Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synchrotron-based infrared microspectroscopy study on the radiosensitization effects of Gd nanoparticles at megavoltage radiation energies
Immaculada
Martínez-Rovira
 *ab,
Olivier
Seksek
*ab,
Olivier
Seksek
 cd,
Josep
Puxeu
e,
Joan
Gómez
b,
Martin
Kreuzer
a,
Tanja
Dučić
a,
Maria Josep
Ferreres
f,
Manel
Artigues
e and
Ibraheem
Yousef
cd,
Josep
Puxeu
e,
Joan
Gómez
b,
Martin
Kreuzer
a,
Tanja
Dučić
a,
Maria Josep
Ferreres
f,
Manel
Artigues
e and
Ibraheem
Yousef
 *a
*a
aALBA-CELLS Synchrotron, MIRAS Beamline, Carrer de la Llum 2-26, 09290 Cerdanyola del Vallès, Spain. E-mail: immamartinez@gmail.com; iyousef@cells.es
bIonizing Radiation Research Group (GRRI), Physics Department, Universitat Autònoma de Barcelona (UAB), Avinguda de l'Eix Central, Edifici C. Campus de la UAB, 08193 Cerdanyola del Vallès, Spain
cImagerie et Modélisation en Neurobiologie et Cancérologie (IMNC), CNRS, Univ Paris Sud, Université Paris-Saclay, F-91400 Orsay, France
dUniversité de Paris, IMNC, F-91400 Orsay, France
eHospital Universitari Sant Joan de Reus, Institut d'Investigació Sanitària Pere Virgili, Avinguda del Dr. Josep Laporte 2, 43204 Reus, Spain
fLaboratori ASPCAT Tortosa, Avinguda Santa Teresa 2–12, 43590 Tortosa, Spain
First published on 31st July 2019
Abstract
The outcome of radiotherapy can be further improved by combining radiotherapy with nanoparticles. Previous biological studies showed a significant amplification of the biological damage in cells charged with nanoparticles prior to radiotherapy treatments. The rationale has been based on the physical dose enhancement. However, this subject is still a matter of controversy and there are clear indications that biochemical effects may play a key role in the radiosensitization effects of nanoparticles. Within this context, the main goal of our study was to provide new insights into the radiosensitization effects of F98 glioma cells exposed to gadolinium nanoparticles combined with clinical megavoltage beams, and compare them with respect to kilovoltage radiotherapy (commonly used in combination with nanoparticles). For this purpose, we used synchrotron-based Fourier transform infrared microspectroscopy (SR-FTIRM) to provide relevant information on the treatment-induced biochemical changes of the main cell biomolecules. Biochemical differences were evaluated after the treatments to assess cellular damage. Multivariate analysis revealed nanoparticle-dependent changes in megavoltage treated cells. The main spectral variations were related to conformational changes in the protein secondary structures, which might be induced by radiation damage and by changes or rearrangements in the nucleic acid structures due to the initiation of DNA repair mechanisms. We also observed significant changes in the phosphate I and II bands, which concerns DNA damage, while few changes were detected in the lipid region. Spectroscopic data showed that these changes increased as a function of the dose. Finally, PCA analysis did not discriminate clearly between megavoltage and kilovoltage groups treated with nanoparticles, indicating that megavoltage radiosensitization effects might not differ significantly from those in kilovoltage radiotherapy.
1 Introduction
The use of nanoparticles (NP) as tumour radiosensitizers was proposed by Hainfeld and collaborators in 2004, when they first reported the control of a malignant tumour (EMT-6) in vivo through the preferential absorption of X-rays by 1.9 nm Au NP.1 This pioneering experiment, along with the results obtained in subsequent biological studies (see ref. 2–7, among others), boosted the use of NP as a promising strategy to increase the efficiency of radiotherapy.The basis for NP use is their selective accumulation within tumours due to the enhanced permeability and retention (EPR) effect,8 leading to a higher cell penetration and less adverse effects with respect to conventional radiosensitizers.9 Therefore, the goal of combining NP with radiotherapy is to improve tumour cell killing, and thus, the differential effect between healthy and tumour tissues. An increase in the radiosensitivity of tumour cells treated with NP has been observed in vitro and in vivo, using kilovoltage and also megavoltage photon beams.10–12
The biological findings have been basically described in terms of physics since the presence of (high-Z) NP increases the probability of photoelectric and Compton interactions (and, thus, the generation of short-range secondary electrons).13 This results in a local dose enhancement in the tumour. However, the radiosensitization observed in some biological experiments is greater than the predicted increase in the physical dose,14–16 indicating that the biological response may not be driven only by dose enhancement (physical) effects. Other mechanisms such as the production of reactive oxygen species (ROS) and oxidative stress, DNA damage induction, cell cycle effects and potential interference with the bystander effects may play a major role.6,14–19 These mechanisms might be amplified in combination with radiotherapy, and thus increase the anticancer efficacy of NP. In addition, different radiosensitizing effects are expected according to the NP size, type, chemical nature, concentration, intracellular localization, radiotherapy configuration (energy) and cell line used.17 That is why the biological mechanisms of nanoparticle radiosensitization still remain unknown.
