A highly selective and sensitive red-emitting fluorescent probe for visualization of endogenous peroxynitrite in living cells and zebrafish†
Zilu
Li
a,
Caiyun
Liu
 a,
Chen
Yu
a,
Yanan
Chen
a,
Pan
Jia
a,
Hanchuang
Zhu
a,
Xue
Zhang
a,
Yamin
Yu
a,
Baocun
Zhu
a,
Chen
Yu
a,
Yanan
Chen
a,
Pan
Jia
a,
Hanchuang
Zhu
a,
Xue
Zhang
a,
Yamin
Yu
a,
Baocun
Zhu
 *a and
Wenlong
Sheng
*b
*a and
Wenlong
Sheng
*b
aSchool of Water Conservancy and Environment, University of Jinan, Shandong Provincial Engineering Technology Research Center for Ecological Carbon Sink and Capture Utilization, Jinan 250022, China. E-mail: lcyzbc@163.com; Fax: +86-531-82767617; Tel: +86-531-82767617
bBiology Institute, Qilu University of Technology (Shandong Academy of Sciences), Jinan 250103, China. E-mail: 15618694162@163.com
First published on 10th April 2019
Abstract
Peroxynitrite (ONOO−) has been proven to participate in various physiological and pathological processes, and may also be a contributing factor in many diseases. In view of this, there is a need to develop detection tools for unambiguously tracking a small amount of endogenous ONOO− to reveal its exact mechanisms. In this paper, a colorimetric and red-emitting fluorescent probe Red-PN, based on a rhodamine-type fluorophore and hydrazide reactive site is described. The probe Red-PN possesses the advantages of rapid response (within 5 s), visual color change (from colorless to pink), preeminent sensitivity (detection limit = 4.3 nM) and selectivity. Because of these outstanding performances, it was possible to accurately detect endogenous ONOO−. It was encouraging that the probe Red-PN could be used effectively for tracking the relatively low levels of endogenous and exogenous ONOO− in living cells and zebrafish. Thus, it is envisioned that the probe Red-PN would have promising prospects in applications for imaging ONOO− in a variety of biological settings.
1. Introduction
Peroxynitrite (ONOO−) is a type of typical reactive oxygen/nitrogen species (RO/NS) which is generated from the rapid diffusion control reactions of nitric oxide and superoxide anionic radicals in vivo,1–3 which are associated with all sorts of physiological and pathological processes. Peroxynitrite with strong oxidability and nucleophilicity could participate in certain reactions with biomolecules including proteins, lipids and nucleic acids, which could lead to cell necrosis and apoptosis eventually.4–6 In addition, many serious diseases are being linked more closely with ONOO−, such as diabetes, Alzheimer's disease, inflammatory diseases, autoimmune diseases and cancer.7–10 However, ONOO− also plays a positive role in living systems. An example is, that it could fight invading pathogens in an immune response or be used as a signaling molecule.11,12 Taking all this into consideration, these findings illustrate that ONOO− may have dual physiological and pathological effects. Therefore, the development of an effective method for monitoring ONOO− is absolutely paramount for the diagnosis of diseases and the research on various pathophysiological functions.In recent years, because of the advantages of using fluorescent probes, for example, their easy operation, high sensitivity, real-time detection and non-invasiveness,13–15 an increasing number of fluorescent probes for ONOO− have been constructed.16–37 For example, Li et al. have reported an ONOO− fluorescent probe for exploring hepatotoxicity,38 which was designed by positioning ketoamide onto the fluorophore, but other researchers noted that the ketoamide recognition receptor was also used in the detection of hydrogen peroxide (H2O2).39 Fluorescent probes based on boronic acid pinacol ester faced a similar situation, and there were potential problems because the boronic acid pinacol ester recognition receptor could recognize relevant reactive oxygen species (ROS) including H2O2 and hypochlorite (OCl−).40,41 Despite this encouraging progress has been achieved,42–56 but the exploitation of fluorescent probes with high selectivity still presents significant challenges in distinguishing between ONOO− and the relevant ROS, especially in physiological conditions where multiple ROS co-existed. However, long wavelength type fluorescent probes have been the subject of growing attention. Yu et al. have published details about reversible near-infrared fluorescent probes for imaging ONOO− in living cells and in vivo,57,58 which exhibited the characteristics of the long wavelength fluorescent probes, such as low phototoxicity to biological samples, high imaging depth and low biological background interference. However, these fluorescent probes with long wavelength emission are still scarce and need to be urgently explored in order to expand widespread applications in complex biological systems. At the same time, because of the difficulty in detecting ONOO−, because of its complex biological characteristics, and its short half-life, plus the low concentration of ONOO−, at homeostasis,1 it is still necessary to optimize its sensitivity and response time. In order to compensate for the previously mentioned flaws, it is still necessary to research and develop valuable tools for ONOO− detection, which will make it possible to monitor endogenous ONOO− in biological systems so its pathophysiological functions can be explored.
