Recent advances in nanomaterial-based electrochemical and optical sensing platforms for microRNA assays
Yi-Han
Wang
,
Liu-Liu
He
,
Ke-Jing
Huang
 *,
Ying-Xu
Chen
,
Shu-Yu
Wang
,
Zhen-Hua
Liu
and
Dan
Li
*,
Ying-Xu
Chen
,
Shu-Yu
Wang
,
Zhen-Hua
Liu
and
Dan
Li
College of Chemistry and Chemical Engineering, Xinyang Normal University, Xinyang 464000, China. E-mail: kejinghuang@163.com; Tel: +86-376-6390611
First published on 13th March 2019
Abstract
MicroRNA (MiRNA) plays a crucial role in biological cells to enable assessment of a cancer's development stage. Increasing evidence has shown that the accurate and sensitive detection of miRNA holds the key toward correct disease diagnosis. However, some characteristics of miRNAs, such as their short chains, low concentration, and similar sequences, make it difficult to detect miRNA in biological samples. Nanomaterials usually have good optical, electronic, and mechanical properties and therefore provide new possibilities for improving the performance of miRNA assays. Many different sorts of nanomaterials, including metal nanomaterials, carbon nanomaterials, quantum dots, and transition-metal dichalcogenides, have been used to construct optical and electrochemical assays for miRNA and have shown attractive results. This review describes recent efforts in the application of nanomaterials as sensing elements in electrochemical and optical miRNA assays. The analytical figures of merit of various methods for the detection of miRNA are compared in the present article. The current capabilities, limitations, and future challenges in miRNA detection and analysis based on nanomaterials are also addressed.
 Dan Li | Dan Li is a scholar at the School of Chemistry and Chemical Engineering, Xinyang Normal University. Her major is in Chemistry. Her research focuses mainly on the testing of nanomaterials. |
1. Introduction
MicroRNAs (miRNAs) are a naturally-occurring family of short endogenous nanocoding RNA molecules (typically 18–22 nucleotides long) with higher stability than messenger RNA (mRNA), and they serve as important regulators in the expression and function of eukaryotic genomes in blood serum, urine, saliva cells, and tissues.1 The basic process and expression of mature miRNA are outlined in Fig. 1A.2 MiRNAs as the pivotal factor in gene regulation and dysregulation can lead to the aberrant expression of cancer, hepatitis, malignancies, and neuro-degeneration. Their regulation makes a great contribution to chromatin structure, chromosome segregation, transcription, RNA processing, RNA stability, and so on.3–5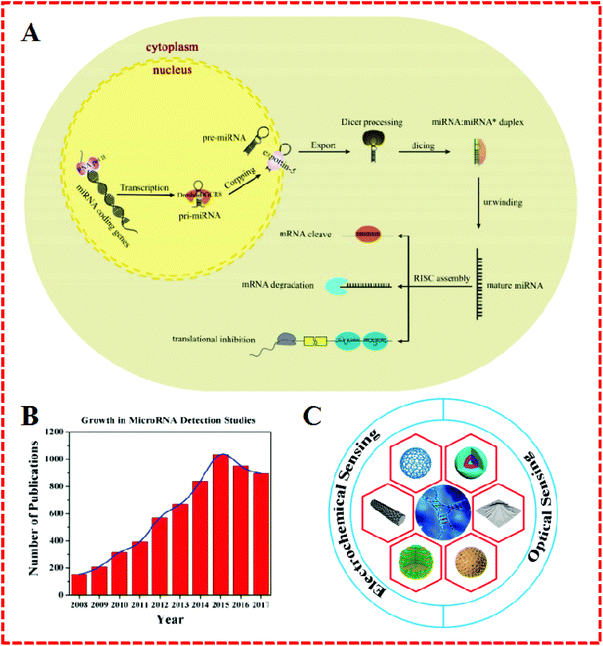 | ||
| Fig. 1 (A) Schematic illustration of miRNA biogenesis and functions. Reproduced with permission from ref. 2, Copyright 2017, Elsevier B.V. (B) Number of publications regarding the detection of miRNA since 2008. These results were obtained from a SciFinder search using the keywords “microRNA detection”. (C) Schematic illustration of nanomaterials as signal amplification elements in miRNA-based electrochemical and optical sensing. | ||
Therefore, the exploration of miRNAs has recently attracted the attention of many chemists, biochemists, physicists, and medical experts, which has led to a rapid increase in the number of publications on “miRNA detection” (Fig. 1B). However, the determination of miRNAs has not yet been achieved in some areas when compared to DNA identification due to their unique characteristics, including short size (e.g., let-7 miRNA), sequence homology among family members, susceptibility to degradation, and low abundance in total RNA samples.6,7 In order to resolve these problems, a number of strategies have been developed, such as northern blotting with radio-labeled probes, sequencing quantitative assessment, in situ hybridization, oligonucleotide (OND) microarrays, and the use of deep-sequencing quantitative polymerase chain reaction (qPCR).8,9 Being regarded as conventional detection methods, these are the mainstream procedures applied for the determination of miRNA. Nonetheless, everything has two sides and so these techniques also display some limitations. For example, northern blotting, as the most commonly used in early miRNA profiling research, shows some drawbacks in terms of low-throughput analysis limitations and semiquantitation issues. Similarly, while qPCR can achieve superior sensitivity and operates over a broad dynamic range, nevertheless, the laboratory skills required and operating conditions are complex. Likewise, the major concern in the use of the microarray is the labeling procedure, despite its high efficiency and effectiveness as a practical economic strategy.
Fortunately, progress has emerged in the field of nanomaterial-based biosensors.10–12 Nanomaterials with a highly specific recognition ability for biomolecules can be subdivided into more subtle types, such as metal nanoparticles (NPs), graphene (GR), quantum dots (QDs), carbon-based nanomaterials, magnetic nanoparticles (MNPs), and polymeric nanoparticles.13,14 Due to their brilliant characteristics, such as large surface area, active binding sites, biological compatibility, chemical stability, nontoxicity, and excellent catalytic and conductivity properties, nanomaterials have been used in the electrochemical detection of miRNA. Research has demonstrated that nanomaterials offer many unique physical and chemical advantages when applied in different contexts, such as serving as: (1) electrode materials for the construction of sensing platforms, (2) carriers for signal elements to assist a molecular probe to complete a series of complex hybridization reactions, (3) tracers based on their direct electrochemistry, (4) separators and collectors (in the case of magnetic nanomaterials) to screen for the presence of magnetic substances, (5) chemical reaction catalysts to aid the building of a biosensor chemical reaction for difficult to easy conversions.