Within this context, the objective of this study is to use synchrotron-based Fourier transform infrared microspectroscopy (SR-FTIRM) to disentangle radiosensitization effects in the F98 glioma rat cell line treated with GdNP (AGuIX®) combined with radiotherapy. In addition to its radiosensitization properties, the use of Gd and its chemical derivatives is particularly interesting since it is the most widely used contrast agent for magnetic resonance imaging (MRI); this makes Gd a perfect tool for theragnosis (providing imaging and therapy at the same time).20 Thanks to this dual capacity, the use of GdNP is increasing.
FTIRM is a vibrational technique for the biochemical analysis of cells on a microscopic scale. FTIRM methods have been used to study the cell cycle, apoptosis, differentiation, and proliferation of different cell lines and tissues (see, for instance ref. 21–24). Analysis of the infrared spectra provides relevant information on the treatment-induced biochemical modifications of the main cell biomolecules (nucleic acids, proteins and lipids). In our previous work,25 we studied the biochemical structure alterations of biomolecules inside cells induced by low energy X-ray irradiations and GdNP using FTIRM. In particular, we observed important spectral signature alterations in the fingerprint (DNA and protein) and lipid regions related to changes in the cellular function and cell death processes. The main goal of the present study is to use SR-FTIRM to gain new insights into the radiosensitization effects involved when using GdNP at megavoltage (MV) photon energies (normally used in clinics) with respect to kilovoltage (kV) photons (commonly used in combination with NP). To the best of our knowledge, this is the first study that uses synchrotron-based FTIRM to evaluate NP-based radiotherapy approaches. The biochemical information provided by the highly brilliant infrared light source produced at ALBA Synchrotron leads to a clear advantage in spectral quality at a cellular level and will allow better characterization of NP-induced damage in cells.
2 Materials and methods
2.1 F98 cells and cell culture
The F98 rat glioma cell line (ATCC-CRL-2397) was purchased from LGC Standards (Molsheim, France). This type of cell has a highly invasive growth pattern and is weakly immunogenic in syngeneic Fischer rats.26The cells were cultured in high glucose DMEM medium (Gibco) supplemented with 10% fetal calf serum, 1% penicillin–streptomycin (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 units per mL each), 2 mM L-glutamine, 1 mM sodium pyruvate and 10 mM HEPES in an incubator at 37 °C. The cells were seeded in 6-well plates on the day prior to the irradiation; 2 mL of a 5 × 104 cells per mL suspension were placed in each well and incubated overnight in order to reach a 75% confluence rate on the day of irradiation.
000 units per mL each), 2 mM L-glutamine, 1 mM sodium pyruvate and 10 mM HEPES in an incubator at 37 °C. The cells were seeded in 6-well plates on the day prior to the irradiation; 2 mL of a 5 × 104 cells per mL suspension were placed in each well and incubated overnight in order to reach a 75% confluence rate on the day of irradiation.
2.2 Nanoparticles
Gadolinium NP (AGuIX®; Activation and Guiding of Irradiation by X-ray) were purchased from Nano-H (Lyon, France). These nanoparticles consist of a polysiloxane network surrounded by around 10 gadolinium chelates (diethylenetriaminepentaacetic acid, DTPA) covalently grafted to the polysiloxane inorganic matrix. The nanoparticles present a diameter of 3.0 ± 1.0 nm.27 The effectiveness of the AGuIX® NP has been proved in previous biological studies with several types of cell lines or animal models, and irradiation conditions.28–30A nanoparticle stock solution of 100 mM was obtained by solubilization in ultrapure Millipore (DirectQ 8) water and kept at 4 °C. For cell treatment, 3 mL of 1 mM nanoparticle solution in fresh supplemented medium was added into the wells and incubated for 6 hours at 37 °C. At such concentrations, NP are not toxic.9,31 A recent study with the same type of NP and cell line showed that the uptake of GdNP was time-dependent and reached a plateau after a 5-hour incubation time.19
2.3 Irradiation
Kilovoltage and megavoltage irradiations were performed at the Hospital Universitari Sant Joan de Reus. Kilovoltage irradiations were performed with a Gulmay D3300 kilovoltage X-ray therapy unit (Gulmay Ltd, Byfleet, UK) with an accelerating potential voltage of 60 kV (HVL = 1.19 mm Al). Dose conformation was performed with a circular 4 cm-diameter cone at 30 cm from the source. The cells were irradiated at a dose rate of 2.1 Gy min−1. Megavoltage irradiations were performed using a 6 MV photon beam from a Varian 2100 linear accelerator (Varian Medical System, Palo Alto, USA). The dose rate at the cells was also 2.1 Gy min−1 at a source to surface distance (SSD) of 95 cm and at 5 cm-depth (cell location). In both cases, several irradiation doses were delivered to the cells: 0 Gy (no irradiation), 5, 10 and 20 Gy.2.4 SR-FTIRM sample preparation
