Open Access Article
Open Access ArticleHigh-throughput quantitation of bovine milk proteins and discrimination of commercial milk types by external cavity-quantum cascade laser spectroscopy and chemometrics†
Milagros
Montemurro
ab,
Andreas
Schwaighofer
 a,
Anatol
Schmidt
c,
María J.
Culzoni
b,
Helmut K.
Mayer
c and
Bernhard
Lendl
a,
Anatol
Schmidt
c,
María J.
Culzoni
b,
Helmut K.
Mayer
c and
Bernhard
Lendl
 *a
*a
aInstitute of Chemical Technologies and Analytics, Vienna University of Technology, Getreidemarkt 9/164-UPA, 1060 Vienna, Austria. E-mail: bernhard.lendl@tuwien.ac.at
bLaboratorio de Desarrollo Analítico y Quimiometría (LADAQ), Cátedra de Química Analítica I, Facultad de Bioquímica y Ciencias Biológicas, Universidad Nacional del Litoral-CONICET, Ciudad Universitaria, 3000 Santa Fe, Argentina
cDepartment of Food Science and Technology, Food Chemistry Laboratory, BOKU – University of Natural Resources and Life Sciences, Muthgasse 11, 1190 Vienna, Austria
First published on 12th August 2019
Abstract
Analysis of bovine milk proteins is crucial in many food and non-food industrial applications, nevertheless labour-intensive wet-chemical, low-throughput methods are still routinely used. In this work, external cavity-quantum cascade laser (EC-QCL) mid-infrared spectroscopy is employed as a rapid method for protein analysis of commercial bovine milk. Combined analysis of the amide I and II bands enabled quantitation of individual proteins (casein, β-lactoglobulin, α-lactalbumin) and total protein content. IR spectra of spiked and diluted milk samples were employed for calibration of the target analytes in the presence of a complex matrix by partial least squares (PLS) regression modelling. A sample set of different milk types (pasteurized; differently processed extended shelf life, ESL; ultra-high temperature, UHT) was analysed, and results agreed well with reference methods. Quantitation of temperature sensitive proteins enables detailed distinction between milk types experiencing different heat loads during processing, and discrimination between diverse bovine milk types is successfully demonstrated.
Introduction
Milk is an essential nutritional resource providing many health benefits with a production volume of 818 × 106 tonnes per year, 83% of which is bovine.1 Its main constituents are water, lipids, lactose and proteins. In bovine milk, the total protein content is ∼32 g L−1. The two major protein groups are casein (Cas) and whey proteins with relative shares of 80% and 20%, respectively. Most abundant whey proteins are β-lactoglobulin (β-LG) and α-lactalbumin (α-LA) with concentrations in raw milk of approx. 3.5 g L−1 and 1.2 g L−1, respectively.2Milk and milk product consumption is recommended by most nutritional guidelines, since these food items contain a combination of essential nutrients.3 Commercially available milk is subject to different thermal processing steps that influence parameters such as storage life and milk quality, i.e. nutritional composition and organoleptic characteristics. Currently, commercially available milk types comprise pasteurized milk, extended shelf life (ESL) milk and ultra-high temperature milk. Pasteurized milk is subjected to the mildest thermal treatment (commonly 72 °C for 15 s) and can be stored at 2–8 °C for up to 10 days.4,5 ESL milk has acquired a substantial market share in recent years, because it allows longer storage time than pasteurized milk (up to 24 days under cooled conditions), while upholding flavour and nutritive properties of fresh foods.6 The processing conditions for manufacturing ESL milk can be classified into non-thermal and thermal treatments. Non-thermal milk treatment belongs to the rather soft processing methods. Examples are bactofugation and microfiltration. Processing of ESL milk involves more severe thermal conditions than pasteurization but less intensive than ultra-high temperature (UHT) manufacturing. Two heating processes are used to produce high temperature short time (HTST) milk, where milk is subjected to approx. 125 °C for 2–3 s. In direct heating processes, the milk is heated at a fast rate by direct contact to dry steam, while indirect heating involves the use of heat exchangers. The slower heating and cooling rates of the indirect method induce more chemical changes in the milk.5,7 Finally, UHT milk is processed by thermal treatment at a minimum temperature of 135 °C for at least 1 s that introduces a characteristic “cooked” taste, and is then storable at ambient conditions for 6 months.7
Exposure to intensive heat loads influences the quality of the final milk product and leads to sensorial (e.g. cooked flavour), nutritional (e.g., protein denaturation, vitamin loss) and chemical (unfolding of proteins, Maillard reaction products) modifications. Analysis of heat load indicators that are related to heat treatment (degradation or formation) enables a direct and quantitative assessment of the heat load impact without knowledge of the preceding thermal history.8,9 Bovine milk comprises multiple heat sensitive components that can be utilized for this kind of evaluation, such as the whey proteins α-LA and β-LG. Denaturation begins at approx. 60 °C for β-LG and at approx. 75 °C for α-LA, consequently the remaining concentration of undenatured fractions of these proteins in the final milk product provides information about the experienced heat load and enables discrimination between different milk types.10 The International Dairy Federation (IDF) suggests a minimum β-LG content of 2.6 g L−1 for pasteurized milk, 2.0 g L−1 for high-pasteurized (ESL) milk, and 0.05 g L−1 for UHT milk.11 For liquid ESL milk, these limits are not obligatory, in contrast to pasteurized/UHT milk. In Austria, however, since July 2018 a minimum β-LG content of 1.8 g L−1 has been introduced for ESL milk in order to minimize the actual heat load of this upcoming milk type.