On the other hand, similar to electrochemical sensors, the optical sensors based on a series of nanomaterials that demonstrate outstanding sensitivity and selectivity, fast analysis, low cost, easy operation, remote control, and biocompatibility have attracted interest in terms of their potential use for miRNA assays. Well-defined nanomaterial quenchers, including Au nanoparticles (AuNPs), carbon nanotubes (CNTs), silicon nanowires, carbon nitride nanosheets, and GO, have been reported possessing a distinguished adsorption capacity for single stranded DNA (ssDNA) and for their ultra-efficient fluorescence quenching capability. Besides the function of fluorescence quenching, nanomaterials have seen widespread utilization in fluorescence enhancement, colorimetry, lateral flow assays, surface-enhanced Raman scatting, and surface plasmon resonance systems.3,11
In summary, nanomaterials have been widely used in electrochemical and fluorescent biosensors for miRNA detection and have displayed good performance and great potential. In this review, the importance of miRNA and the advantages of nanomaterials are first discussed. Then, recent advances in nanomaterials-based miRNA biosensors, including electrochemical and optical sensors, are highlighted (Fig. 1C). Finally, possible challenges and potential opportunities for these assays are proposed.
2. Electrochemical assays for miRNA detection
2.1. Nanomaterials as electrode materials to construct sensing platforms
Due to their unique properties, such as large surface area and active binding sites, special magnetism, electrical conductibility, and catalytic activity, a great number of nanomaterials, such as carbon based-materials, metal nanoparticles, and layered transition-metal dichalcogenides (TMDCs), have been used to construct electrochemical sensing platforms for miRNA.15Recently, AuNPs have been widely used as an electrode surface substance as they possess high surface energy and offer fine catalytic efficiency, good biocompatibility, and electrical conductivity and consequently have even been applied for miRNA detection.16–18 For example, Zouari et al. developed a competitive RNA/RNA hybridization configuration for miRNA determination, as shown in Fig. 2A.1 They structured an electrochemical biosensor by integrating the AuNPs with a screen-printed carbon electrode (AuNPs–SPCE). Biotinylated miRNAs with the same sequence as target miRNA were mixed with samples to form competition with the target miRNA for a thiolated RNA probe assembled onto the surface of AuNPs–SPCE. With the help of AuNPs, the miRNA detection avoided the necessity for multiple reagents, bulky instruments, and the use of time-consuming working protocols. A very low detection limit (25 fM) for miRNA-21 was accomplished without redundant steps. In addition, Yuan's group proposed an enzyme-free electrochemical sensor for the highly sensitive detection of miRNA by coupling with GR-coated AuNPs (Au–GR) as an electrode modifier (Fig. 2B).19 The good electrical conductivity and abundant binding sites of Au–GR enhanced the effective surface area of the electrode to promote the reaction rate. This miRNA sensor achieved a detection limit of 3.3 fM and showed a wide dynamic range spanning six orders of magnitude.
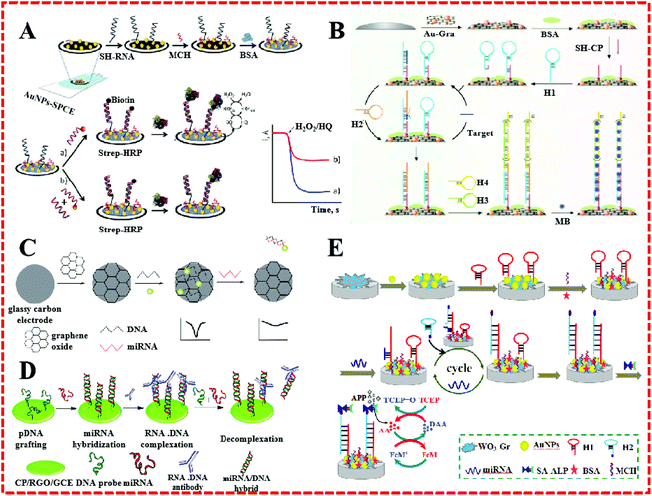 | ||
| Fig. 2 (A) Schematic illustration of the preparation of SH–RNA/MCH–AuNPs–SPCEs and a direct competitive hybridization assay developed for miRNA determination. Reproduced with permission from ref. 1, Copyright 2016, Elsevier B.V. (B) Schematic illustration of an electrochemical miRNA biosensor based on the enzyme-free dual signal amplification of CHA and HCR. Reproduced with permission from ref. 19, Copyright 2014, Elsevier B.V. (C) Scheme of an electrochemical biosensor for the detection of miRNA. Reproduced with permission from ref. 25, Copyright 2015, The Royal Society of Chemistry. (D) A simple and sensitive label-free immunosensor for the detection of microRNAs. Reproduced with permission from ref. 26, Copyright 2013, American Chemical Society. (E) Schematic illustration of biosensor detection of miRNA based on CHA target miRNA recycling and signal amplification of ALP coupled with ECC redox cycling. Reproduced with permission from ref. 27, Copyright 2016, Elsevier B.V. | ||
GR has shown excellent properties, such as a large and specific surface area, super conductive capacity, high mechanical strength, and low cost efficiency. GR-based materials are generally applied to micro-nanocomponents, sensors, and capacitors, and used in hydrogen storage and other fields.20–24 For example, Sun et al. fabricated a GO-modified electrode for the quantitative detection of miRNA.25 Because of its special structure and characteristics, GR was directly bonded to the GCE to construct a sensing plane with multiple AgNPs–labeled DNA probe sites (Fig. 2C). In the presence of miRNA, complementary miRNA sequences were hybridized with DNA. Afterward, the AgNPs–labeled DNA probes were released from the electrode surface into the solution. This miRNA sensor achieved an ultrahigh selectivity and required no further enzymes or amplification. Meanwhile, a high stability was proved. Pham described a label-free electrochemical sensor based on a conducting polymer/reduced GO-modified electrode to detect miR-29b-1 and miR-141, as shown in Fig. 2D.26 On account of the surface activity and increased reliability of the GR composite, the sensor worked as a hybridized assay for the specific binding of the anti-DNA–RNA antibody with DNA–RNA. Simultaneously, a low detection limit of 5 fM was obtained. Huang's group described an electrochemical biosensor for miRNA based on tungsten oxide–GR composites combined with catalyzed hairpin assembly target recycling and enzyme signal amplification, as shown in Fig. 2E.27 A GCE was modified by WO3–GR composites and AuNPs for the immobilization of a thiol-terminated capture probe. The extensive surface area of the WO3–GR composites facilitate they functioning as a fantastic sensing substrate that achieved an admirable lower detection limit of 0.05 fM.