SR-FTIRM sample preparation was performed following our previous protocol.25 In order to evaluate immediate treatment-induced effects, the cells were fixed two hours after the irradiation. For this purpose, the medium was removed (having previously collected floating cells) and 100 μL of a 0.05% trypsin–EDTA solution (Lonza) was added to each well in order to detach the cells; the floating cells were added back to the trypsinised cells to take into account the whole cell population that was treated. Then, 500 μL of supplemented DMEM medium was added and the cell suspension was centrifuged at 1500 rpm for 5 minutes at 4 °C. The cellular pellet was rinsed with PBS, and re-centrifuged at 1500 rpm for 5 minutes. The pellet was then re-suspended in 10% formalin neutral buffered solution (Sigma-Aldrich) and incubated for 1 hour at room temperature. Then, the samples were centrifuged at 1500 rpm for 5 minutes, and the pellet was rinsed 3 times in Millipore water to wash out the residual phosphate ions. Dried cells, previously deposited on 0.5 mm-thick infrared transparent calcium fluoride (CaF2), were then analyzed by SR-FTIRM.2.5 SR-FTIRM measurements
SR-FTIRM measurements (transmission mode) were performed at the MIRAS beamline of ALBA Synchrotron using a Hyperion 3000 microscope coupled to a Vertex 70 spectrometer (Bruker, Germany). The FTIR microscope is supplied with a liquid nitrogen-cooled mercury cadmium telluride 50 μm MCT detector. This microscope operates with a 36× Schwarzschild magnification objective (NA = 0.65) coupled to a 36× magnification condenser. All spectra were obtained using a single masking aperture size of 7 μm × 7 μm, covering the full cell size. Single point maps of individual spectra were collected in the 3800–900 cm−1 mid-infrared range at 4 cm−1 resolution with 256 co-added scans per spectrum. For each condition, between 60 and 80 cells were randomly collected. A background measurement was performed under the same acquisition parameters every 10 samples.2.6 SR-FTIRM data treatment and statistical analysis
Spectral features to assess the differences between the control and treated cells were highlighted by Principal Component Analysis (PCA) using Unscrambler X (CAMO Software AS, Norway). PCA was performed on vector normalized second derivative spectra (11 smoothing points, Savitzky–Golay algorithm). Vector normalization and PCA were applied in the 3000–2800 & 1760–950 cm−1 spectral regions, as performed in previous FTIRM studies.21,32Raw spectra were corrected in the 3100–950 cm−1 spectral region following the rubber band method (32 baseline points) using OPUS 7.5 (Bruker Optik GmbH, Germany). Then, the area of the following bands/regions was evaluated: methylene asymmetric stretching, 2945–2900 cm−1 (CH2); methyl asymmetric stretching, 2980–2945 cm−1 (CH3); lipid spectral region, 3000–2835 cm−1 (Lip); amide I, 1710–1598 cm−1 (AI); amide II, 1590–1483 cm−1 (AII); phosphate I, 1270–1186 cm−1 (PhI); and phosphate II, 1146–1004 cm−1 (PhII). The total area (3000–2835 and 1760–950 cm−1 regions) was associated with the total cell biomass (Cell) following previous studies.32,33 Violin plots showing the probability density of the data for different ratios (PhI/AII, PhII/AII, AII/AI, Lip/Cell, CH2/Cell, and CH2/CH3) were generated for the different configurations. These ratios have been reported to provide valuable information on cell response after treatments.32,34
3 Results and discussion
Details of the SR-FTIRM analysis are presented in this section for several irradiation modalities (megavoltage and kilovoltage), for several doses (from 0 to 20 Gy), and in the presence (+NP) and absence (−NP) of GdNP. Section 3.1 shows the effects of GdNP at 0 Gy (without irradiation), while megavoltage radiotherapy results are presented in section 3.2. In section 3.3, megavoltage radiosensitization effects by GdNP are compared with respect to kilovoltage radiation energies.3.1 Radiosensitization effects of GdNP
The effects of the nanoparticles at 0 Gy (without irradiation) are evaluated in this section. Fig. 1 shows the score plots (left) and the loading plots (right) in the presence (+NP) and absence (−NP) of nanoparticles in the 3000–2800 & 1760–950 cm−1 spectral regions. In the PCA score plot, each single point represents a cell spectrum. The PCA plot shows clusters of samples based on their similarity, while the loading plot expresses the influence of the variables (wavenumbers) in each principal component.The PC1–PC2 score plot shows the separation in the two clusters (−NP and +NP groups) with some overlapping. The group separation is mostly along PC-1, which explains 41% of the variance. The loading plot indicates that the largest biochemical changes induced by the NP are associated with protein modifications in the region 1700–1500 cm−1. These results indicate changes in the protein conformational structures (amide I and amide II) from α-helix toward β-sheet.25,34,35