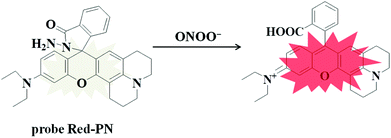
So a novel probe Red-PN was devised based on a red-emitting rhodamine-type fluorophore with hydrazide as the reactive site (Scheme 1). The probe Red-PN displayed a relatively long wavelength emission (∼585 nm) which could minimize auto-fluorescence and biological damage, which was beneficial to realize the wide number of potential applications in biological systems. It is also important to note that the probe Red-PN also showed outstanding properties, including ultrafast response speed towards ONOO− (within 5 s) and an obvious color change from colorless to pink. Additionally, the probe Red-PN showed outstanding selectivity towards ONOO− in preference to various relevant analytes including ROS and reactive nitrogen species (RNS). More striking was its high sensitivity (the detection limit was calculated to be 4.3 nM), and the very low detection limit indicated that the probe Red-PN would allow the detection of endogenous ONOO−. Furthermore, its toxicity was also negligible so that the probe Red-PN could be successfully used in the imaging of low concentrations of both exogenous and endogenous ONOO− in living cells and zebrafish.
2. Experimental
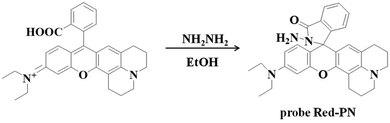
2.1 Synthesis of probe Red-PN
The rhodamine derivative fluorophore (467 mg, 1 mmol) and hydrazine hydrate (NH2NH2·H2O, 500 mg, 10 mmol) were dissolved in 10 mL of anhydrous ethanol (EtOH), and the mixed solution was stirred under reflux for 8 h. The resulting solution was concentrated under reduced pressure and purified using silica column chromatography (dichloromethane containing 15% methanol as eluent) to obtain probe Red-PN (337 mg) with a 70% yield. Proton nuclear magnetic resonance spectroscopy (1H-NMR) (400 MHz, DMSO-d6) δ (ppm): 1.123 (t, J = 7.2 Hz, 6H), 2.083–2.200 (m, 4H), 2.354 (t, J = 6.0 Hz, 2H), 3.075 (t, J = 7.2 Hz, 2H), 3.346–3.398 (m, 4H), 6.388–6.468 (m, 3H), 6.660 (d, J = 2.0 Hz, 1H), 7.049 (d, J = 6.0 Hz, 1H), 7.105 (d, J = 2.0 Hz, 1H), 7.226 (dd, J = 2.0, 9.2 Hz, 1H), 7.271 (d, J = 8.8 Hz, 1H), 7.492–7.552 (m, 2H), 7.823–7.843 (m, 1H), 8.387 (d, J = 9.2 Hz, 1H); 13C-NMR (100 MHz, DMSO-d6) δ (ppm): 12.92, 41.28, 44.15, 47.42, 51.24, 64.69, 98.03, 105.04, 109.36, 109.73, 110.47, 117.91, 119.09, 121.97, 122.87, 123.88, 124.29, 124.79, 128.77, 129.04, 131.00, 133.25, 135.93, 147.48, 149.07, 152.60, 153.63, 157.36, 167.64. [HRMS with electron spray ionization]: Calc'd for C30H33N4O2 [M + H]+, 481.2598; Found, 184.2599.2.2 Live subject statement
All procedures were performed in strict accordance with the National Institute of Health (NIH) guidelines for the Care and Use of Laboratory Animals and the regulations of Qilu University of Technology on the ethical use of animals. All procedures used were approved by the faculty Ethical Committee of the Biology Institute of the Shandong Academy of Sciences. All efforts were made to minimize the number of animals used and their suffering.3. Results and discussion
3.1 The design and synthesis of probe Red-PN for recognizing ONOO−