CNTs are seamless hollow nanoscale coaxial cylinders rolled from lamellar graphite. According to the number of layers of CNTs, they can be divided into single-walled carbon nanotubes (SWCNTs) and multi-walled carbon nanotubes (MWCNTs). CNTs can load a large number of signal molecules, speed up the reaction rate, and achieve signal amplification, thereby they can improve the sensitivity of sensors. CNTs also have been used to qualitatively and quantitatively detect miRNA.28–30 Tran's group employed screen-printed gold electrodes modified with reduced GO and CNTs to form an electrochemical sensor for miRNA detection.31 A detection limit of 10 fM was obtained. Similarly, Li et al. proposed an electrochemical signal-amplification biosensor for miRNA based on MWCNTs’ great potential.32 This assay was based on the guanine oxidation subsequent to the hybridization between the target miRNA and the complementary DNA capture probe. Because CNTs have an excellent electrochemical property for guanine oxidation, a voltammetric signal change was generated, which could be measured by differential pulse voltammetry (DPV).
Layered TMDCs in the construction of electrochemical sensors also play an equally important role.33–35 The most used TMDCs are WS2, MoS2, SnS2, and VS2. These have exhibited fascinating performance, such as large surface area, metallic and semi-conducting electrical capabilities, intercalatable morphologies, and eminent catalytic properties.2 The marriage of TMDCs and electrochemical biosensors has created many productive sensing strategies for miRNA. Our group integrated hollow MoS2 microcubes and AuNPs as an electrode supporting substrate.36 An ultrasensitive miRNA-21 electrochemical sensor was fabricated based on the hollow MoS2 microcubes, duplex-specific nuclease (DSN), and enzyme signal amplification, which demonstrated a remarkable low detection limit (0.086 fM), and high selectivity and sensitivity.
As for the construction of the nanomaterial-based bio-interfaces, it is important to accurately and efficiently immobilize aptamers on the surface of nanomaterials. These nanomaterials, such as AuNPs, GR, CNTs, and TMDCs, with a large surface area and good biocompatibility can provide more binding sites for the immobilization of larger amounts of aptamers. The amount of immobilized aptamers will directly affect the sensitivity of the sensor. Due to their poor electrical conductivity, transition metal oxides are rarely used as electrode materials. As sensing electrode materials, nanomaterials face many challenges, one of which is how to prepare nanomaterials with aptamer and enzyme functions in a gentle manner.
2.2. Nanomaterials as carriers for signal elements
Nanomaterials have been used as carriers in miRNA electrochemical biosensors due to their many superior properties, such as large surface area, easy functionalization, and good biocompatibility, and hence they are generally applied to load enzymes, ONDs, dyes, and other proteins.2,37–39AuNPs are widely used because they can form stable Au–S bonds with the functional mercapto groups to form functionalized AuNPs when using functional groups containing these immobilized ligands or other parts of the ligand, such as nucleic acids and proteins.4,7,40–45 For example, Kavosi et al. developed an impedimetric miRNA assay based on DNAzyme tag-initiated deposition of an insulating film on a gold electrode, as shown in Fig. 3A and B. Here, AuNPs were used as the carrier for the DNAzymes and reporter DNA. An increase in the loading of the DNAzyme on the electrode by using the reDNA–AuNP conjugates, combined with the enzymatic reactions and cumulative nature of the protocol significantly enhanced the sensitivity of the sensor, and lowered the detection limit to subfemtomolar levels.46
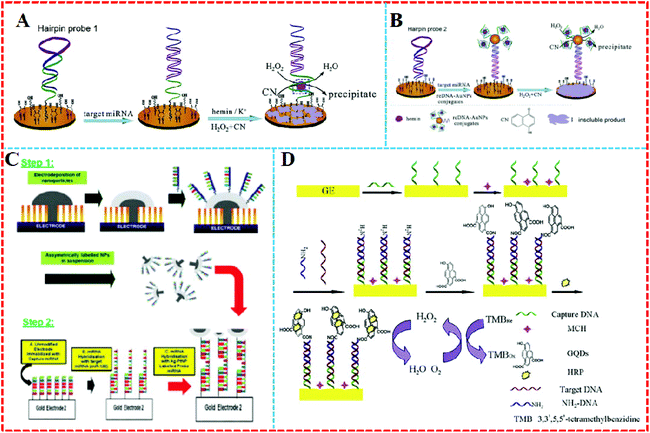 | ||
| Fig. 3 (A) Single DNAzyme signal amplification strategy and (B) multiple-DNAzyme signal amplification strategy. Reproduced with permission from ref. 46, Copyright 2015, Elsevier B.V. (C) Hemispherical platinum: silver core: shell nanoparticles formation and regioselective miRNA functionalization. Reproduced with permission from ref. 47, Copyright 2017, The Royal Society of Chemistry. (D) Principle of the enzyme catalytic amplification of miRNA-155 detection with a graphene quantum dot-based electrochemical biosensor. Reproduced with permission from ref. 52, Copyright 2015, Elsevier B.V. | ||
Except for AuNPs, other noble metal nanoparticles have also been used in miRNA electrochemical sensors as carriers. For example, a layer of silver was modified on the surface of shaped platinum nanoparticles (PtNPs) to create a hemispherical platinum silver core shell nanoparticle (Fig. 3C). The upper surface of silver was decorated with thiolated probe strand miRNA and the device realized linear faradaic current detection from 1 aM to 1 μM without the need for chemical amplification of the target.47
Graphene quantum dots (GQDs) are single or few-layer GR with a tiny size of only several nanometers.48 They possess many fantastic properties, such as chemical stability, excellent water solubility, and suitability.49 GQDs also have abundant carboxyl groups, in smaller volumes than GR, as well as great electrical conductivity.50 Most importantly, GQDs’ carboxylic acid moieties make it easy to functionalize them with various organic, polymeric, inorganic, or biological species.51 For example, Wang's group proposed a quantitative assay for miRNA based on loading horseradish peroxidase (HRP) on GQDs (Fig. 3D).52 As the GQDs have a large surface-to-volume ratio and excellent compatibility, they could combine a large amount of HRP by a noncovalent assembly to trigger high electrochemical reduction signals.53 The system was able to detect miRNA-155 from 1 fM to 100 pM in human serum.
