molecular weight of PA allows it to maintain a sufficient number of entanglements of the polymer chains, hence ensuring a sufficient level of solution viscosity to yield a uniform jet during electrospinning and restrain effects of surface tension, which plays a major role in bead formation.70 Despite this, PA loaded scaffolds resulted in bead formations on the fibres. Although this might be considered as a hindrance, it could actually serve as an advantage of visualizing the intersection between the layer of PA only fibres and PA loaded fibres for double layer PA scaffolds. As mentioned before,27 beads can be used as drug reservoirs of SDF-1 and G-SCF and provide an explanation on such a significant effect on migration in both single and double layer PA scaffolds on both direct and transwell types of seeding. Furthermore, Gaharwar et al. showed that the encapsulation of dexamethasone within the beaded structure resulted in sustained release of the drug over a period of 28 days.71 In spite of the improvement of cellular infiltration, the presence of a bead is a distortion of the original shape of the scaffold, possibly changing its mechanical properties, resulting in a change of the cellular behaviour. Moreover, one could argue that physical changes through the presence of beads resulting in a smaller fibre diameter, but a similar if not higher porosity, could be the reason for higher cellular infiltration regardless of the chemokines present. Nevertheless, Zander et al. used sacrificial layers in a PEO combined PCL scaffold, but no difference was observed in cellular infiltration possibly due to the bead or cluster formation of PEO fibres. Here, we observed bead formation, yet with successful cell migration into the scaffold. Other methods can be used to avoid bead formation during electrospinning, such as adjusting the different parameter setups, as well as adding a salt to improve the charge carrying capacity of the solution creating bead-free fibres.72 Exposing hMSCs to G-CSF can significantly enhance the homing capacity of these stem cells towards SDF-1.73 Further studies can be envisioned to engineer a dual incorporation of G-SCF and SDF-1 into the electrospun scaffolds. Moreover, SDF-1 and G-SCF have been known not only to promote hMSC migration but also angiogenesis and revascularization of the tissue.74,75 Hence, this strategy would provide a promising template for many tissue engineering applications.
5. Conclusion
Electrospinning is indeed a notable and convenient way to fabricate scaffolds for tissue engineering purposes. Optimization of electrospinning is crucial to provide a biocompatible, electrospun mimicking ECM with proper infiltration for successful tissue regeneration, without reducing its original simplicity. Our study optimised three different polymers, PCL, PLA and PA, and investigated their biocompatibility showing high and stable metabolic activity and proliferation, viable cells, and modulation of glucose and lactate concentrations. The scaffolds showed potential in stimulating cells toward osteogenic lineage with PCL providing most early osteogenic upregulation and PLA early chondrogenesis. We introduced a one-step electrospinning technique using the stable fibre producing PA scaffolds to incorporate the chemokines G-SCF and SDF-1 to enhance hMSC infiltration and proved through cell migratory studies their significant potential in improving cellular infiltration. In conclusion, this study has provided a potential and simple way to overcome the challenges in cellular infiltration in highly packed dense electrospun scaffolds for 3D tissue regeneration.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research forms part of the Project P3.03 DialysisXS of the research program of the BioMedical Materials Institute, co-funded by the Dutch Ministry of Economic Affairs, Agriculture and Innovation. The financial contribution of the Nierstichting Nederland is gratefully acknowledged. This research project has been made possible with the support of the Dutch Province of Limburg.References
- D. H. Reneker and I. Chun, Nanotechnology, 1996, 7, 216–223 CrossRef CAS.
- J. M. Deitzel, J. Kleinmeyer, D. Harris and N. C. Beck Tan, Polymer, 2001, 42, 261–272 CrossRef CAS.
- A. K. Haghi and M. Akbari, Phys. Status Solidi A, 2007, 204, 1830–1834 CrossRef CAS.
- Z. M. Huang, Y. Z. Zhang, M. Kotaki and S. Ramakrishna, Compos. Sci. Technol., 2003, 63, 2223–2253 CrossRef CAS.
- E. Eggenhofer, F. Luk, M. H. Dahlke and M. J. Hoogduijn, Front. Immunol., 2014, 5, 148 Search PubMed.
- E. Eggenhofer, V. Benseler, A. Kroemer, F. C. Popp, E. K. Geissler, H. J. Schlitt, C. C. Baan, M. H. Dahlke and M. J. Hoogduijn, Front. Immunol., 2012, 3, 297 CAS.