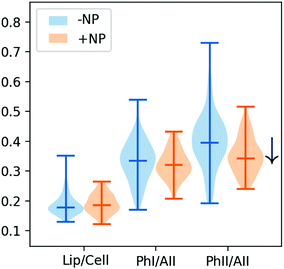
Fig. 2 and 3 show the second derivative for the two controls (−NP and +NP) and the distribution of the relative intensities of several spectral bands (Lip/Cell, PhI/AII and PhII/AII) described in section 2.6, respectively. We observe changes in the phosphodiester vibrations of the DNA. In particular, a shift in the PO2− asymmetric band (1238 cm−1) is detected in the presence of NP. We also observe a decrease in the ratio of PhII/AII in the NP-treated cells. These findings are in agreement with the ones obtained in our previous work, which was carried out with an internal source of infrared radiation.25
Protein and DNA modifications could be due to the biological interactions of NP within the cells. Previous studies have shown that NP can induce conformational changes in cell proteins.36–38 Moreover, Taupin et al. reported proliferation arrest and cell cycle modifications when using similar NP to those used in our work.19 Thus, these FTIRM spectra changes could also be related to initial modifications in the cells induced by the NP.39,40
No significant differences were observed in the lipid region in the PCA scores. However, the violin plots show a slight increase in the total lipid content (Lip/Cell ratio) for the population of NP-treated cells. It is important to note that the cells were fixed two hours after the irradiation to assess direct treatment-induced effects and that important lipid modifications are typically detected at longer times after the treatment.19,25
3.2 Radiosensitization effects of GdNP at megavoltage radiation energies
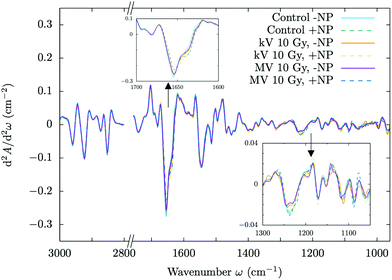
Fig. 4 and 5 show the overall changes in the averaged absorbance spectra and the corresponding Savitzky–Golay second derivative for several investigated groups, respectively. PCA scores and loadings are shown in Fig. 6, which illustrates sample grouping for the control and cells treated with megavoltage radiotherapy with/without GdNP for several doses (5, 10 and 20 Gy) in the 3000–2800 & 1760–950 cm−1 spectral regions. After the treatments (MV and/or NP), several infrared bands (mostly located in the protein and DNA regions) showed intensity changes or frequency shifts.The PCA scores do not discriminate clearly among the three treated groups (+NP, MV −NP, and MV + NP) for 5 Gy. Discrimination between the treated and control (−NP) groups is mainly associated with PC-1 and, in particular, with changes in the protein spectral region in the range 1700–1500 cm−1 (amide I and amide II). The separation increases in the presence of nanoparticles, as the MV + NP groups are clearly separated from the other treated groups, especially for 10 Gy and 20 Gy. The separation of the PCA scores for treated cells increases as a function of the dose. The shift in amide I in the treated cells with respect to the control is clearly seen in the Savitzky–Golay second derivative plot (see the inset in Fig. 5).
This shift in amide I indicates a change in the overall conformational state in the secondary structure of proteins. In particular, it might be related to an increase in the amount of β-sheet secondary structures, which is typically associated with unfolding and/or denaturation of proteins or apoptosis.35,39 Immediately after the radiotherapy treatments, this protein damage might be related to direct damage of the ionizing radiation,41–44 and specifically to changes in the protein secondary structures due to changes or rearrangements in the DNA structure.34,45 Moreover, in the violin plots of Fig. 7 (middle), we observed a slight increase in the absorbance intensity of proteins (amide II) for the MV-treated cells with respect to controls, as in previous studies.34 This might indicate that the DNA repairing processes had started since they normally involve a large quantity of enzymes which recognize and correct physical damage in DNA.46
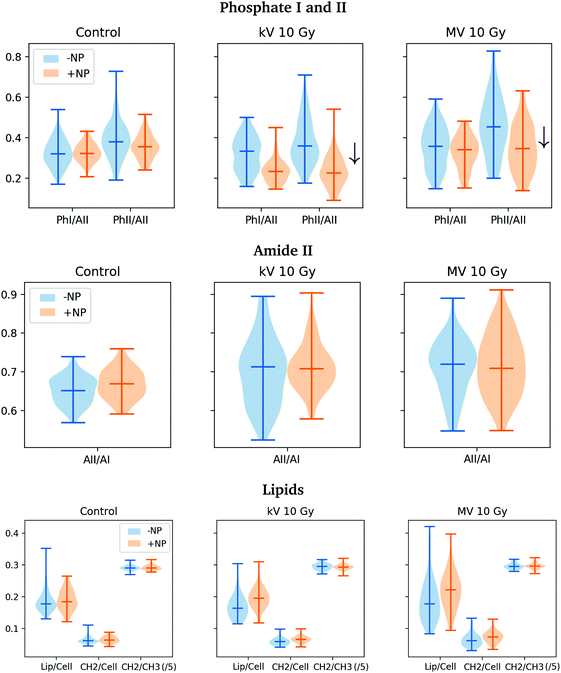 | ||
| Fig. 7 Violin plots showing the distribution of the relative intensities of several spectral bands (PhI/AII, PhII/AII, AII/AI, Lip/Cell, CH2/Cell and CH2/CH3 scaled by a factor 5) for controls, megavoltage (MV) and kilovoltage (kV) radiotherapy. The specific intervals for each ratio are described in section 2.6.32 For clarity, one dose (10 Gy) is presented. ‘+NP’ labels the presence of nanoparticles and ‘−NP’ the absence of nanoparticles. | ||