Rhodamine dyes have been widely used for developing numerous fluorescent probes, which invariably hold two dimethylamino or diethylamino structures according to previous reports. Based on this making these structures into a ring could improve their structure,59 and trigger the increase of fluorescence quantum efficiency, thus the detection sensitivity of the probe could be optimized by using this property. Not only that, the other flash point was, that rhodamine exhibited a relatively long wavelength emission spectra, which was more suitable for the construction of a fluorescent probe that could be used for bioimaging applications. Therefore, rhodamine was chosen as the fluorophore for constructing the long wavelength fluorescent probe Red-PN.The fluorophore was critical for the construction of a novel ONOO− fluorescent probe, but of equal importance was the appropriate recognition receptor. Recently, several studies have shown that hydrazide might be useful as a favorable recognition unit for recognizing ONOO−.60–62 As an example, Liu et al. reported a fluorescent probe based on a hydrazide recognition unit, which demonstrated its rapid response to ONOO− and had excellent selectivity.60 This encouraged the further use of this recognition unit to design and synthesize a ONOO− fluorescent probe with the best performance. Inspired by these studies, probe Red-PN was put forward, which was based on a rhodamine-type fluorophore and hydrazide as the reactive site. The hydrazide moiety was conjugated to the fluorophore which could lead to the formation of a non-fluorescent spirocyclic ring system, and the fluorescence could be recovered when the probe Red-PN reacted with ONOO− to open the corresponding spiral ring structure (Scheme 2).60–63 This design strategy was also confirmed using high resolution mass spectrometry (HRMS, a new peak at 467.2326 was obtained, which corresponded to the peak of the fluorophore) in this work. The synthetic procedures are described detail in the Experimental section and the synthetic route for probe Red-PN is illustrated in Scheme 1.
3.2 Sensitivity and quantification of ONOO−
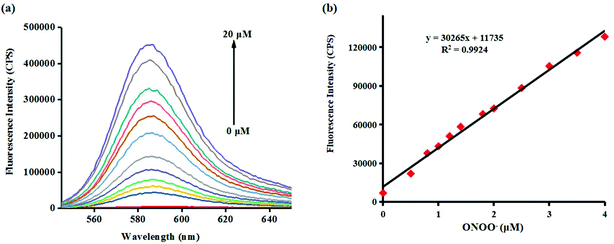
Investigating the fluorescence and absorption responses of probe Red-PN for ONOO− was essential in order to determining the sensing ability of probe Red-PN. The fluorescence responses of probe Red-PN for detecting ONOO− were assessed in 20 mM phosphate buffered saline [PBS, pH 7.4, 10% dimethyl sulfoxide (DMSO)]. As can be seen in Fig. 1a, the free probe Red-PN (5 μM) solution exhibited a fantastically low background fluorescence, and then an emission peak at about 585 nm was observed upon addition of ONOO− (0–20 μM), and the fluorescence intensity increased continuously. It was noteworthy, that a good linear relationship between the fluorescence intensities at 585 nm and the concentrations of ONOO− in the range of 0–4 μM was achieved (linear equation: y = 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 265 × [ONOO−] (μM) + 11
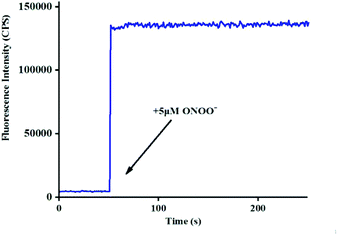
265 × [ONOO−] (μM) + 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 735, R2 = 0.9924) (Fig. 1b), and the detection limit was calculated to be 4.3 nM. More importantly, the reaction of probe Red-PN with ONOO− was completed within 5 s (Fig. 2). These results implied that probe Red-PN could quantitatively and sensitively capture ONOO− as had been expected and was convenient for the rapid monitoring of ONOO−.
735, R2 = 0.9924) (Fig. 1b), and the detection limit was calculated to be 4.3 nM. More importantly, the reaction of probe Red-PN with ONOO− was completed within 5 s (Fig. 2). These results implied that probe Red-PN could quantitatively and sensitively capture ONOO− as had been expected and was convenient for the rapid monitoring of ONOO−.