Nowadays, the traditional Kjeldahl method for determination of organic nitrogen in food and beverages continues to be the employed standard analytical technique for quantification of total protein content in milk, even though it is a fairly labour-intensive wet-chemical approach with low throughput. For quantitation of individual milk proteins including Cas, α-LA and β-LG, diverse techniques based on chromatographic11–13 and electrophoretic13 methods can be employed, all of them involving time-consuming, wet-chemical sample preparation steps. Mid-infrared (IR) spectroscopy has been applied for rapid and non-destructive analysis of quality and composition of dairy products due to its high-throughput capacity, simplicity and low cost.14–16 Furthermore, an mid-IR spectroscopic approach was approved by the Association of Official Analytical Chemists (AOAC International) for the analysis of liquid milk, resulting in the development of several commercially available FTIR milk analysers for quantification of total protein and casein content, among other parameters.17 Apart from mid-IR spectroscopy, also near-IR spectroscopy was applied for analysis of the total protein content of milk.18–20
Mid-IR spectroscopy is a well-established analytical technique that detects the fundamental vibrations of covalent bonds in molecules in a label free manner. For quantitative analysis of total protein content in milk, typically the amide II (1500–1600 cm−1) band is evaluated at path lengths of approx. 50 μm for FTIR transmission measurements, most commonly combined with multivariate calibration techniques.14,15,21,22 Qualitative discrimination between proteins is preferably performed by evaluation of the amide I (1600–1700 cm−1) band, that is particularly sensitive to the protein secondary structure. However, it was shown that additional and more in-depth information about protein secondary structure can be gained by collective analysis of both spectral regions, particularly with chemometric analysis.23,24 Application of FTIR transmission spectroscopy in aqueous solution in the 1600–1700 cm−1 spectral region is cumbersome due to the strong absorbance of H2O centered at 1645 cm−1 and the low emission powers of thermal light sources in conventional FTIR spectrometers. For this reason, path lengths of typically <10 μm are employed in order to prevent total IR absorption in this region, which impairs sensitivity levels necessary for the analysis of biologically relevant concentrations, and considerably reduces the robustness of the application. Consequently, custom-built setups for IR spectroscopy based on quantum cascade lasers were developed to overcome the disadvantages of FTIR instruments, and already diverse applications in mid-IR spectroscopy were reported.25 In IR transmission spectroscopy of proteins, the transmission paths could be considerably increased by using an external cavity-quantum cascade laser (EC-QCL) light source that provides significantly higher emission powers.26 Laser-based IR transmission measurements were successfully performed for examination of the protein secondary structure by evaluation of the amide I band.27,28 Further, the viability of protein discrimination and quantitation in commercial bovine milk samples was successfully demonstrated.29,30 Most recently, a new and improved EC-QCL based IR transmission setup was introduced for analysis of the protein amid I and amide II regions, allowing more sensitive quantitative and more detailed qualitative analysis of proteins.31
One of the challenges faced in the analysis of complex samples is the quantitation of an individual analyte in a multicomponent system. Partial least-square (PLS) regression is a well-known algorithm applied to first-order multivariate calibration, which allows rapid determination of multiple components usually without the need for prior separation.32 When applying PLS, calibration can be performed considering only the concentration of the analytes of interest. However, all the expected components of the matrix must be present in the calibration step, even though their concentration can be ignored. Furthermore, if the sample contains non-calibrated interferences, it can be identified as an outlier because of the unusually large spectral residuals, a property known as first-order advantage.33 PLS has been widely applied to the analysis of complex systems, such as for fluorescence data showing inner filter effects,34 due to the flexible structure of the algorithm that allows considering these effects by including additional latent variables in the model.35
In this work, the fast and direct quantitation of individual proteins (i.e. Cas, α-LA and β-LG) as well as total bovine milk protein content is presented by employing a latest generation EC-QCL setup for analysis of a sample set comprising different milk types. The obtained results were validated by comparison with the standard reference methods. It was illustrated that the obtained concentration values can be used for discrimination of the most prevalent commercial milk types.