In the second derivative plot (Fig. 5), we can observe significant changes between the ‘MV + NP’ and ‘MV − NP’ groups in the DNA signatures in the region 1330–950 cm−1. Several changes are found simultaneously (changes in the intensity and shifts), which implies a number of different variations in the DNA organization. The most clear changes appear in the vibrations of the phosphodiester groups; there are modifications in the PO2− asymmetric band (1238 cm−1), especially in the case of NP-treated cells. These modifications might be associated with local conformational changes in the DNA,47,48 such as the formation of pyrimidine dimers or DNA–DNA and DNA–protein cross-links.48 There is also a shift in the PO2− symmetric stretching (around 1085−1) in the NP-treated cells. We also observed treatment-dependent absorbance variations in the phosphate II band, as it can be seen in the (top right) violin plots of Fig. 7. Taken together, these features could point to an increase in DNA strand breaks, base cleavage reactions and chromatin fragmentation34,45,48–51 for cells charged with nanoparticles prior to MV radiotherapy irradiations.
An analysis of the PCA loading plots reveals less contribution from the lipid region (3000–2800 cm−1). We observe a slight increase in the total quantity of lipids for the treated cells in Fig. 7 (bottom right). This is due to a slight increase in the absorbance in the CH2 and CH3 stretching modes in the treated cells (especially in the NP-treated group), indicating some changes following lipid peroxidation or changed lipid metabolism due to initial cell death processes.52,53 We do not observe a significant increase in the ratio of CH2/CH3, which is normally observed several hours after the treatment due to oxidative processes occurring in response to treatment and consistent with cell death.54,55
Finally, it is important to note that there is a clear increase in the variance of the biochemical content of the cell population after both treatments (nanoparticle and radiation), as can be seen in Fig. 7, probably as a result of the initiation of several DNA repair processes or cell death.50,56
3.3 Comparison of the megavoltage radiosensitization effects of GdNP with respect to kilovoltage radiation energies
PCA scores and loadings are shown in Fig. 8 for cells treated with megavoltage and kilovoltage radiotherapy with/without GdNP for several doses (5, 10 and 20 Gy) in the 3000–2800 & 1760–950 cm−1 spectral regions.PCA does not show significant differences between the MV and kV groups in the absence of NP. Moreover, in the presence of nanoparticles, both the ‘kV + NP’ and ‘MV + NP’ groups are equally separated (along PC-1) from the ‘−NP’ groups. Discrimination of NP-treated groups is mainly associated with PC-1 and, in particular, with changes in the protein spectral region (amide I and amide II). As has been discussed in the previous section, these spectral variations may be related to conformational changes of the protein secondary structures due to radiation damage or due to initial changes or rearrangements in the nucleic acid structures. We do not observe significant changes in the content of proteins (amide II) between megavoltage and kilovoltage groups (see Fig. 7, middle), which has been previously related to DNA repair mechanisms.46,48
The results presented here indicate that similar damage occurs in the F98 glioma cells for both types of irradiation (MV and kV) since PCA analysis does not allow clear discrimination between the two ‘+NP’ groups. However, in the Savitzky–Golay second derivative plot (Fig. 5) and the DNA-related violin plots (Fig. 7, top), we can observe some differences in the phosphate I and II infrared bands (1270–1186 cm−1 and 1146–1004 cm−1, respectively). On average, there is a decrease in the intensities of these bands in the ‘kV + NP’ group with respect to the ‘MV + NP’ group, which may indicate some differences in DNA damage in both treatments.
Our SR-FTIRM analysis, supported by multivariate analysis, shows clear radiosensitization effects for megavoltage radiotherapy, which do not differ significantly from the ones observed in kilovoltage radiotherapy. Despite Monte Carlo simulations at the nanoscale predicting that dose enhancement should be much higher using kV energies (compared to MV beams),57 previous biological studies have observed significant radiosensitization in studies where no dose enhancement was expected.14 Butterworth et al. has reported that for most of the experimental results, the observed enhancement is higher than the overall physical dose increase.14 Our results, in agreement with these observations, indicated that the biochemical component in the nanoparticle radiosensitization plays a major role.