- W. Cui, Y. Zhou and J. Chang, Sci. Technol. Adv. Mater., 2010, 11, 014108 CrossRef PubMed.
- W. Friess, Eur. J. Pharm. Biopharm., 1998, 45, 113–136 CrossRef CAS.
- N. G. Rim, C. S. Shin and H. Shin, Biomed. Mater., 2013, 8, 014102 CrossRef.
- L. Moroni, J. R. de Wijn and C. A. van Blitterswijk, J. Biomed. Mater. Res., Part A, 2005, 75, 957–965 CrossRef CAS PubMed.
- M. A. Woodruff and D. W. Hutmacher, Prog. Polym. Sci., 2010, 35, 1217–1256 CrossRef CAS.
- D. Garlotta, J. Polym. Environ., 2001, 9, 63–84 CrossRef CAS.
- S. Khorshidi, A. Solouk, H. Mirzadeh, S. Mazinani, J. M. Lagaron, S. Sharifi and S. Ramakrishna, J. Tissue Eng. Regener. Med., 2016, 10, 715–738 CrossRef CAS PubMed.
- B. A. Blakeney, A. Tambralli, J. M. Anderson, A. Andukuri, D. J. Lim, D. R. Dean and H. W. Jun, Biomaterials, 2011, 32, 1583–1590 CrossRef CAS PubMed.
- S. J. Eichhorn and W. W. Sampson, J. R. Soc., Interface, 2005, 2, 309–318 CrossRef PubMed.
- A. K. Ekaputra, G. D. Prestwich, S. M. Cool and D. W. Hutmacher, Biomacromolecules, 2008, 9, 2097–2103 CrossRef CAS PubMed.
- B. M. Baker, A. O. Gee, R. B. Metter, A. S. Nathan, R. A. Marklein, J. A. Burdick and R. L. Mauck, Biomaterials, 2008, 29, 2348–2358 CrossRef CAS PubMed.
- M. F. Canbolat, C. Tang, S. H. Bernacki, B. Pourdeyhimi and S. Khan, Macromol. Biosci., 2011, 11, 1346–1356 CrossRef PubMed.
- J. T. Seil and T. J. Webster, Int. J. Nanomed., 2011, 6, 1095–1099 CAS.
- A. Townsend-Nicholson and S. N. Jayasinghe, Biomacromolecules, 2006, 7, 3364–3369 CrossRef CAS PubMed.
- X. Liu, B. Duan, Z. Cheng, X. Jia, L. Mao, H. Fu, Y. Che, L. Ou, L. Liu and D. Kong, Protein Cell, 2011, 2, 845–854 CrossRef CAS PubMed.
- Y. Wu and R. C. Zhao, Stem Cell Rev., 2012, 8, 243–250 CrossRef CAS PubMed.
- S. K. Both, A. J. C. v. d. Muijsenberg, C. A. v. Blitterswijk, J. d. Boer and J. D. d. Bruijn, Tissue Eng., 2007, 13, 3–9 CrossRef CAS PubMed.
- F. F. Damanik, T. C. Rothuizen, C. van Blitterswijk, J. I. Rotmans and L. Moroni, Sci. Rep., 2014, 4, 6325 CrossRef CAS PubMed.
- S. Lavenus, M. Berreur, V. Trichet, P. Pilet, G. Louarn and P. Layrolle, Eur. Cells Mater., 2011, 22, 84–96 CrossRef CAS ; discussion 96.
- M. J. Biggs and M. J. Dalby, Proc. Inst. Mech. Eng., Part H, 2010, 224, 1441–1453 CrossRef CAS.
- H. Qi, P. Hu, J. Xu and A. Wang, Biomacromolecules, 2006, 7, 2327–2330 CrossRef CAS PubMed.
- C. T. Laurencin, A. M. Ambrosio, M. D. Borden and J. A. Cooper Jr., Annu. Rev. Biomed. Eng., 1999, 1, 19–46 CrossRef CAS PubMed.
- F. Ahmed and D. E. Discher, J. Controlled Release, 2004, 96, 37–53 CrossRef CAS PubMed.
- E. Suihko, R. T. Forbes, O. Korhonen, J. Ketolainen, P. Paronen, J. Gynther and A. Poso, J. Pharm. Sci., 2005, 94, 745–758 CrossRef CAS PubMed.
- Y. He, W. Wang and J. Ding, Chin. Sci. Bull., 2013, 58, 2404–2411 CrossRef CAS.