Experimental
Standards, reagents and solutions
Ethanol (96% v/v, EtOH), aqueous sodium hydroxide (NaOH) solution (50%), hydrochloric acid 37% (HCl) ACS reagent, lyophilized powders of α-LA (≥85%), β-LG (≥90%) and Cas sodium salt, all from bovine milk, were obtained from Sigma-Aldrich (Steinheim, Germany). Sodium phosphate monobasic dihydrate p.a. (NaH2PO4·2H2O) was purchased from Fluka (Buchs, Switzerland). A Merck Millipore (Darmstadt, Germany) Milli-Q water purification system was used for generating ultrapure water. Solvents used for chromatographic analysis were of HPLC-grade. Individual stock solutions of α-LA (40.0 mg mL−1), β-LG (75.0 mg mL−1) and Cas (36.0 mg mL−1) were prepared by weighing and dissolving appropriate amounts of lyophilized protein in 45 mmol L−1 phosphate buffer at pH 6.6. Nineteen homogenized, commercial bovine milk samples of six different milk types (pasteurized, ESL bactofugated, ESL filtered, ESL HTST directly heated, ESL HTST indirectly heated, and UHT) were obtained from local grocery stores and processed as received, i.e. without performing any sample pretreatment.EC-QCL setup
A detailed description of the custom-made EC-QCL setup can be found elsewhere.31 Briefly, a thermoelectrically-cooled external-cavity quantum cascade laser (Hedgehog, Daylight Solutions Inc., San Diego, USA) was used operating at a repetition rate of 100 kHz and a pulse width of 5000 ns. All spectra were recorded in the spectral tuning range between 1730–1470 cm−1, covering the amide I and amide II region of proteins, at a scan speed of 1200 cm−1 s−1. The MIR light was focused on the detector element by a gold plated off-axis parabolic mirror with a focal length of 43 mm. A thermoelectrically-cooled MCT detector operating at −78 °C (PCI-10.6, Vigo Systems S.A., Poland) was used as IR detector, as shown in Fig. 1. To reduce the influence of water vapor, the setup was placed in a housing of polyethylene foil and constantly flushed with dry air. | ||
| Fig. 1 Schematic of the EC-QCL based IR transmission setup. A path length of 31 μm was employed for measurements of milk. | ||
The measured signal was processed by a lock-in amplifier (Stanford Research Systems, CA, USA) and digitized by a NI DAQ 9239 24-bit ADC (National Instruments Corp., Austin, USA). Each single beam spectrum consisting of 6000 data points was recorded during the tuning time for one scan of approx. 250 ms. A total of 100 scans were recorded for background and sample single beam spectra at a total acquisition time of 53 s. All measurements were carried out using a custom-built, temperature-controlled flow cell equipped with two MIR transparent CaF2 windows and 31 μm-thick spacer, at 20 °C. For spectra acquisition, 1 mL of the sample liquid were applied to the transmission cell either by a suitable syringe or by an automated flow injection sampling system.26 Reference spectra were recorded after measurement of 10 samples. Prior to acquisition of the reference spectrum, the transmission cell was cleaned with ethanol and 1% sodium dodecyl sulfate (SDS). The laser was controlled by Daylight Solution driver software; data acquisition and temperature control was performed using a custom-made LabView-based GUI (National Instruments Corp., Austin, USA).
Data processing
Data processing was performed in MATLAB 2013b (Mathworks Inc., Nattick, USA) using an in-house written routine. A pre-processing routine for EC-QCL raw data was implemented to sort out scans (approx. 3%) based on similarity index evaluation, that are shifted more than ±0.1 cm−1 due to mechanical imperfections and triggering issues. A detailed description of the procedure is given elsewhere.31 An interface for data input and parameters setting was used for PLS implementation.36Calibration and validation samples
In the present study, the analytes of interest are embedded in a complex matrix containing a large number of compounds. QCL-IR spectra of the pure analytes α-LA, β-LG and Cas are shown in Fig. 2A. In order to build a PLS model suitable for the analysis of the samples, a calibration step must be performed considering all the sources of spectral variability present in the unknown samples. In case the composition of the matrix is perfectly known, artificial mixtures could be prepared and used for modelling in the calibration step. However, this information is not usually available or, even if it is, the complex matrix composition cannot be mimicked by a simple mixture of the individual components, which is the situation encountered in the analysis of milk. For this reason, even though the composition of bovine milk is known, a different strategy had to be applied in this work. The calibration samples were prepared using two kinds of milk samples (UHT and ESL HTST) with known concentrations of proteins determined by reference methods (see below). These milk types were selected for the calibration set because they allowed spanning most of the expected spectral variability by addition of protein stock solutions. Three calibration sets, shown in Table S1 (ESI†), were built for α-LA (0.10–1.75 mg mL−1), β-LG (0.20–4.40 mg mL−1) and Cas (20.0–31.0 mg mL−1) in seven concentration levels, four of which were prepared with UHT milk, while the other three were prepared with HTST milk, in triplicate. Further, to model the effect of varying degrees of water content in different milk samples on the IR spectra in the amide I region,29 each replicate was prepared with increasing amounts of milk. Fig. 2B shows QCL-IR spectra of milk samples of the calibration set that contained high (3.8 mg mL−1) and low (0.2 mg mL−1) β-LG levels at varying dilution. For milk samples with high β-LG content, a shoulder at 1633 cm−1, characteristic for the β-sheet secondary structure of this protein (compare Fig. 2A), is recognizable compared to the QCL-IR spectra of milk samples with low β-LG content at the respective dilution. The increasing amide II band with lower degree of dilution indicates higher protein concentrations in the samples. However, in the amide I region this straight forward relation is not given because of variations in the absorbance intensity of the HOH bending band at 1645 cm−1 due to the displacement of water caused by milk matrix compounds relative to the H2O background spectra.29 Hence, the final concentrations of each protein were reached by spiking a portion of the selected milk with appropriate aliquots of standard solution, considering the exact concentration determined by HPLC and the amount added in each replicate, and completing to final volume with buffer solution. A total of 21 calibration samples were obtained for each set, and a total of 63 samples for the complete calibration set after combining the three sets. In order to determine the content of total proteins, a separate calibration set consisting of 18 ternary mixtures of α-LA, β-LG and Cas and varying amounts of milk was prepared, resulting in a concentration range of total protein of 29.0–36.0 mg mL−1 (Table S2, ESI†). | ||
| Fig. 2 (A) QCL-IR spectra of 10 mg mL−1 Cas, β-LG and α-LA. (B) QCL-IR spectra of milk at dilution levels (75%, 50%, 25% milk) with high (3.8 mg mL−1) and low (0.2 mg mL−1) β-LG levels. | ||
In addition, a validation set was prepared applying the same strategy used for the calibration samples. Here, 18 validation samples, 9 for each type of milk (UHT and ESL HTST), were prepared by spiking the samples with ternary mixtures of the proteins in three concentration levels, in triplicate. Different aliquots of milk were added in each replicate, differing from those used in the calibration step.