2.3. Nanomaterials as catalysts
Compared with enzymes, nanocatalysts have plentiful active sites on their surface, which can aid them to produce a large number of amplified signals.54 Therefore, nanocatalysts can be used in biosensors to solve some of the remaining problems related to the thermal and environmental instability inherent in biological materials. Nanomaterials as nanozyme-assisted signal amplification strategies have been widely used recently in biomolecular detection. The association of the nanoparticle enzyme to mimic miRNA resulted in the formation of a catalytic system on the biosensor surface, which catalyzed the polymerization.As is well known, NPs have the ability to catalyze the decomposition of hydrogen peroxide, and AgNPs are no exception. However compared to silver nanoclusters (Ag-NCs), the catalytic ability of AgNPs is slightly inferior. Recently, Zhang's group55 fabricated a functional OND probe with both a recognition sequence for hybridization and a template sequence for the in situ synthesis of Ag-NCs, as shown in Fig. 4A. Using the OND-encapsulated Ag-NCs as an effective electrochemical label, the MB probe was immobilized on the surface of gold electrodes. The MB probe subsequently hybridized with the target and functional probe. Then the OND-encapsulated Ag-NCs were tied to the electrode surface and produced a detection signal in response to H2O2 reduction. This electrochemical miRNA biosensing strategy achieved 67 fM with a linear range of 5 orders of magnitude. Similarly, a layer of silver was modified on the surface of platinum nanoclusters to produce hemispherical platinum silver core shell nanoparticles (Pt–AgNPs), which were subsequently used for an miRNA electrochemical biosensor.47
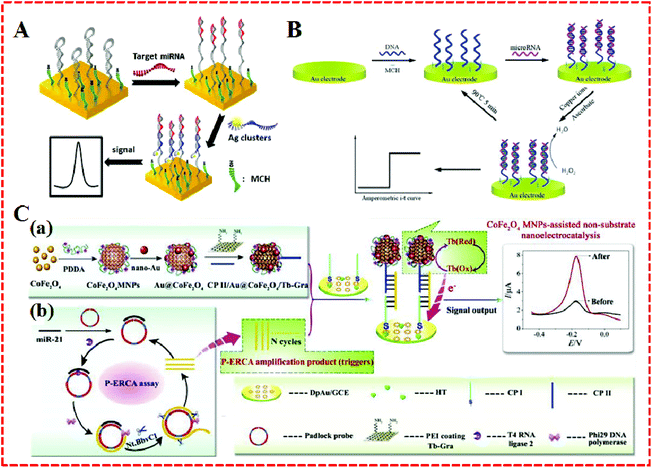 | ||
| Fig. 4 (A) Illustration of the electrochemical detection of MiRNA using oligonucleotide-encapsulated Ag-NCs. Reproduced with permission from ref. 55, Copyright 2012, American Chemical Society. (B) Principle of a reusable miRNA biosensor based on the electrocatalytic property of DNA–miRNA heteroduplexes-templated CuNCs. Reproduced with permission from ref. 56, Copyright 2015, The Royal Society of Chemistry. (C) Schematic illustration of a P–ERCA assay and CoFe2O4 MNPs-assisted no-substrate nanoelectrocatalysis for miR-21 detection. (a) Fabrication of a CP II/Au@CoFe2O4/Tb–Gra probe (CP II bioconjugate) and (b) P–ERCA reaction. Reproduced with permission from ref. 58, Copyright 2017, Elsevier B.V. | ||
Similar to AgNPs, metallic copper nanoclusters (CuNCs) have also been used for miRNA detection. Simply running several rounds of CuNCs formation, a repeated detection of miRNA was further realized, which is important in miRNA biosensors. Here, the CuNCs served as templates to fabricate a structure comprising a recycled miRNA biosensor (Fig. 4B).56 The CuNCs were developed by taking DNA–RNA heteroduplexes as templates. With the help of ascorbic acid (Vc) and Cu2+, the CuNCs could be synthesized on the electrode surface. They could then catalyze H2O2 reduction, leading to steady and amplified electrochemical signals.
In recent years, metal oxide nanoparticles have been found to provide a highly conductive oxide and resistant material toward harsh conditions together with strong catalytic activity, which makes such metal oxides as good candidates for applications in electrochemical sensing.10 A sensitive electrochemical microRNA biosensor based on a ruthenium oxide’ combination was established by Peng et al.57 They applied the ruthenium oxide nanoparticles (RuO2 NPs)-initiated polymerization of 3,3′-dimethoxybenzidine (DB) and miRNA-templated deposition of an insulating poly (3,3′-dimethoxybenzidine) (PDB) film in an miRNA biosensor. The ruthenium oxide was modified as a catalyst at the end of the target miRNA, decorating the target capture probe miRNA, and then the capture probe was hybridized. The RuO2 NPs provided effective catalytic ability toward the polymerization of DB, while the hybridized miRNA strands and free capture probe strands guided the deposition of PDB. This biosensor afforded an excellent possibility as a low-to-medium density electrical sensor array in miRNA expression profiling.
The current nanotechnology enables many metal ions to obtain artificial enzymatic potency as magnetic nanoparticles. Because of the merits of better magnetic separation, nontoxicity, and easy synthesis compared to conventional artificial enzymes, CoFe2O4 magnetic nanoparticles (CoFe2O4 MNPs) are particularly special. They have not only displayed an intrinsic peroxidase-like activity with H2O2 substrates, but have also served as nanoelectrocatalysts (e.g., thionine, methylene blue) for signal amplification even in the absence of H2O2.58 For example, Yu et al. took advantages of CoFe2O4 MNPs to construct an electrochemical biosensor for miR-21 detection, as shown in Fig. 4C. A nanocatalyst and redox molecule (Tb) were co-immobilized onto the GR surface to form a Au@CoFe2O4/Tb–GR composite. It exhibited high performance Tb catalysis was based on the reduced interaction distance between CoFe2O4 MNPs and Tb. The Au@CoFe2O4/Tb–GR not only narrowed the interaction distance between the catalyst (CoFe2O4 MNPs) and Tb, but also averted the need to add any substrate in the test solution, which showed good stability and sensitivity as well as accuracy for miRNA electrochemical detection.