4 Conclusions
This synchrotron-based infrared microspectroscopy investigation showed infrared spectral modifications in F98 cells exposed to Gd nanoparticles combined with megavoltage (normally used clinically) and kilovoltage radiotherapy (commonly used in combination with nanoparticles). Biochemical differences were evaluated two hours after the irradiation to assess cellular damage.PCA results showed NP-dependent changes in megavoltage treated cells. The main spectral variations were related to conformational changes of the protein secondary structures (amide I and amide II), which were probably related to radiation damage and to changes or rearrangements in the nucleic acid structures due to the initiation of DNA repair mechanisms. We also observed significant changes in the phosphate I and II bands (1270–1186 cm−1 and 1146–1004 cm−1, respectively), while minimal changes were observed in the lipids region. Spectroscopic data showed that these changes increased as a function of the dose, especially at 10 and 20 Gy.
PCA analysis did not discriminate clearly between megavoltage and kilovoltage groups in the presence of nanoparticles, indicating that GdNP radiosensitization effects in megavoltage radiotherapy do not differ significantly from those in kilovoltage radiotherapy. We observed some differences between the two types of radiation in the average intensities of the phosphate I and II bands, which might indicate higher DNA damage in kilovoltage irradiation. However, few differences were observed in the protein region.
Finally, it is important to note that it is difficult to draw overall conclusions for other types of nanoparticles or cell lines, since radiosensitization effects depend on the nanoparticle size and type, surface functionalization, concentration and intracellular localization, cell type and irradiation configuration. Thus, further work is required to shed more light on the mechanisms involved in the radiosensitization effects of nanoparticles. Comprehensive characterization studies of different cellular responses to GdNP are required for the further development of this promising technique.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors acknowledge the granted beamtime at CELLS-ALBA Synchrotron, and at the Hospital Universitari Sant Joan de Reus (Institut d'Investigació Sanitària Pere Virgili).This project has received funding from the European Union's Horizon 2020 Research and Innovation Programme under the Marie Sklodowska-Curie grant agreement number 748889, as well as from the Secretaria d'Universitats i Recerca del Departament d'Economia i Coneixement de la Generalitat de Catalunya.
Finally, we also acknowledge Pere, Esther, Guillermina, Maria Cinta and Marta from ASPCAT Laboratory (Tortosa) for their support.
References
- J. F. Hainfeld, D. N. Slatkin and H. M. Smilowitz, Phys. Med. Biol., 2004, 49, 309–315 CrossRef PubMed
.
- J. Hainfeld, F. Dilmanian, D. Slatkin and H. Smilowitz, J. Pharm. Pharmacol., 2008, 60, 977–985 CrossRef CAS PubMed
.
- N. Chattopadhyay, Z. Cai, J. Pignol, B. Keller, E. Lechtman, R. Bendayan and R. Reilly, Mol. Pharm., 2010, 7, 2194–2206 CrossRef CAS PubMed
.
- S. Jain, D. Hirst and J. O'Sullivan, Br. J. Radiol., 2009, 85, 101–113 CrossRef PubMed
.
- D. Joh, L. Sun, M. Stangl, A. Al Zaki, S. Murty, P. Santoiemma, J. Davis, B. Baumann, M. Alonso-Basanta, D. Bhang, G. Kao, A. Tsourkas and J. Dorsey, PLoS One, 2013, 8, e62425 CrossRef CAS PubMed
.
- Y.-W. Jiang, G. Gao, H.-R. Jia, X. Xhang, J. Zhao, N. Ma, J.-B. Liu, P. Liu and F.-G. Wu, ACS Biomater. Sci. Eng., 2019, 5, 1569–1579 CrossRef CAS
.
- N. Ma, Y.-W. Jian, X. Zhang, H. Wu, J. N. Myers, P. Liu, H. Jin, N. Gu, N. He, F.-G. Wu and Z. Chen, ACS Appl. Mater. Interfaces, 2016, 8, 28480–28494 CrossRef CAS PubMed
.
- Y. Matsumura and H. Maeda, Cancer Res., 1986, 46, 6387–6392 CAS
.
- E. Porcel, S. Liehn, H. Remita, N. Usami, K. Kobayashi, Y. Furusawa, C. Le Sech and S. Lacombe, Nanotechnology, 2010, 21, 085103 CrossRef PubMed
.
- R. I. Berbeco, W. Ngwa and G. M. Makrigiorgos, Int. J. Radiat. Oncol., Biol., Phys., 2011, 81, 270–276 CrossRef PubMed
.
- S. Jain, J. A. Coulter, A. R. Hounsell, K. T. Butterworth, S. J. McMahon, W. B. Hyland, M. F. Muir, G. R. Dickson, K. M. Prise, F. J. Currell, J. M. O'Sullivan and D. G. Hirst, Int. J. Radiat. Oncol., Biol., Phys., 2011, 79, 531–539 CrossRef CAS PubMed
.