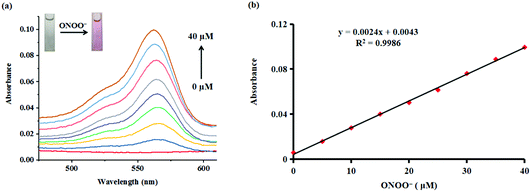
Subsequently, the absorption responses of probe Red-PN towards ONOO− were also investigated under the same experimental conditions. As can be seen in Fig. 3, probe Red-PN (10 μM) showed a weaker absorption intensity in the absence of ONOO−. As expected, the absorption spectra of probe Red-PN exhibited an obvious increase at 563 nm and a distinct color change from colorless to pink in the presence of ONOO−. In addition, the absorption intensities at 563 nm were linearly proportional to the increasing ONOO− concentrations (0–40 μM). Therefore, these results indicated that probe Red-PN could not only quantitatively detect ONOO− using the absorption spectra method, but could also be used for naked eye analysis.
These results demonstrated that probe Red-PN not only provided a convenient analytical method for detecting ONOO− by the naked eye, but also offered a rapid quantitative testing tool for detecting ONOO− at low concentrations, implying that there was a great potential for monitoring intracellular ONOO− levels (Table 1).
| Probe | λ em | Detection limit | Time | Imaging | Ref. |
|---|---|---|---|---|---|
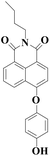
|
550 nm | 16 nM | 6 min | Living cells | 19 |
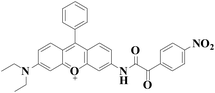
|
558 nm | 43 nM | 30 min | Living cells | 27 |
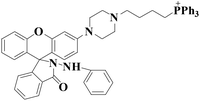
|
578 nm | 53 nM | 20 min | Living cells | 36 |
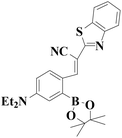
|
522 nm | 0.83 nM | 50 s | Living cells | 45 |
|
380/485 nm | 40 nM | 5 min | Living cells, zebrafish, high-fat diet animal model | 47 |
|
545 nm | 4 nM | Within 5 s | Living cells | 64 |
|
585 nm | 4.3 nM | Within 5 s | Living cells and zebrafish | This work |
3.3 Selectivity of probe Red-PN for ONOO−
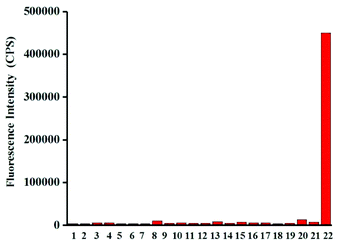
Next, the selectivity of probe Red-PN towards various relevant analytes including: Cl−, Cu2+, cysteine (Cys), Fe2+, Fe3+, glutathione (GSH), homocysteine (Hcy), H2O2, K+, Mg2+, Na+, NO, OCl−, ˙OtBu, ˙OH, ONOO−, 1O2, O2−, SO42−, t-butyl hydroperoxide (TBHP), and Zn2+, were tested under the conditions mentioned previously. It was encouraging, that hardly any conspicuous fluorescence change was observed when other analytes were added to the free probe Red-PN solution (Fig. 4). Comparatively, the fluorescence intensity at 585 nm exhibited great enhancement upon the addition of ONOO−. This demonstrated that the probe Red-PN could detect ONOO− with high selectivity. In addition, the effects of pH on the recognition property of the probe Red-PN for ONOO− were also studied. As shown in Fig. 5, probe Red-PN was relatively stable over the pH range of 5.5–10 and the responses of probe Red-PN to ONOO− were excellent at the biological pH range. These combined results indicated that probe Red-PN could specifically recognize ONOO− without the interference of other bioactive molecules and pH.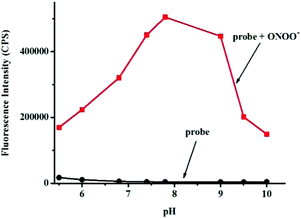 | ||
| Fig. 5 The fluorescence intensities at 585 nm of probe Red-PN (5 μM) in the presence and absence of ONOO− (20 μM) at various pH values. | ||
3.4 Bioimaging of ONOO− in living cells
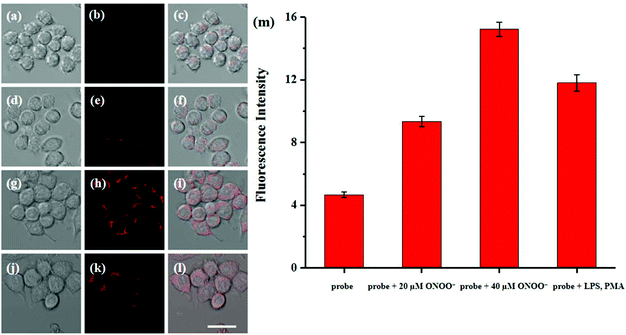