Determination of nitrogen content using the Kjeldahl method
Total nitrogen (TN) was determined by the Kjeldahl method according to the corresponding IDF/ISO standard,37 non-casein N (NCN) according to IDF/ISO,38 and non-protein N (NPN) according to IDF/ISO.39 Whey protein N was calculated from the difference between NCN and NPN, and casein N from TN and NCN, respectively. Protein equivalents were calculated from nitrogen data using the factor 6.38.Reversed phase – high performance liquid chromatography (RP-HPLC) analysis of α-LA and β-LG
Sample preparation for the determination of α-LA and β-LG as well as RP-HPLC analysis were carried out according to the corresponding IDF/ISO standard40 and as recently described in the literature.8,11 Briefly, caseins and denatured whey proteins were precipitated at pH 4.6 by dropwise addition of HCl (2 mol L−1). Acid whey containing the acid-soluble whey proteins was separated by centrifugation and diluted (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, or 1
10, or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 in the case of UHT milk) with sodium phosphate buffer solution (0.1 mol L−1, pH 6.7). Samples were filtered through 0.20 μm Minisart RC 4 filters (Sartorius, Goettingen, Germany).
5 in the case of UHT milk) with sodium phosphate buffer solution (0.1 mol L−1, pH 6.7). Samples were filtered through 0.20 μm Minisart RC 4 filters (Sartorius, Goettingen, Germany).
RP-HPLC was performed on a Waters chromatography system using a model 600E multisolvent delivery system, a Rheodyne 7725i injector, guard column (Sentry Guard, Symmetry™ C18, 3.5 μm, 2.1 × 10 mm) and a Symmetry™ 300 C18 column (3.5 μm, 2.1 × 150 mm) (Waters Corporation, Milford, MA, USA). Column eluates were monitored at 205 nm using a Waters 2489 UV/Vis detector interfaced with a PC running Waters Millennium chromatography software for data acquisition and management.
Gradient separation was carried out within 18 min, followed by column equilibration leading to sample injection intervals of 35 min.11 Solvent A consisted of 0.1% trifluoroacetic acid (TFA) in ultrapure water, solvent B of 0.1% TFA in acetonitrile. Solvents for HPLC analysis were freshly prepared weekly and vacuum filtered (Whatman™, ME24ST Membrane Filters White, 0.2 μm, diameter 47 mm) before use. Gradient elution was carried out by increasing solvent B linearly from 36% to 50% over 14 min, followed by increasing to 100% B within 0.5 min, and finally holding at 100% B for 3.5 min. Separation was performed at a column temperature of 40 °C with a flow rate of 0.35 mL min−1, and the injection volume was set to 10 μL.11
Results and discussion
PLS data modelling
For direct protein quantitation by EC-QCL based IR spectroscopy in bovine milk samples, PLS-1 was applied to obtain a calibration model for the individual proteins of interest. PLS modelling involves a calibration step, in which the relation between the response and the analyte concentration is described, and a prediction step, in which the calibration model is applied to estimate the concentration of the analyte in unknown samples. The parameters applied to the direct absorption QCL-IR spectra used for each model are described in Table 1. Data pre-processing tools, such as mean centering and second derivative, were applied. In addition, different spectral regions were selected and used in order to improve the predictive ability of the model. Determination of the number of latent variables was performed by the well-known leave-one-out cross-validation procedure.33 The large optimum number of latent variables indicates that there were many different sources of spectral variability in the system under study. This result is reasonable, because the calibration samples contain not only the standard of each calibrated protein, but also various components of the milk matrix, as discussed above.| α-LA | β-LG | Cas | Total protein | |||
|---|---|---|---|---|---|---|
| Full range | Low level range | Full range | Low level range | |||
| LV – latent variable, 2nd Der. – 2nd derivative, MC – mean centering, RMSECV – root mean squared error of cross validation, LOD – limit of detection, LOQ – limit of quantitation. | ||||||
| Concentration range (mg mL−1) | 0.10–1.75 | 0.10–0.75 | 0.20–4.40 | 0.20–1.80 | 20–31 | 29–36 |
| Spectral region (cm−1) | 1696–1624/1576–1504 | 1672–1504 | 1648–1600 | 1638–1614 | 1720–1495 | 1566–1518 |
| Pre-processing | 2nd Der./MC | 2nd Der./MC | MC | MC | MC | MC |
| LVs | 9 | 7 | 9 | 6 | 9 | 6 |
| RMSECV (mg mL−1) | 0.14 | 0.19 | 0.14 | 0.09 | 0.32 | 0.34 |
| Exp. var. (%) | 99.967 | 99.847 | 99.989 | 99.936 | 99.998 | 99.940 |
| LODmin | 0.077 | 0.052 | 0.071 | 0.11 | 0.28 | 0.22 |
| LODmax | 0.17 | 0.13 | 0.21 | 0.30 | 0.52 | 0.35 |
| LOQmin | 0.23 | 0.16 | 0.22 | 0.33 | 0.82 | 0.67 |
| LOQmax | 0.52 | 0.38 | 0.66 | 0.93 | 1.6 | 1.06 |
The models were validated with a set of validation samples, for which the concentrations of α-LA, β-LG and Cas were predicted (Table 2). It is important to mention that two calibration ranges were used for α-LA and β-LG due to the large difference in their expected concentrations among the studied types of milks. Consequently, models for prediction of low protein concentration samples were built by restricting the calibration range to 0.10–0.75 mg mL−1 and 0.20–1.80 mg mL−1 for α-LA and β-LG, respectively, by selecting the corresponding calibration samples from the complete set. This model was employed when the results of either the validation or the milk samples obtained by the primary model (containing the entire calibration range) were in the low concentration range. Employing this approach, the models were suitable for concentration prediction of the target proteins in the studied ranges, despite the presence of signal variations due to the effect of the sample matrix. This fact represents an important advantage to the standard methods for individual protein quantitation in milk, because it allows processing the sample in its original state, i.e. omitting time-consuming sample pre-treatments, or spectra correction steps to deal with the matrix effect.