2.4. Nanomaterials as tracers based on their direct electrochemistry
Some nanoparticles, such as AuNPs, QDS, and nanomaterials–enzyme complexes, possess special properties whereby amplified signals come into being due to their own redox reactions. They can act as tracers in the amplification process of electrochemical signals,59 which contributes to the generation of high sensitivity, facile, cost-effective, and good compatibility electrochemical approaches that have found widespread use in the electrochemical detection of miRNA.As a universal tracer, AuNPs have been popularly employed in the construction of miRNA sensors. For example, a sensitive electrochemical sensor was applied to detect miRNAs based on the triple signal amplification of AuNPs, alkaline phosphatase (ALP), and p-aminophenol redox cycling (Fig. 5A).60 The strategy was developed based on the difference in the structures of RNA and DNA. First, the pre-immobilized DNA probes that were modified on the gold electrode captured the target miRNA with the cis-diol group of ribose sugar at the end of the miRNAs chain, which attracted 3-aminophenylboronic acid/biotin-modified multifunctional AuNPs by the formation of a boronate ester covalent bond to facilitate the capture of streptavidin-conjugated alkaline phosphatase (SA-ALP) via the biotin–streptavidin (SA) interaction. The biotinylated AuNPs as a tracer completed the amplification of the triple signal. This method took advantage of AuNPs tracking and signal amplification, which were enhanced linearly with the miRNAs concentration over a range of 10 fM–5 pM, with a detection limit of 3 fM realized. Similarly, a label-free method for the detection of miRNAs was developed based on the formation of boronate ester covalent bonds and the dual-amplification of AuNPs, as shown in Fig. 5B and C.61 Here, 4-mercaptophenylboronic acid (MBA) was used to wrap AuNPs to form a compound (MBA–AuNPs) at the same time as other AuNPs were wrapped by electrochemically active dopamine (DA–AuNPs). The sandwich-type system was completed by the specific identification of anti-miRNAs probes on the surface of the gold electrode toward miRNAs, followed by the successive attachment of MBA–AuNPs and DA–AuNPs via the formation of boronate ester covalent bonds. A composite of MBA–AuNPs and DA–AuNPs bound at the end of the probe resulted in a double-amplification of the signal. The proposed strategy not only verified the feasibility and sensitivity but also the detection limit was shown to be equal to (or even lower than) other amplified electrochemical approaches. A recent strategy with Pt/Sn–In2O3 as electrochemical tracer and hairpin capture probe was developed by Zhang's group.62 They doped tin into the structure of indium oxide as a support for Pt, with the system possessing high conductivity. The marriage between Pt and the surface Sn led to a distinguished oxygen reduction reaction electrochemical catalytic ability and stability of the Pt/Sn–In2O3. Using the Pt/Sn–In2O3 hybrids as electrochemical tracer and hairpin capture probe, a sensitive and selective electrochemical miRNA biosensor was developed, as shown in Fig. 5D. This biosensor obtained a linear range from 5 pM to 0.5 fM and the limit of detection was down to 1.92 fM.
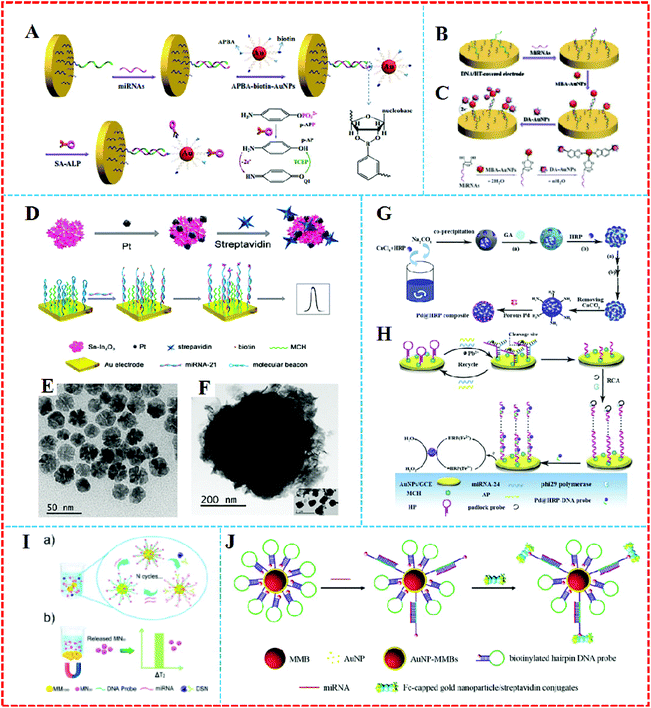 | ||
| Fig. 5 (A) Schematic representation of the label-free detection of miRNAs based on the triple signal amplification of APBA–biotin–AuNPs, SA–ALP, and the p-AP redox-cycling reaction. Reproduced with permission from ref. 60, Copyright 2013, Elsevier B.V. (B) Schematic representation showing the capture of miRNAs by the immobilized DNA probes and the detection of miRNAs by the attachment of MBA–AuNPs and DA–AuNPs and (C) The formation of boronate ester bonds between MBA–AuNPs and miRNAs as well as DA–AuNPs. Reproduced with permission from ref. 61, Copyright 2013, Elsevier B.V. (D) An ultrasensitive and selective electrochemical miRNA biosensor. Reproduced with permission from ref. 62, Copyright 2016, American Chemical Society. (E) TEM images of the porous Pd, (F) TEM images of the HRP spheres, (G) synthesis of Pd@HRP composite and (H) electrochemical miRNA sensing based on Pd@HRP as a signal tag and electrocatalyst coupled with DNAzyme-aided target recycling amplification. Reproduced with permission from ref. 63, Copyright 2016, Elsevier B.V. (I) A quantitative detection of miRNA in one step via next-generation magnetic-relaxation switch-based sensing. Reproduced with permission from ref. 67, Copyright 2016, American Chemical Society. (J) Schematic representation of voltammetric detection of miRNA amplified by AuNP–MMBs and Fc-capped gold nanoparticle/streptavidin conjugates. Reproduced with permission from ref. 68, Copyright 2016, Elsevier B.V. | ||
It is generally believed that rolling circle amplification (RCA) make it highly sensitive. Such formed DNA concatamers are able to bind more signal probes. However, there are potential risks of sample contamination, which can lead to processing complexity and limited efficiency. In order to solve this problem, horseradish peroxidase (HRP) is usually selected as a redox probe. For example, Wu et al. proposed an electrochemical strategy for miRNA detection on the basis of porous palladium-modified HRP spheres (Pd@HRP) (Fig. 5E and F) and a target-induced assembly of DNAzyme as emerging tracing tags (Fig. 5G and H).63 CaCO3 was covalently self-assembled to manufacture the highly loaded HRP sphere tracer, which first served as the sacrificial template that was decorated by porous Pd. The assay reached a low detection of 0.2 fM limit for miRNA analysis with a linear range of 3 fM–1 nM.