Cytotoxicity estimation of probe Red-PN was carried out in live RAW 264.7 mouse macrophage cells using a Cell Counting Kit-8 (CCK-8) assay. The experimental results showed that the viability of the cells was better than 90%, thus, demonstrating that probe Red-PN possessed low cytotoxicity (Fig. 6). Next experiments were done to determine whether probe Red-PN could be successfully used in live cells for the visualization of ONOO−. The macrophage cells were pre-incubated with probe Red-PN (10 μM) for 30 min, and subsequently were incubated with different concentrations of ONOO− (20 μM, 40 μM) for another 30 min. Next, the macrophage cells were stimulated with phorbol myristate acetate (PMA, 1.0 μg mL−1) and lipopolysaccharide (LPS, 1.0 μg mL−1) for 1 h to produce endogenous ONOO−, and were subsequently loaded with probe Red-PN (10 μM). As can be seen from Fig. 7, the cells loaded with probe Red-PN showed a faint red fluorescence without external stimulation, and exhibited continuous increase in fluorescence brightness because of the addition of ONOO− (20 μM, 40 μM). As expected, the cells stimulated with PMA and LPS also exhibited an intense red intracellular fluorescence. The previous information suggested that the probe Red-PN could be used to effectively visualize the relatively low ranges of endogenous and exogenous ONOO− in living cells.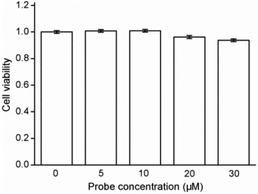 | ||
| Fig. 6 Results of cytotoxicity assays with probe Red-PN at different concentrations in RAW 264.7 macrophage cells. | ||
3.5 Bioimaging of ONOO− in zebrafish
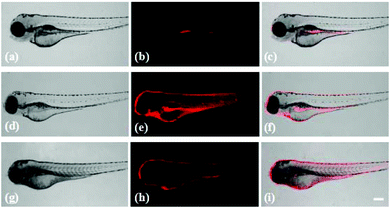
Probe Red-PN has demonstrated its feasibility for the bioimaging of ONOO− in living cells, and it was important to further evaluate the feasibility of using probe Red-PN for monitoring endogenous and exogenous ONOO−in vivo. Following this idea, zebrafish were used as an model to study the distribution of endogenous and exogenous ONOO− (Fig. 8). The four day old zebrafish were pre-incubated with probe Red-PN for 30 min, and then were fed with ONOO− (20 μM) for another 30 min. The zebrafish changed from having a faint fluorescence to a largely enhanced red fluorescence. In another experiment, the zebrafish pre-treated with PMA (1.0 μg mL−1) and LPS (1.0 μg mL−1) also exhibited a remarkable red fluorescence enhancement. These results indicated that probe Red-PN not only was suitable for imaging ONOO− in living cells, but could also be used for monitoring endogenous and exogenous ONOO−in vivo.4. Conclusions
In summary, a novel colorimetric and long wavelength fluorescent probe Red-PN for ultrasensitive detection of ONOO− was synthesized. The probe Red-PN possessed many outstanding features, such as extremely high sensitivity (detection limit down to 4.3 nM), ultra-fast response (within 5 s), naked-eye analysis possible because of marked color changes from colorless to pink, excellent selectivity against other ROS/RNS. Not only that, the negligible toxicity of probe Red-PN was advantageous for utilization in biological systems. The satisfactory imaging demonstrated the feasibility of probe Red-PN for visualization of ONOO− in macrophage cells and zebrafish. Thus, it is anticipated that probe Red-PN will be a promising imaging tool for tracking ONOO− in biological systems and for further revealing its pathophysiological functions.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge financial support from the National Natural Science Foundation of China (21777053 and 21607053), Shandong Provincial Natural Science Foundation (ZR2017MB014 and ZR2018PC013), a Project of Shandong Province Higher Educational Science and Technology Program (J16LD01), and the Youth Fund of Shandong Academy of Science (2019QN001).References
- G. Ferrer-Sueta and R. Radi, ACS Chem. Biol., 2009, 4, 161–177 CrossRef CAS PubMed.