| Validation sample | Milk sample added (%) | α-LA (mg mL−1) | β-LG (mg mL−1) | Cas (mg mL−1) | |||
|---|---|---|---|---|---|---|---|
| Nominal | Predicted | Nominal | Predicted | Nominal | Predicted | ||
| V1: prepared with UHT milk; V2: prepared with HTST milk.a Calculated with low-level calibration. | |||||||
| V1-01 | 40 | 0.90 | 0.92 | 2.0 | 2.5 | 24 | 24.3 |
| V1-02 | 60 | 0.90 | 0.93 | 2.0 | 2.2 | 24 | 24.3 |
| V1-03 | 80 | 0.90 | 0.94 | 2.0 | 2.2 | 24 | 23.8 |
| V1-04 | 40 | 0.15 | 0.20a | 0.25 | 0.25a | 28 | 28.0 |
| V1-05 | 60 | 0.15 | 0.24a | 0.25 | 0.33a | 28 | 28.7 |
| V1-06 | 80 | 0.15 | 0.30a | 0.25 | 0.34a | 28 | 28.3 |
| V1-07 | 40 | 1.25 | 1.22 | 3.4 | 3.8 | 22 | 22.3 |
| V1-08 | 60 | 1.25 | 1.23 | 3.4 | 3.9 | 22 | 22.6 |
| V1-09 | 80 | 1.25 | 1.24 | 3.4 | 3.8 | 22 | 22.8 |
| V2-01 | 40 | 0.75 | 1.20 | 2.7 | 2.9 | 28 | 28.4 |
| V2-02 | 60 | 0.75 | 1.30 | 2.7 | 2.8 | 28 | 28.7 |
| V2-03 | 80 | 0.75 | 1.39 | 2.7 | 2.8 | 28 | 28.7 |
| V2-04 | 40 | 1.25 | 1.00 | 2.0 | 2.6 | 30 | 29.9 |
| V2-05 | 60 | 1.25 | 1.10 | 2.0 | 2.7 | 30 | 30.1 |
| V2-06 | 80 | 1.25 | 1.19 | 2.0 | 1.8 | 30 | 30.3 |
| V2-07 | 40 | 1.75 | 1.50 | 3.5 | 3.3 | 26 | 25.3 |
| V2-08 | 60 | 1.75 | 1.60 | 3.5 | 3.8 | 26 | 25.8 |
| V2-09 | 80 | 1.75 | 1.69 | 3.5 | 3.7 | 26 | 26.2 |
| RMSEP (mg mL−1) | 0.25 | 0.33 | 0.45 | ||||
| REP (%) | 25.6 | 13.8 | 1.7 | ||||
Further, LODs and LOQs were calculated for α-LA, β-LG and Cas. These figures of merit were obtained in the form of intervals (min-max) applying the approach proposed by Allegrini et al.,41 which considers the multivariate calibration scenario and whose values depend on the variability of the background composition. The limits obtained for the models, shown in Table 1, are suitable for detection and quantitation of the target analytes in low heat load samples with higher protein concentration. However, according to the expected concentration of α-LA and β-LG in samples treated with high temperatures, there were samples for which the proteins were detectable but non-quantifiable or, in some cases, non-detectable. The validated models were implemented for the quantitation of proteins in commercial milk samples, in order to further evaluate the performance of the method and its applicability to the routine analysis of this kind of samples.