- S. J. McMahon, W. B. Hyland, M. F. Muir, J. A. Coulter, S. Jain, K. T. Butterworth, G. Schettino, G. R. Dickson, A. R. Hounsell, J. M. O'Sullivan, K. M. Prise, D. G. Hirst and F. J. Currell, Radiother. Oncol., 2011, 100, 412–416 CrossRef CAS PubMed
.
- I. Martínez-Rovira and Y. Prezado, Med. Phys., 2011, 38, 4430–4439 CrossRef PubMed
.
- K. Butterworth, S. McMahon, F. Currel and K. Prise, Nanoscale, 2012, 4, 4830–4838 RSC
.
- S. Rosa, C. Connolly, G. Schettino, K. T. Butterworth and K. M. Prise, Cancer Nanotechnol., 2017, 8, 1–25 CrossRef PubMed
.
- I. Martínez-Rovira and Y. Prezado, Med. Phys., 2015, 42, 6703–6710 CrossRef PubMed
.
- N. Ma, F.-G. Wu, X. Zhang, Y.-W. Jiang, H.-R. Jia, H.-Y. Wang, Y.-H. Li, P. Liu, N. Gu and Z. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 13037–13048 CrossRef CAS PubMed
.
- D. Choudhury, P. Xavier, K. Chaudhari, R. John, A. Dasgupta, T. Pradeep and G. Chakrabarti, Nanoscale, 2013, 5, 4476–4489 RSC
.
- F. Taupin, M. Flaender, R. Delorme, T. Brochard, J.-F. Mayol, J. Arnaud, P. Perriat, L. Sancey, F. Lux, R. Barth, M. Carriere, J.-L. Ravanat and H. Elleaume, Phys. Med. Biol., 2015, 60, 4449–4464 CrossRef PubMed
.
- F. Lux, L. Sancey, A. Bianchi, Y. Cremillieux, S. Roux and O. Tillement, Nanomedicine, 2015, 10, 1801–1815 CrossRef CAS PubMed
.
- M. J. Baker, J. Trevisan, P. Bassan, R. Bhargava, H. J. Butler, K. M. Dorling, P. R. Fielden, S. W. Fogarty, N. J. Fullwood, K. A. Heys, C. Hughes, P. Lasch, P. L. Martin-Hirsch, B. Obinaju, G. D. Sockalingum, J. Sulé-Suso, R. J. Strong, M. J. Walsh, B. R. Wood, P. Gardner and F. L. Martin, Nat. Protoc., 2014, 9, 1771–1791 CrossRef CAS PubMed
.
-
M. Diem, P. Griffiths and J. Chalmers, Vibrational spectroscopy for medical diagnosis, John Wiley & Sons, New York, 2008 Search PubMed
.
- I. Yousef, J. Breard, N. SidAhmed-Adrar, A. Maamer-Azzabi, C. Marchal, P. Dumas and F. Le Naour, Analyst, 2011, 136, 5162–5168 RSC
.
- L. Buriankova, Z. Nadova, D. Jancura, M. Refregiers, I. Yousef and M. J. P. Miskovsky, Laser Phys. Lett., 2010, 7, 613 CrossRef CAS
.
- I. Yousef, O. Seksek, Y. Prezado, J. Sulé-Suso and I. Martínez-Rovira, Analyst, 2016, 141, 2238–2249 RSC
.
- R. Barth and B. Kaur, J. Neurooncol., 2009, 94, 299–312 CrossRef PubMed
.
- R. Di Corato, F. Gazeau, C. Le Visage, D. Fayol, P. Levitz, F. Lux, N. Letourneur, D. Luciani, O. Tillement and C. Wilhelm, ACS Nano, 2013, 7, 7500–7512 CrossRef CAS PubMed
.
- L. Sancey, F. Lux, S. Kotb, S. Roux, S. Dufort, A. Bianchi, Y. Cremillieux, P. Fries, J. Coll, C. Rodriguez-Lafrasse, M. Janier, M. Dutreix, M. Barberi-Heyob, F. Boschetti, F. Denat, C. Louis, E. Porcel, S. Lacombe, G. Le Duc, E. Deutsch, J. Perfettini, A. Detappe, C. Verry, R. Berbeco, K. Butterworth, S. McMahon, K. Prise, P. Perriat and O. Tillement, Br. J. Radiol., 2014, 87, 20140134 CrossRef CAS PubMed
.
- M. Luchette, H. Korideck, M. Makrigiorgos, O. Tillement and R. Berbeco, Nanomedicine, 2014, 10, 1751–1755 CrossRef CAS PubMed
.
- L. Sancey, S. Kotb, C. Truillet, F. Appaix, A. Marais, E. Thomas, B. van der Sanden, J. Klein, B. Laurent, M. Cottier, R. Antoine, P. Dugourd, G. Panczer, F. Lux, P. Perriat, V. Motto-Ros and O. Tillement, ACS Nano, 2015, 9, 2477–2488 CrossRef CAS PubMed
.