- S. Goldstein, J. Lind and G. Merényi, Chem. Rev., 2005, 105, 2457–2470 CrossRef CAS PubMed.
- C. Nathan, J. Clin. Invest., 2003, 111, 769–778 CrossRef CAS PubMed.
- B. Alvarez and R. Radi, Amino Acids, 2003, 25, 295–311 CrossRef CAS PubMed.
- R. Radi, J. Biol. Chem., 2013, 288, 26464–26472 CrossRef CAS PubMed.
- L. Virág, E. Szabó, P. Gergely and C. Szabó, Toxicol. Lett., 2003, 140–141, 113–124 CrossRef.
- H. Ischiropoulos and J. S. Beckman, J. Clin. Invest., 2003, 111, 163–169 CrossRef CAS PubMed.
- D. A. Wink, Y. Vodovotz, J. Laval, F. Laval, M. W. Dewhirst and J. B. Mitchell, Carcinogenesis, 1998, 19, 711–721 CrossRef CAS PubMed.
- P. Pacher, J. S. Beckman and L. Liaudet, Physiol. Rev., 2007, 87, 315–424 CrossRef CAS PubMed.
- C. Szabó, H. Ischiropoulos and R. Radi, Nat. Rev. Drug Discovery, 2007, 5, 662–680 CrossRef PubMed.
- A. Weidinger and A. V. Kozlov, Biomolecules, 2015, 5, 472–484 CrossRef CAS PubMed.
- L. Liaudet, G. Vassalli and P. Pacher, Front. Biosci., 2009, 14, 4809–4814 CrossRef.
- Q. Duan, H. Zhu, C. Liu, R. Yuan, Z. Fang, Z. Wang, P. Jia, Z. Li, W. Sheng and B. Zhu, Analyst, 2019, 144, 1426–1432 RSC.
- W. Sun, S. Guo, C. Hu, J. Fan and X. Peng, Chem. Rev., 2016, 116, 7768–7817 CrossRef CAS PubMed.
- Q. Duan, P. Jia, Z. Zhuang, C. Liu, X. Zhang, Z. Wang, W. Sheng, Z. Li, H. Zhu, B. Zhu and X. Zhang, Anal. Chem., 2019, 91, 2163–2168 CrossRef CAS PubMed.
- H. Zhu, J. Fan, J. Du and X. Peng, Acc. Chem. Res., 2016, 49, 2115–2126 CrossRef CAS PubMed.
- Z. Li, S. H. Yan, C. Chen, Z.-R. Geng, J. Y. Chang, C. X. Chen, B. H. Huang and Z. L. Wang, Biosens. Bioelectron., 2017, 90, 75–82 CrossRef CAS PubMed.
- J. Cao, W. An, A. G. Reeves and A. R. Lippert, Chem. Sci., 2018, 9, 2552–2558 RSC.
- B. Zhu, Z. Wang, Z. Zhao, W. Shu, M. Zhang, L. Wu, C. Liu, Q. Duan and P. Jia, Sens. Actuators, B, 2018, 262, 380–385 CrossRef CAS.
- H. Xiao, C. Wu, P. Li and B. Tang, Anal. Chem., 2018, 90, 6081–6088 CrossRef CAS PubMed.
- L. Li, M. Zan, X. Qie, J. Yue, P. Miao, M. Ge, Z. C. Wang, F.-Q. Bai, H. X. Zhang, J. K. Ferri and W. F. Dong, J. Phys. Chem. A, 2018, 122, 217–223 CrossRef CAS PubMed.
- J. Zhang, X. Zhen, J. Zeng and K. Pu, Anal. Chem., 2018, 90, 9301–9307 CrossRef CAS PubMed.