Quantitation of proteins in commercial bovine milk samples
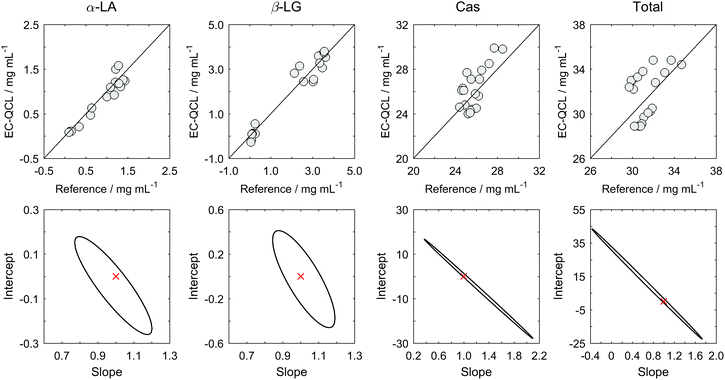
Concentrations of α-LA, β-LG, Cas and total protein in commercial bovine milk samples were determined by EC-QCL based IR spectroscopy coupled to multivariate calibration and compared with the values obtained by the reference methods described in the Experimental section. Inspection of the predicted concentrations for the analyzed samples presented in Table 3 suggests that the developed IR based method is appropriate for the determination of total protein content and for the quantitation of the individual target proteins. A more detailed review allowed noticing that the predictions for the proteins present in high concentration, such as Cas and α-LA and β-LG for low heat load samples, were more similar to those provided by the reference methods than those for the proteins present in lower concentrations, such as α-LA and β-LG for milk samples with high heat load. Besides, it is relevant to mention that two of these concentration values were below the LOD and could not be determined. In order to evaluate if the developed technique could be accepted as a potential alternative method, a statistical comparison between the results achieved by the novel QCL-IR method and the reference method was carried out.42 Therefore, the concentrations obtained by EC-QCL based IR spectroscopy were plotted against those provided by the reference methods to obtain the regression parameters (see Fig. 3, top). Additionally, the elliptical joint confidence region (EJCR) test was applied, in which the linear regression of predicted vs. nominal analyte concentrations is performed (see Fig. 3, bottom).43 The EJCR is calculated considering the obtained slope and intercept and their corresponding confidence intervals. The predictions are considered accurate if the ideal point (slope = 1, intercept = 0) is included in the EJCR. The fact that the ideal point is located within the obtained ellipses for α-LA, β-LG, Cas and total protein indicates that there is no statistical difference between the results provided by both methods in the quantitation of total and individual proteins in milk.| Milk type | Milk sample | α-LA (mg mL−1) | β-LG (mg mL−1) | Cas (mg mL−1) | Total (mg mL−1) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Reference | EC-QCLa | Reference | EC-QCLa | Reference | EC-QCLa | Reference | EC-QCLa | ||
| a Recovery (%) between parenthesis. b Calculated with low-level calibration. | |||||||||
| Pasteurized | M-01 | 1.36 | 1.20 (88) | 3.41 | 3.64 (107) | 25.6 | 24.4 (95) | 31.1 | 29.7 (96) |
| M-02 | 1.43 | 1.24 (87) | 3.61 | 3.54 (98) | 26.0 | 24.5 (94) | 31.7 | 30.2 (95) | |
| M-03 | 1.21 | 1.18 (98) | 3.28 | 3.59 (110) | 25.2 | 24.0 (95) | 30.6 | 28.9 (94) | |
| M-04 | 1.33 | 1.11 (84) | 3.55 | 3.73 (105) | 25.4 | 24.3 (96) | 30.9 | 29.1 (94) | |
| ESL – filtered | M-05 | 1.20 | 1.21 (101) | 3.02 | 2.44 (81) | 25.4 | 24.1 (95) | 30.8 | 28.9 (94) |
| M-06 | 1.39 | 1.26 (91) | 3.46 | 3.06 (88) | 26.2 | 25.6 (98) | 31.9 | 30.5 (96) | |
| M-07 | 1.21 | 1.50 (124) | 3.04 | 2.54 (84) | 24.9 | 24.8 (100) | 30.2 | 28.9 (96) | |
| M-08 | 1.28 | 1.58 (123) | 3.34 | 3.28 (98) | 25.9 | 25.8 (100) | 31.5 | 30.1 (96) | |
| ESL – bactofugated | M-09 | 1.00 | 0.88 (88) | 2.57 | 2.44 (95) | 24.7 | 26.3 (107) | 30.0 | 33.0 (110) |
| M-10 | 1.29 | 1.18 (92) | 3.56 | 3.80 (107) | 26.3 | 27.1 (103) | 32.0 | 34.8 (109) | |
| ESL – HTST direct | M-11 | 1.18 | 0.92 (78) | 2.38 | 3.14 (132) | 25.5 | 27.1 (106) | 31.0 | 33.8 (109) |
| M-12 | 1.09 | 1.09 (100) | 2.13 | 2.82 (132) | 24.4 | 24.6 (101) | 29.7 | 32.4 (110) | |
| ESL – HTST indirect | M-13 | 0.61 | 0.47 (77) | 0.25 | 0.55b (220) | 24.6 | 26.1 (106) | 29.9 | 33.0 (110) |
| M-14 | 0.34 | 0.21b (62) | 0.16 | 0.13b (81) | 24.8 | 26.1 (105) | 30.1 | 32.2 (107) | |
| M-15 | 0.64 | 0.63 (98) | 0.26 | 0.11b (42) | 25.1 | 27.7 (110) | 30.5 | 33.3 (109) | |
| UHT | M-16 | 0.12 | 0.11b (92) | 0.12 | 0.11b (92) | 28.5 | 29.8 (105) | 34.7 | 34.4 (99) |
| M-17 | 0.15 | 0.11b (73) | 0.08 | ND | 26.5 | 27.9 (105) | 32.2 | 32.8 (102) | |
| M-18 | 0.15 | 0.11b (73) | 0.10 | 0.09b (90) | 27.2 | 28.5 (105) | 33.1 | 33.7 (102) | |
| M-19 | 0.10 | 0.09b (90) | 0.05 | ND | 27.7 | 29.9 (108) | 33.7 | 34.8 (103) | |
Mean recovery (![[R with combining macron]](https://www.rsc.org/images/entities/i_char_0052_0304.gif) %) %) |
90 | 104 | 102 | 102 | |||||
Discrimination of commercial milk types by evaluation of α-LA and β-LG concentrations