2.5. Magnetic nanomaterials as separators and collectors
It has been reported that the emergence of magnetic nanomaterials can solve the issue of needing to construct complex steps and needing the use of expensive instruments for the detection of electrochemical signals. Also, magnetic nanomaterials possess the same excellent performance as other nanoparticles, such as large surface area and good electrical conductivity. Furthermore, magnetic nanomaterials can be easily separated from a solution accompanied by the appearance and removal of magnets.64 Separation and enrichment technologies based on magnetic nanomaterials have become quite efficient methods to purify and enrich biomolecules.65,66 A sensor based on magnetic nanomaterials can respond to the free targets in the liquid phase with a higher efficiency than those immobilized on a solid electrode.Lu et al. designed a quantitative assay for miRNA in one step via a magnetic relaxation switch (Fig. 5I)67 with magnetic microparticle (1 μm in diameter, MM1000)–DNA probemagnetic nanoparticle (30 nm in diameter, MN30) conjugates (MM1000–DNA–MN30). With the help of DSN enzyme, DNA in the formed DNA–RNA heteroduplexes could be specifically cleaved by DSN, leading to the release of MN30, while a variance of the transverse relaxation time was induced by the aggregation or dissociation of magnetic particles in the presence of the target miRNA. The use of magnets could separate different magnetic substances, which afforded quantitative and sensitive detection of miRNA-21 in urine samples and tumor cells without the conversion of miRNA to cDNA or the need for multiple assay steps.
On the other hand, magnetic nanoparticles can also be combined with AuNPs to obtain a large amplified signal. For example, Wang's group68 constructed a voltammetric sensor based on AuNPs-coated magnetic microbeads (AuNPs–MMBs) (Fig. 5J). With AuNPs over the surface of the magnetic microbeads, a higher loading density of hairpin DNA sequences was realized. The hybridization complex of target miRNA with the loop region led to the opening of the hairpin DNA probe, and then a novel assembly was formed via linking biotinylated hairpin DNA probe-covered AuNPs–MMBs with ferrocene-capped AuNPs/SA conjugates. The AuNPs–MMBs were collected on the surface of magnetic electrode to obtain the oxidation peak current in voltammetric detection in the presence of the target miRNAs. In their work, the employment of AuNPs–MMBs not only served as collectors to separate miRNAs conveniently but they also enhanced the conductivity of the magnetic electrode. The magnetic complex electrochemical sensor brought down the detection sensitivity limit to 0.14 fM. Cai et al. fabricated an electrochemical biosensor based on the PO43−-induced in situ formation of PMo12O403−as an electronic mediator for miRNA-155 detection.69 AuNPs-modified polystyrene magnetic microspheres (PSC@AuNPs) were used for immobilizing ALP and SA to form ALP–PSC@AuNPs–SA bioconjugates as a signal tracing probe. Compared to traditional electrochemical methods, this approach utilized the byproduct of the ALP-catalyzed reaction to in situ generate an electronic mediator for the direct quantitative analysis of targets, avoiding the need for fussy labeling steps and complicated operational processes.
3. Optical-based assays for miRNA
Due to their many advantages, such as simplicity of operation, convenience, and sensitivity, optical assays have also been used in the detection of miRNA. Recently, nanomaterials, such as AuNPs and GO, have been applied in the construction of optical sensors.70–72 These assays mainly consist of four types: fluorescent, colorimetric, chemiluminescent, and surface plasmon resonance (Table 1).| Sensor type | MiRNA | Nanomaterials | LOD | Highlights | Ref. |
|---|---|---|---|---|---|
| Fluorescence | MiRNA-205 | AuNPs | 3.8 pM | Label-free; AuNPs quenching-based competition assay system | 11 |
| RLS | MiRNA-21 | AuNPs | 2.21 pM | Analyte-induced aggregation of AuNPs | 74 |
| ERET | MiRNA-15a | CdTe/AuNCs | 21.7 fM | The quenching mechanism between CdTe/AuNCs is verified | 75 |
| Colorimetric | MiRNA-122 | AuNPs | ∼16 pM | DSN-assisted signal amplification; AuNPs aggregation | 77 |
| Colorimetric | MiRNA-155; miRNA-196a; miRNA-210 | AuNPs/polyA | 10 pM | Fast pH-assisted assembly method; multicolor detection | 78 |
| Fluorescence | MiRNA-21 | GO | 160 fM | DSN-induced target recycling | 82 |
| Fluorescence | MiRNA-126 | GO | ∼3.0 fM | Fluorescence recovery of the FAM-labeled probe | 83 |
| Fluorescence | MiRNA-16; miRNA-21; miRNA-26a | GO | 0.17 fM | Multiple miRNAs detection methods; ISDPR and GO fluorescence quenching | 84 |
| Fluorescence | MiRNA-21 | QDs | 37 fM | Double quenching; CHA amplification | 87 |
| Fluorescence | MiRNA-20a | Ag2S QDs | 12 fM | Without need to employ time-consuming radioactive labeling or complex amplification strategies | 88 |
| RLS | MiRNA-122 | CdTe QDs | 9.4 pM | CdTe QDs combined with RLS technology | 89 |
| Photoelectro-chemical | MiRNA-155 | CH3NH3PbI3 QDs | 0.005 fM | CH3NH3PbI3 quantum dots were first used for biosensing | 91 |
| Fluorescence | MiRNA-141 | AgNCs | 2 aM | Simultaneous quantitative analysis of multiple miRNAs in tissues or cells | 105 |
| Colorimetric | MiRNA-21 | GR/AuNPs | 3.2 nM | Spontaneous absorption of ssPNA-21 on GR/AuNPs surfaces | 113 |
| SPR | MiRNA-16 | GNRs | 0.045 pM | Streptavidin-functionalized gold nanorods as a tag | 114 |
3.1. Au nanomaterial-based optical sensing