- D. Lee, C. Lim, G. Ko, D. Kim, M. K. Cho, S.-J. Nam, H. M. Kim and J. Yoon, Anal. Chem., 2018, 90, 9347–9352 CrossRef CAS PubMed.
- X. Jia, Q. Chen, Y. Yang, Y. Tang, R. Wang, Y. Xu, W. Zhu and X. Qian, J. Am. Chem. Soc., 2016, 138, 10778–10781 CrossRef CAS PubMed.
- Z. Li, R. Liu, Z.-l. Tan, L. He, Z. L. Lu and B. Gong, ACS Sens., 2017, 2, 501–505 CrossRef CAS.
- D. Cheng, Y. Pan, L. Wang, Z. Zeng, L. Yuan, X. Zhang and Y. T. Chang, J. Am. Chem. Soc., 2017, 139, 285–292 CrossRef CAS PubMed.
- D. Cheng, W. Xu, L. Yuan and X. Zhang, Anal. Chem., 2017, 89, 7693–7700 CrossRef CAS PubMed.
- J.-B. Li, L. Chen, Q. Wang, H.-W. Liu, X.-X. Hu, L. Yuan and X.-B. Zhang, Anal. Chem., 2018, 90, 4167–4173 CrossRef CAS PubMed.
- A. C. Sedgwick, H.-H. Han, J. E. Gardiner, S. D. Bull, X. P. He and T. D. James, Chem. Commun., 2017, 53, 12822–12825 RSC.
- J. Li, C. S. Lim, G. Kim, H. M. Kim and J. Yoon, Anal. Chem., 2017, 89, 8496–8500 CrossRef CAS PubMed.
- T. Peng, X. Chen, L. Gao, T. Zhang, W. Wang, Ji. Shen and D. Yang, Chem. Sci., 2016, 7, 5407–5413 RSC.
- D. Li, J. Cheng, C.-K. Wang, H. Ying, Y. Hu, F. Han and X. Li, Chem. Commun., 2018, 54, 8170–8173 RSC.
- P. Cheng, J. Zhang, J. Huang, Q. Miao, C. Xu and K. Pu, Chem. Sci., 2018, 9, 6340–6347 RSC.
- J. Miao, Y. Huo, H. Shi, J. Fang, J. Wang and W. Guo, J. Mater. Chem. B, 2018, 6, 4466–4473 RSC.
- B. Zhu, L. Wu, Y. Wang, M. Zhang, Z. Zhao, C. Liu, Z. Wang, Q. Duan and P. Jia, Sens. Actuators, B, 2018, 259, 797–802 CrossRef CAS.
- H. Li, X. Li, X. Wu, W. Shi and H. Ma, Anal. Chem., 2017, 89, 5519–5525 CrossRef CAS.
- Z. Wang, L. Wu, Y. Wang, M. Zhang, Z. Zhao, C. Liu, Q. Duan, P. Jia and B. Zhu, Anal. Chim. Acta, 2019, 1049, 219–225 CrossRef CAS.
- Y. Li, X. Xie, X. Yang, M. Li, X. Jiao, Y. Sun, X. Wang and B. Tang, Chem. Sci., 2017, 8, 4006–4011 RSC.
- X. Xie, X. Yang, T. Wu, Y. Li, M. Li, Q. Tan, X. Wang and B. Tang, Anal. Chem., 2016, 88, 8019–8025 CrossRef CAS PubMed.
- B. C. Dickinson, C. Huynh and C. J. Chang, J. Am. Chem. Soc., 2010, 132, 5906–5915 CrossRef CAS.
- J. Chan, S. C. Dodani and C. J. Chang, Nat. Chem., 2012, 4, 973–984 CrossRef CAS PubMed.
- D.-Y. Zhou, J. Yang, Y. Lia, W.-L. Jiang, Y. Tian, Z.-M. Yi and C.-Y. Li, Dyes Pigm., 2019, 161, 288–295 CrossRef CAS.
- F. Yu, P. Song, P. Li, B. Wang and K. Han, Analyst, 2012, 137, 3740–3749 RSC.
- B. Wang, F. Yu, P. Li, X. Sun and K. Han, Dyes Pigm., 2013, 96, 383–390 CrossRef CAS.
- J. Zhang, Y. Li, J. Zhao and W. Guo, Sens. Actuators, B, 2016, 237, 67–74 CrossRef CAS.