The quantitation of α-LA and β-LG concentrations offers the possibility of discriminating between different commercial milk types due to the chemical changes associated with the experienced heat processing. In a previous work, classification of milk samples by EC-QCL based IR spectroscopy according to the experienced heat load could be successfully demonstrated by quantitation of β-LG.30 Employing a new and improved experimental setup, in this work not only β-LG, but also α-LA could be quantitated, enabling a more detailed discrimination between different types of milk.Fig. 4 provides an overview of the differences between milk types considering the concentrations of α-LA and β-LG. The evaluation allows conducting three discriminations. Firstly, there is a clear discrimination between low and high heat load samples (indicated by blue and red ellipses, calculated at a confidence level of 99%). The low heat load samples include the milk types produced with soft processing methods such as pasteurized, ESL filtered, ESL bactofugated and ESL HTST direct milk. High heat load samples contain ESL HTST indirect and UHT milk. The threshold for discrimination is 1.25 mg mL−1 β-LG and 0.75 mg mL−1 α-LA. Secondly, it is also possible to distinguish between the different heating technologies within one milk type. ESL HTST milk can be produced by direct and indirect heating systems which have influence on the experienced chemical modifications. Direct heating is a milder method leading to less protein degradation; consequently, ESL HTST direct milk contains higher levels of both β-LG and α-LA. Thirdly, successful separation could also be achieved between ESL HTST indirect and UHT milk. Both heating procedures are rather intensive, leading to an excessive degeneration of the temperature sensitive β-LG. However, for the analyzed samples within this study, the evaluation of α-LA enables to discriminate between ESL HTST indirect (light red) and UHT (dark red) milk with a threshold of 0.2 mg mL−1. This fast discrimination method of commercial bovine milk types holds large potential for application in the milk industry. Since no time consuming pre-processing steps are required in contrast to currently employed HPLC methods, this QCL-IR method represents a useful tool for at-line quality control and process analytical applications.
Conclusions
A rapid method for the quantitative determination of Cas, β-LG, α-LA and total protein content in bovine milk was presented. The combination of an improved EC-QCL setup and multivariate analysis of the amide I and amide II bands allowed the fast screening of commercial bovine milk samples without any sample pretreatment. Comparison between the concentration results obtained by the developed method and those received from standardized reference methods confirmed the good performance of the proposed approach. The reported method holds high potential as an alternative to current methods, which involve labor-intensive sample preparation and long analysis times. With the achieved low limits of detection, the presented high-throughput method allows the quantification of temperature sensitive proteins α-LA and β-LG that enable the discrimination between different commercial bovine milk types. The feasibility of milk differentiation using a fast method is relevant in food processing, technology and policy, and therefore of particular interest in the milk industry.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Iris Biedermann for performing Kjeldahl analyses. This work has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No 780240. M. M. gratefully acknowledges the financial support provided by CONICET and the Austrian Agency for International Cooperation in Education and Research (OeAD). M. M. and M. J. C. are grateful to Universidad Nacional del Litoral (Project CAI+D 2016-50120150100110LI), CONICET (Consejo Nacional de Investigaciones Científicas y Técnicas, Project PIP-2015 N° 0111) and ANPCyT (Agencia Nacional de Promoción Científica y Tecnológica, Project PICT 2014-0347) for financial support. The authors acknowledge the TU Wien University Library for financial support through its Open Access Funding Program.References
- IDF/ISO, Bulletin of the international dairy federation 485/2016. The world dairy situation 2016, B. I. D. F. Brussels, 2016 Search PubMed.
- P. F. Fox, T. Uniacke-Lowe, P. L. H. McSweeney and J. A. O'Mahony, in Dairy Chemistry and Biochemistry, Springer International Publishing, Cham, 2015, pp. 145–239, DOI:10.1007/978-3-319-14892-2_4.
- S. Rozenberg, J. J. Body, O. Bruyere, P. Bergmann, M. Brandi, C. Cooper, J. P. Devogelaer, E. Gielen, S. Goemaere, J. M. Kaufman, R. Rizzoli and J. Y. Reginster, Calcif. Tissue Int., 2016, 98, 1–17 CrossRef CAS PubMed.
- L. I. Boitz and H. K. Mayer, Dairy Sci. Technol., 2016, 96, 677–692 CrossRef CAS PubMed.
- H. Deeth, Foods, 2017, 6, 102 CrossRef PubMed.
- V. S. J. Schmidt, V. Kaufmann, U. Kulozik, S. Scherer and M. Wenning, Int. J. Food Microbiol., 2012, 154, 1–9 CrossRef PubMed.
- G. Rysstad and J. Kolstad, Int. J. Dairy Technol., 2006, 59, 85–96 CrossRef.
- L. I. Boitz and H. K. Mayer, LWT–Food Sci. Technol., 2017, 79, 384–393 CrossRef CAS.
- A. Schmidt and H. K. Mayer, Food Control, 2018, 91, 123–127 CrossRef CAS.
- L. I. Boitz, G. Fiechter, R. K. Seifried and H. K. Mayer, J. Chromatogr. A, 2015, 1386, 98–102 CrossRef CAS PubMed.
- H. K. Mayer, B. Raba, J. Meier and A. Schmid, Dairy Sci. Technol., 2010, 90, 413–428 CrossRef CAS.
- G. Bordin, F. Cordeiro Raposo, B. de la Calle and A. R. Rodriguez, J. Chromatogr. A, 2001, 928, 63–76 CrossRef CAS PubMed.
- E. D. Strange, E. L. Malin, D. L. Van Hekken and J. J. Basch, J. Chromatogr. A, 1992, 624, 81–102 CrossRef CAS PubMed.