AuNPs possess a distinctive phenomenon termed as surface plasmon resonance, in which any change/alteration in the size, shape, or geometry of particles alters the local electron confinement, which is thereby reflected in the absorption maxima and color of a colloidal solution.73 Fluorescence analysis with nanomaterials as quenchers has been developed. For example, a AuNPs quenching-based competition assay was proposed by Cheng's team.11 With the help of their conspicuous plasmonic and biocompatible properties, AuNPs could be reliably and readily aggregated with various biomolecules and targeting agents. Thanks to the establishment of synthesis techniques, size and shape-tunable AuNPs can now be easily achieved. Furthermore, the advantage of high efficiency over a wide range of wavelengths permitted them to quench several fluorophores. In the presence of high concentrations of miRNAs, AuNPs quenching-based competition assay have generated a low limit of detection of 3.8 pM and a detection range ranging from 3.8 pM to 10 nM.On the other hand, AuNPs can not only serve as a fluorescence quencher but also could trigger a total resonance light enhancement. For example, a resonance light scattering (RLS) sensor for miRNA based on the analyte-induced aggregation of AuNPs was constructed.74,114 The sensor was fabricated by specifically modifying AuNPs with two strands of cDNA sequence, to which target miRNAs could be complemented to the sequence. When target miRNA was added, the cDNAs could complement the target miRNAs, thereby forming a cDNA/RNA heteroduplex. The construction of such a cDNA/RNA heteroduplex resulted in the aggregation of AuNPs, therefore promoting RLS enhancement.7 Owing to the electronic coupling between the localized plasmon of the AuNPs and the surface plasmon wave, as well as the enhancement of the refractive index of the medium over the Au film induced by DNA supersandwich structure, the SPR response was significantly enhanced.
Another optical sensor was constructed based on CdTe nanocrystals and Au nanoclusters (AuNCs) by Cheng's group.75 With the assistance of ligase, a distance-dependent electrochemiluminescence resonance energy transfer (ERET) system was developed for the highly selective detection of miRNA. Owing to its various advantages, such as its high sensitivity and wide dynamic concentration response range as well as allowing potential and spatial control in particular, electrochemiluminescence (ECL) has been great potential for application in the determination of miRNA.76 For example, AuNCs-functionalized hairpin DNA was synthesized via Au–S chemistry. The hairpin DNA–AuNCs composite could react with carboxylated–CdTe nanocrystals through an amide reaction on a glassy carbon electrode. The ECL quenching of CdTe nanocrystals arose by the strong interaction between CdTe nanocrystals and AuNCs. With the help of assistant DNA and miRNA, the ligase could selectively ligate both of them on the strand of the hairpin DNA to fabricate a long DNA–RNA complex, which largely enhanced the ECL signal owing to an inhibition of the ERET process.
Colorimetric strategies offer some advantages too, such as being simple to use, rapid, cost-effective, and highly selective. Recently, AuNPs were used in the colorimetric detection of miRNA. Wang et al. proposed a simple system for specific sequence miRNA analysis.77 Their colorimetric approach was designed based on DSN-assisted signal amplification and AuNPs. For high extinction coefficients and strong distance-dependent optical properties, they described a probe complex transit for two partly complementary DNA probes, which could prevent the hydrolysis of DNA enzyme, which afterwards was disintegrated by the invasion of target miRNA. A DNA–RNA heteroduplex was formed by the strand one of the probe complex becoming hybridized and becoming the active matrix for the DSN enzyme. The other strand served as the linker for single stranded DNA-modified AuNPs to trigger AuNPs aggregation, resulting in a concomitant color change from red to blue. This assay allowed the quantitative detection of miR-122 in the target scope from 20 pM to 1 nM with a detection limit of ∼16 pM. Wang et al. constructed multicolor nanoprobes based on AuNPs and polyadenine (polyA)-mediated nanoscale molecular beacon (MB) probes and a hairpin block for the detection of multiple miRNAs.78
3.2. Graphene-based optical sensing
GO and GR have attracted great interests in the biosensors field due to their brilliant physical and chemical properties.79–81 The fluorescence quenching of GO and GR is one of the most significant and classical properties of GO and GR, and is consequently widespread utilized in optical amplified signal sensing. In particular, the extraordinary and distance-dependent fluorescence quenching characteristic of GO and GR, whether in a matrix or a carrier, has been employed to elaborately design sensors to detect miRNA.Guo's group developed an assay for miRNA based on the fluorescence quenching of GO and on DSN-induced target cycling.82 Here, GO acted as a carrier. Fluorophore–labeled DNA strands as probes were cleaved by DSN and absorbed on the surface of GO by the interaction between the fluorophore–labeled DNA sequence and GO, resulting in amplified signal fluorescence quenching. Another experiment involved using GO as a carrier, where it participated in fluorescence quenching, as reported by Tu et al.83 The strategy was established by combining the fluorescence quenching of GO with the site-specific cleavage of an endonuclease to enhance selectivity. The single-stranded probe carried a binding region that took charge of the interaction with GO, which triggered fluorescence quenching of the 5′-terminus-labeled fluorophore, with a sensing area that could specifically recognize the target and hybridize with it. Due to the cleavage of RsaI endonuclease, the duplex was released from the GO surface, leading to fluorescence recovery. The detection limit reached ∼3.0 fM for miR-126. In another aspect, GO could also serve as a fluorescent quenching substrate besides acting as a carrier.84 With the assistance of the extraordinary fluorescence quenching of a GO substrate on multiple organic dyes, the capability to discriminate ssDNA and double-stranded nucleic acid structure permitted such a system to simultaneously and selectively determinate several miRNAs. A colorimetric miRNA sensor for microRNA-21 based on GR/AuNPs hybrid polymer was reported by Zhao's group.14 Here, the hybridization of GR and AuNPs was first carried out. Then, the spontaneous absorption of single-stranded PNA-21 (ssPNA-21) on GR/AuNPs hybrid surfaces caused the peroxidase-like catalytic activity of hybrids to be almost completely deactivated via p–p stacking interactions between ssPNA and GR, resulting in no color change. Afterward, the PNA/DNA duplex was released from the hybrid by the decrease in the exposed base groups, such that the catalytic ability of the GR/AuNPs hybrids could be recovered with a concomitant colorless-to-blue color change.