- S. Sart, A. C. Tsai, Y. Li and T. Ma, Tissue Eng., Part B, 2014, 20, 365–380 CrossRef PubMed.
- S. N. Rampersad, Sensors, 2012, 12, 12347–12360 CrossRef CAS PubMed.
- T. Omasa, K. Higashiyama, S. Shioya and K. Suga, Biotechnol. Bioeng., 1992, 39, 556–564 CrossRef CAS.
- S. Ghoraishizadeh, A. Ghorishizadeh, P. Ghoraishizadeh, N. Daneshvar and M. H. Boroojerdi, Adv. Regener. Med., 2014, 2014, 1–14 CrossRef.
- D. Howard, L. D. Buttery, K. M. Shakesheff and S. J. Roberts, J. Anat., 2008, 213, 66–72 CrossRef CAS PubMed.
- W. J. Li, R. Tuli, X. Huang, P. Laquerriere and R. S. Tuan, Biomaterials, 2005, 26, 5158–5166 CrossRef CAS PubMed.
- K. Kaveh, R. Ibrahim, B. M. Zuki Abu and T. Azmi Ibrah, J. Anim. Vet. Adv., 2011, 10, 2317–2330 CrossRef CAS.
- J. L. Ifkovits, K. Wu, R. L. Mauck and J. A. Burdick, PLoS One, 2010, 5, e15717 CrossRef CAS PubMed.
- M. J. Dalby, N. Gadegaard, R. Tare, A. Andar, M. O. Riehle, P. Herzyk, C. D. Wilkinson and R. O. Oreffo, Nat. Mater., 2007, 6, 997–1003 CrossRef CAS PubMed.
- G. Kumar, C. K. Tison, K. Chatterjee, P. S. Pine, J. H. McDaniel, M. L. Salit, M. F. Young and C. G. Simon Jr., Biomaterials, 2011, 32, 9188–9196 CrossRef CAS PubMed.
- X. F. Tian, B. C. Heng, Z. Ge, K. Lu, A. J. Rufaihah, V. T. Fan, J. F. Yeo and T. Cao, Scand. J. Clin. Lab. Invest., 2008, 68, 58–67 CrossRef CAS PubMed.
- A. Neve, A. Corrado and F. P. Cantatore, Cell Tissue Res., 2011, 343, 289–302 CrossRef CAS PubMed.
- M. Q. Hassan, R. S. Tare, S. H. Lee, M. Mandeville, M. I. Morasso, A. Javed, A. J. van Wijnen, J. L. Stein, G. S. Stein and J. B. Lian, J. Biol. Chem., 2006, 281, 40515–40526 CrossRef CAS PubMed.
- L. D. Quarles, D. A. Yohay, L. W. Lever, R. Caton and R. J. Wenstrup, J. Bone Miner. Res., 1992, 7, 683–692 CrossRef CAS PubMed.
- J. E. Aubin, Rev. Endocr. Metab. Disord., 2001, 2, 81–94 CrossRef CAS PubMed.
- Z. Huang, E. R. Nelson, R. L. Smith and S. B. Goodman, Tissue Eng., 2007, 13, 2311–2320 CrossRef CAS PubMed.
- N. Logan and P. Brett, Stem Cells Int., 2013, 2013, 361637 Search PubMed.
- S. Watari, K. Hayashi, J. A. Wood, P. Russell, P. F. Nealey, C. J. Murphy and D. C. Genetos, Biomaterials, 2012, 33, 128–136 CrossRef CAS PubMed.
- D. Noel, D. Gazit, C. Bouquet, F. Apparailly, C. Bony, P. Plence, V. Millet, G. Turgeman, M. Perricaudet, J. Sany and C. Jorgensen, Stem Cells, 2004, 22, 74–85 CrossRef CAS PubMed.
- J. M. Curran, Z. Tang and J. A. Hunt, J. Biomed. Mater. Res., Part A, 2009, 89, 1–12 CrossRef CAS PubMed.
- J. Fan, H. Park, S. Tan and M. Lee, PLoS One, 2013, 8, e72474 CrossRef CAS PubMed.
- T. Chen, Y. Zhou and W. S. Tan, Cell Biol. Toxicol., 2009, 25, 573–586 CrossRef CAS PubMed.
- D. H. Kohn, M. Sarmadi, J. I. Helman and P. H. Krebsbach, J. Biomed. Mater. Res., 2002, 60, 292–299 CrossRef CAS PubMed.