- P. Mowat, A. Mignot, W. Rima, F. Lux, O. Tillement, C. Roulin, M. Dutreix, D. Bechet, S. Huger, L. Humbert, M. Barberi-Heyob, M. Aloy, E. Armandy, C. Rodriguez-Lafrasse, G. Le Duc, S. Roux and P. Perriat, J. Nanosci. Nanotechnol., 2011, 11, 7833–7839 CrossRef CAS PubMed
.
- G. Birarda, D. E. Bedolla, E. Mitri, S. Pacor, G. Grenci and L. Vaccari, Analyst, 2014, 139, 3097 RSC
.
- A. Kretlow, Q. Wang, M. Beekes, D. Naumann and L. M. Miller, Biochim. Biophys. Acta, Mol. Basis Dis., 2008, 1782, 559–565 CrossRef CAS PubMed
.
- N. Gault and J. Lefaix, Radiat. Res., 2003, 160, 238–250 CrossRef CAS PubMed
.
- A. Barth, Biochim. Biophys. Acta, 2007, 1767, 1073–1101 CrossRef CAS PubMed
.
- N. Volkova, O. Pavlovich, O. Fesenko, O. Budnyk, S. Kovalchuk and A. Goltsev, J. Nanomater., 2017, 16934757, 1–9 Search PubMed
.
- S. R. Saptarshi, A. Duschl and A. L. Lopata, J. Nanobiotechnol., 2013, 11, 1–12 CrossRef PubMed
.
- D. Sahoo, P. Bhattacharya, H. K. Patra, P. Mandal and S. Chakravorti, J. Nanopart. Res., 2011, 13, 6755–6760 CrossRef CAS
.
- H. Holman, M. Martin, E. Blakely, K. Bjornstad and W. McKinney, Biopolymers, 2000, 57, 329–335 CrossRef CAS PubMed
.
- S. Boydston-White, M. Romeo, T. Chernenko, A. Regina, M. Miljkovic and M. Diem, Biochim. Biophys. Acta, 2006, 1758, 908–914 CrossRef CAS PubMed
.
- J. Reisz, N. Bansal, J. Qian, W. Zhao and C. Furdui, Antioxid. Redox Signaling, 2014, 21, 260–292 CrossRef CAS PubMed
.
- H. Fricke, Nature, 1952, 169, 965–966 CrossRef CAS PubMed
.
- M. J. Daly, DNA Repair, 2012, 11, 12–21 CrossRef CAS PubMed
.
- A. Singh and H. Singh, Prog. Biophys. Mol. Biol., 1982, 39, 69–107 CrossRef CAS PubMed
.
- N. Gault, O. Rigaud, J. Poncy and J. Lefaix, Int. J. Radiat. Biol., 2005, 81, 767–779 CrossRef CAS PubMed
.
- A. Sancar and G. B. Sancar, Annu. Rev. Biochem., 1988, 57, 29–67 CrossRef CAS PubMed
.
- D. Whelan, K. Bambery, P. Heraud, M. Tobin, M. Diem, D. McNaughton and B. Wood, Nucleic Acids Res., 2011, 39, 5439–5448 CrossRef CAS PubMed
.
- E. Lipiec, K. Bambery, P. Heraud, C. Hirschmugl, J. Lekki, W. Kwiatek, M. J. Tobin, C. Vogel, D. Whelan and B. Wood, J. Mol. Struct., 2014, 1073, 134–141 CrossRef CAS
.
- D. Dovbeshko, N. Gridina, E. Kruglova and O. Pashchuk, Talanta, 2000, 53, 233–246 CrossRef PubMed
.
- A. Meade, C. Clarke, H. Byrne and F. Lyng, Radiat. Res., 2010, 173, 225–237 CrossRef CAS PubMed
.
- N. Gault, O. Rigaud, J. Poncy and J. Lefaix, Radiat. Res., 2007, 167, 551–562 CrossRef PubMed
.
- B. Vileno, S. Jeney, A. Sienkiewicz, P. Marcoux, L. Miller and L. Forro, Biophys. Chem., 2010, 152, 164–169 CrossRef CAS PubMed
.
- E. Albi, S. Cataldi, G. Rossi and M. Viola, Arch. Biochem. Biophys., 2008, 478, 52–58 CrossRef CAS PubMed
.
- G. Zhang, R. Zhu, Y. Xu, Y. Yan and Y. Dai, Acta. Bot. Sin., 2004, 46, 711–718 CAS
.
- G. Blankenberg, P. Katsikis, R. Storrs, C. Beaulieu, D. Spielman, J. Chen, L. Naumovski and J. Tait, Blood, 1997, 89, 3778–3786 Search PubMed
.
- K. M. Prise, G. Schettino, M. Folkard and K. D. Held, Lancet Oncol., 2005, 6, 520–528 CrossRef CAS PubMed
.
- J. C. Roeske, L. Nunez, M. Hoggarth, E. Labay and R. R. Weichselbaum, Annu. Rev. Biochem., 2007, 6, 395–401 Search PubMed
.
| This journal is © The Royal Society of Chemistry 2019 |