- Y. Etzion, R. Linker, U. Cogan and I. Shmulevich, J. Dairy Sci., 2004, 87, 2779–2788 CrossRef CAS PubMed.
- B. Aernouts, E. Polshin, W. Saeys and J. Lammertyn, Anal. Chim. Acta, 2011, 705, 88–97 CrossRef CAS PubMed.
- S. Mazurek, R. Szostak, T. Czaja and A. Zachwieja, Talanta, 2015, 138, 285–289 CrossRef CAS PubMed.
- K. E. Kaylegian, J. M. Lynch, G. E. Houghton, J. R. Fleming and D. M. Barbano, J. Dairy Sci., 2006, 89, 2833–2845 CrossRef CAS PubMed.
- A. Melfsen, E. Hartung and A. Haeussermann, Biosyst. Eng., 2012, 112, 210–217 CrossRef.
- S. E. Holroyd, J. Near Infrared Spectrosc., 2013, 21, 311–322 CrossRef CAS.
- B. Aernouts, E. Polshin, J. Lammertyn and W. Saeys, Visible and near-infrared spectroscopic analysis of raw milk for cow health monitoring: Reflectance or transmittance?, 2011 Search PubMed.
- H. J. Luinge, E. Hop, E. T. G. Lutz, J. A. Vanhemert and E. A. M. Dejong, Anal. Chim. Acta, 1993, 284, 419–433 CrossRef CAS.
- J. C. Moore, J. W. DeVries, M. Lipp, J. C. Griffiths and D. R. Abernethy, Compr. Rev. Food Sci. Food Saf., 2010, 9, 330–357 CrossRef CAS.
- F. Dousseau and M. Pezolet, Biochemistry, 1990, 29, 8771–8779 CrossRef CAS PubMed.
- M. R. Alcaráz, A. Schwaighofer, H. Goicoechea and B. Lendl, Spectrochim. Acta, Part A, 2017, 185, 304–309 CrossRef PubMed.
- A. Schwaighofer, M. Brandstetter and B. Lendl, Chem. Soc. Rev., 2017, 46, 5903–5924 RSC.
- M. R. Alcaráz, A. Schwaighofer, C. Kristament, G. Ramer, M. Brandstetter, H. Goicoechea and B. Lendl, Anal. Chem., 2015, 87, 6980–6987 CrossRef PubMed.
- A. Schwaighofer, M. R. Alcaráz, C. Araman, H. Goicoechea and B. Lendl, Sci. Rep., 2016, 6, 33556 CrossRef CAS PubMed.
- M. R. Alcaráz, A. Schwaighofer, H. Goicoechea and B. Lendl, Anal. Bioanal. Chem., 2016, 408, 3933–3941 CrossRef PubMed.
- J. Kuligowski, A. Schwaighofer, M. R. Alcaráz, G. Quintás, H. Mayer, M. Vento and B. Lendl, Anal. Chim. Acta, 2017, 963, 99–105 CrossRef CAS PubMed.
- A. Schwaighofer, J. Kuligowski, G. Quintás, H. K. Mayer and B. Lendl, Food Chem., 2018, 252, 22–27 CrossRef CAS PubMed.
- A. Schwaighofer, M. Montemurro, S. Freitag, C. Kristament, M. J. Culzoni and B. Lendl, Anal. Chem., 2018, 90, 7072–7079 CrossRef CAS PubMed.
- M. J. Culzoni, M. M. De Zan, J. C. Robles, V. E. Mantovani and H. C. Goicoechea, J. Pharm. Biomed. Anal., 2005, 39, 1068–1074 CrossRef CAS PubMed.
- K. S. Booksh and B. R. Kowalski, Anal. Chem., 1994, 66, 782A–791A CrossRef CAS.
- D. B. Gil, A. M. de la Peña, J. A. Arancibia, G. M. Escandar and A. C. Olivieri, Anal. Chem., 2006, 78, 8051–8058 CrossRef CAS PubMed.
- S. Wold, M. Sjöström and L. Eriksson, Chemom. Intell. Lab. Syst., 2001, 58, 109–130 CrossRef CAS.
- A. C. Olivieri, H. C. Goicoechea and F. A. Iñón, Chemom. Intell. Lab. Syst., 2004, 73, 189–197 CrossRef CAS.
- IDF/ISO, Milk – Determination of nitrogen content – Part 1: Kjeldahl method. Standard 20-1, B. I. D. F. Brussels, 2001 Search PubMed.
- IDF/ISO, Milk – Determination of casein-nitrogen content – Part 1: Indirect method (reference method). Standard IDF 29-1, B. I. D. F. Brussels, 2004 Search PubMed.
- IDF/ISO, Milk – Determination of nitrogen content – Part 4: Determination of non-protein-nitrogen content. Standard IDF 20-4, B. I. D. F. Brussels, 2001 Search PubMed.
- IDF/ISO, Liquid milk - Determination of acid-soluble β-lactoglobulin content – Reverse-phased HPLC method. Standard 178, B. I. D. F. Brussels, 2005 Search PubMed.
- F. Allegrini and A. C. Olivieri, Anal. Chem., 2014, 86, 7858–7866 CrossRef CAS PubMed.
- J. M. Miller and J. C. Miller, Statistics Chemometrics for Analytical Chemistry, Prentice-Hall, Harlow, UK, 4th edn, 2000 Search PubMed.
- A. C. Olivieri, Anal. Chim. Acta, 2015, 868, 10–22 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9an00746f |
| This journal is © The Royal Society of Chemistry 2019 |