- T. Okabe, M. Sakamoto, H. Takeuchi and K. Matsushima, J. Endod., 2006, 32, 198–201 CrossRef PubMed.
- M. J. Dalby, D. McCloy, M. Robertson, C. D. Wilkinson and R. O. Oreffo, Biomaterials, 2006, 27, 1306–1315 CrossRef CAS PubMed.
- M. J. Dalby, A. Andar, A. Nag, S. Affrossman, R. Tare, S. McFarlane and R. O. Oreffo, J. R. Soc., Interface, 2008, 5, 1055–1065 CrossRef PubMed.
- H. Mahboudi, B. Kazemi, M. Soleimani, H. Hanaee-Ahvaz, H. Ghanbarian, M. Bandehpour, S. E. Enderami, M. Kehtari and G. Barati, Gene, 2018, 643, 98–106 CrossRef CAS PubMed.
- A. Di Luca, K. Szlazak, I. Lorenzo-Moldero, C. A. Ghebes, A. Lepedda, W. Swieszkowski, C. Van Blitterswijk and L. Moroni, Acta Biomater., 2016, 36, 210–219 CrossRef CAS.
- A.-M. Haaparanta, E. Järvinen, I. F. Cengiz, V. Ellä, H. T. Kokkonen, I. Kiviranta and M. Kellomäki, J. Mater. Sci.: Mater. Med., 2013, 25, 1129–1136 CrossRef PubMed.
- J. Markowski, A. Magiera, M. Lesiak, A. L. Sieron, J. Pilch and S. Blazewicz, J. Nanomater., 2015, 2015, 1–9 CrossRef.
- J. Wu, G. T. Huang, W. He, P. Wang, Z. Tong, Q. Jia, L. Dong, Z. Niu and L. Ni, J. Endod., 2012, 38, 614–622 CrossRef PubMed.
- L. Kyllonen, S. Haimi, B. Mannerstrom, H. Huhtala, K. M. Rajala, H. Skottman, G. K. Sandor and S. Miettinen, Stem Cell Res. Ther., 2013, 4, 17 CrossRef CAS PubMed.
- N. Jaiswal, S. E. Haynesworth, A. I. Caplan and S. P. Bruder, J. Cell. Biochem., 1997, 64, 295–312 CrossRef CAS PubMed.
- L. A. Solchaga, K. J. Penick and J. F. Welter, Methods Mol. Biol., 2011, 698, 253–278 CrossRef CAS PubMed.
- W. J. Li, R. Tuli, C. Okafor, A. Derfoul, K. G. Danielson, D. J. Hall and R. S. Tuan, Biomaterials, 2005, 26, 599–609 CrossRef CAS PubMed.
- X. Xu, J. F. Zhang and Y. Fan, Biomacromolecules, 2010, 11, 2283–2289 CrossRef CAS PubMed.
- N. E. Zander, J. A. Orlicki, A. M. Rawlett and T. P. Beebe, J. Mater. Sci.: Mater. Med., 2012, 24, 179–187 CrossRef PubMed.
- K. Tuzlakoglu, M. I. Santos, N. Neves and R. L. Reis, Tissue Eng., Part A, 2011, 17, 463–473 CrossRef CAS PubMed.
- N. Bhardwaj and S. C. Kundu, Biotechnol. Adv., 2010, 28, 325–347 CrossRef CAS PubMed.
- A. K. Gaharwar, S. M. Mihaila, A. A. Kulkarni, A. Patel, A. Di Luca, R. L. Reis, M. E. Gomes, C. van Blitterswijk, L. Moroni and A. Khademhosseini, J. Controlled Release, 2014, 187, 66–73 CrossRef CAS.
- K. Nartetamrongsutt and G. G. Chase, Polymer, 2013, 54, 2166–2173 CrossRef CAS.
- P. B. Bolno, D. Morgan, A. Wechsler and J. Y. Kresh, J. Am. Coll. Surg., 2004, 199, 33 CrossRef.
- J. Deshane, S. Chen, S. Caballero, A. Grochot-Przeczek, H. Was, S. Li Calzi, R. Lach, T. D. Hock, B. Chen, N. Hill-Kapturczak, G. P. Siegal, J. Dulak, A. Jozkowicz, M. B. Grant and A. Agarwal, J. Exp. Med., 2007, 204, 605–618 CrossRef CAS PubMed.
- Y. Ohki, FASEB J., 2005, 19(14) DOI:10.1096/fj.04-3496fje.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8bm00693h |
| This journal is © The Royal Society of Chemistry 2019 |