- Y. Shen, X. Zhang, Y. Zhang, H. Li, L. Dai, X. Peng, Z. Peng and Y. Xie, Anal. Chim. Acta, 2018, 1014, 71–76 CrossRef CAS PubMed.
- S. Palanisamya, P.-Y. Wu, S.-C. Wu, Y.-J. Chen, S.-C. Tzou, C.-H. Wang, C.-Y. Chen and Y.-M. Wang, Biosens. Bioelectron., 2017, 91, 849–856 CrossRef PubMed.
- D.-Y. Zhou, Y. Li, W.-L. Jiang, Y. Tian, J. Fei and C.-Y. Li, Chem. Commun., 2018, 54, 11590–11593 RSC.
- B. Guo, J. Jing, L. Nie, F. Xin, C. Gao, W. Yang and X. Zhang, J. Mater. Chem. B, 2018, 6, 580–585 RSC.
- Y. Fu, H. Nie, R. Zhang, F. Xin, Y. Tian, J. Jing and X. Zhang, RSC Adv., 2018, 8, 1826–1832 RSC.
- W. Sun, Y.-D. Shi, A.-X. Ding, Z.-L. Tan, H. Chen, R. Liu, R. Wang and Z.-L. Lu, Sens. Actuators, B, 2018, 276, 238–246 CrossRef CAS.
- Q. Li and Z. Yang, Tetrahedron Lett., 2018, 59, 125–129 CrossRef CAS.
- T. Hou, K. Zhang, X. Kang, X. Guo, L. Du, X. Chen, L. Yua, J. Yue, H. Ge, Y. Liu, A. M. Asirie, K. A. Alamrye, H. Yu and S. Wang, Talanta, 2019, 196, 345–351 CrossRef CAS PubMed.
- X. Xie, F. Tang, G. Liu, Y. Li, X. Su, X. Jiao, X. Wang and B. Tang, Anal. Chem., 2018, 90, 11629–11635 CrossRef CAS PubMed.
- T. Peng, N.-K. Wong, X. Chen, Y.-K. Chan, D. Ho, Z. Sun, J. J. Hu, J. Shen, H. El-Nezami and D. Yang, J. Am. Chem. Soc., 2014, 136, 11728–11734 CrossRef CAS PubMed.
- A. C. Sedgwick, W.-T. Dou, J.-B. Jiao, L. Wu, G. T. Williams, A. T. A. Jenkins, S. D. Bull, J. L. Sessler, X.-P. He and T. D. James, J. Am. Chem. Soc., 2018, 140, 14267–14271 CrossRef CAS PubMed.
- F. Yu, P. Li, G. Li, G. Zhao, T. Chu and K. Han, J. Am. Chem. Soc., 2011, 133, 11030–11033 CrossRef CAS PubMed.
- F. Yu, P. Li, B. Wang and K. Han, J. Am. Chem. Soc., 2013, 135, 7674–7680 CrossRef CAS PubMed.
- Y. Zhang, S. Xia, M. Fang, W. Mazi, Y. Zeng, T. Johnstona, A. Papa, R. L. Luck and H. Liu, Chem. Commun., 2018, 54, 7625–7628 RSC.
- D. Liu, S. Feng and G. Feng, Sens. Actuators, B, 2018, 269, 15–21 CrossRef CAS.
- D. Wu, J.-C. Ryu, Y. W. Chung, D. Lee, J.-H. Ryu, J.-H. Yoon and J. Yoon, Anal. Chem., 2017, 89, 10924–10931 CrossRef CAS PubMed.
- B. Zhu, M. Zhang, L. Wu, Z. Zhao, C. Liu, Z. Wang, Q. Duan, Y. Wang and P. Jia, Sens. Actuators, B, 2018, 257, 436–441 CrossRef CAS.
- H. N. Kim, M. H. Lee, H. J. Kim, J. S. Kim and J. Yoon, Chem. Soc. Rev., 2008, 37, 1465–1472 RSC.
- H. Zhang, J. Liu, Y.-Q. Sun, Y. Huo, Y. Li, W. Liu, X. Wu, N. Zhu, Y. Shi and W. Guo, Chem. Commun., 2015, 51, 2721–2724 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9an00347a |
| This journal is © The Royal Society of Chemistry 2019 |