3.3. Quantum dot-based optical sensing
QDs have become powerful tools in the area of biological imaging, sensing, and diagnostics due to their unique optical properties, such as size-tunable emission, broad absorption, narrow and symmetric photoluminescence spectra, strong luminescence, and robust photostability.85,86 Many sensors have been developed based on QDs, such as fluorescence quenching,87 fluorescence emission,88 RLS,89 and fluorescence resonance energy transfer (FRET) sensors.90 QDs-based sensors have also been applied in the determination of miRNA.91For example, target-triggered DNA was nanoassembly-integrated on QDs and then associated with the DNAzyme-modulated double quenching of QDs.87 In the work, the target miRNA induced catalytic hairpin assembly (CHA) amplification, simultaneously accelerating a highly efficient DNA nanoassembly on the surface of QDs, which shortened the distance among numerous G-quadruplexes and the QDs. Then, the luminescence of QDs was quenched via photoinduced electron transfer by hemin related to the particles and the electron acceptor of O2, which was in situ produced with the horseradish peroxidase-mimicked G-quadruplex/hemin DNAzymes toward H2O2. This strategy possessed a brilliant signal amplification ability. The detection limit reached 37 fM with a wide linear detection range of 1 × 10−13 to 1.0 × 10−8 M.
In addition, QDs have also been used in fluorescence-enhancement assays. Near-infrared (NIR) QDs show high biocompatibility, minimal autofluorescence, negligible tissue scattering in NIR region, and bright photoluminescence with an exceptionally large Stokes shift.92 Miao et al.88 designed a DNA logic gate platform based on Ag2S QDs for multiplex low abundant miRNAs detection, utilizing the principle of enzyme-free toehold exchange-mediated strand displacements. Owing to its wide band gap, the light absorption of ZnO was restricted to a small spectral region. However, its light harvesting ability in the near-infrared region needed to be expanded. For this aim, some methods have been adopted to decrease its band gap. Among them, CH3NH3PbI3/QDs has a suitable band gap energy for sunlight absorption to sensitize ZnO to enhance its light-harvesting efficiency.93,94 In Du's work, CH3NH3PbI3 QDs-functionalized ZnO nanosheets were utilized as a light harvester to fabricate a photoelectrochemical (PEC) aptasensor for the determination of miRNA-155.91 The PEC property was obviously improved by the ZnO@CH3NH3PbI3 signal generator, while the incorporation of ZnO nanosheets and CH3NH3PbI3 QDs depressed h+/e− recombination effectively, which improved the charge separation efficiency and charge transfer ability. The proposed aptasensor achieved a low detection limit and broad detection range.
RLS has attracted considerable interest because of its high sensitivity, low cost, stable signals, and simple instrumentation.95,96 Lv et al. reported a QD-based RLS method based on the principle of the analyte-induced aggregation of nanoparticles.89 Different probe sequences P1 and P2 were assembled on the surface of QDs to form functional QDs–P1 and QDs–P2 conjugates (QDs–P). In the lack of miRNA-122, the QDs–P stably coexisted in the solution, leading to a low RLS signal intensity. On the contrary, QDs–P conjugated with miRNA-122 formed a QDs–P–miRNA-122 complex, which enhanced the RLS intensity. The RLS intensity was directly proportional to the concentration of the proportionate aggregation of QDs to the miRNA-122 concentration.
Recently, carbon quantum dots (CQDs) have been attracting considerable attention owing to several favorable attributes they possess, including small size, chemical inertness, low toxicity, size-dependent luminescence emission, and water solubility, especially in the fields of sensors and bioimaging.97,98 For example, Khakbaz et al. synthesized CQDs through a one-step strategy.90 In FRET-based detection, a dye quencher-labeled single-strand DNA for use as a sensing element was first assembled on the CQDs by p–p interaction, which caused a quenching of the fluorescence of CQDs. The existence of miRNA 9-1 triggered the formation of a duplex helix, resulting in releasing double-strand DNA/miRNA from the surface of CQDs, and the fluorescence intensity was thus restored. Except for this fluorescence turn-on strategy, Liu et al. designed an ECL biosensor with DNA-functionalized N–CQDs as signal enhancers for the ultrasensitive detection of miRNA-21.99
3.4. Ag nanomaterial-based optical detection
Recently, there has been an explosion of interest in optical sensors based on fluorescent silver nanomaterials synthesis, which has achieved a wide range of applications particularly in the area of bioassays,100 which typically exhibit outstanding spectral, photophysical properties, high quantum yields, and good biocompatibility.As early as 1998, DNA-scaffolded silver nanoclusters (DNA/AgNCs) were first reported by Braun et al.101 DNA-based AgNCs have admirable performance, such as high brightness, good photostability, and good affordability. Recently, AgNCs were also used for miRNA detection. Shah et al. reported a strategy based on Ag nanoclusters (AgNCs) by adopting DNA/RNA chimera templates for AgNC-based probes.102 A AgNCs sensor was fabricated with a RNA backbone and DNA/RNA chimeras in templates to launch AgNCs encapsulation and efficient target recognition. This sensor showed high emissive intensity and improved sensitivity toward target miRNA when the target sensing part of two non-functional DNA/AgNCs sensors was substituted with an RNA sequence to make DNA/RNA chimera templates. A sensor based on the AgNCs sweeping a target range found that not all the systematically designed DNA/AgNC sensors could efficiently detect miRNAs.
In Shah's recent experiment, bright red emissive AgNPs were applied to detect microRNA.103 Shah's group aimed to construct a DNA sequence (DNA-12nt-RED-160) probe to detect the formed bright red fluorescence within 1 h after the addition of AgNO3 and reduction with a NaBH4 target RNA-miR160 sequence. In the modified sequence, mismatched self-dimers generated bright red emitting AgNCs, which showed a higher emission compared to DNA-12nt-RED-160 at their maximum excitation. In the meanwhile, the thermal stability of the secondary DNA structures as well as the observed AgNC red emission intensity could not be ignored. This research achievement opened up a new research area, where diverse AgNC-based DNA probes sensors could be used for the specific detection of plant and animal miRNAs.
Similar to AuNPs, AgNPs have been used as quenchers in the fabrication of an miRNA sensor.104 Zhang et al. applied OND-encapusulated AgNPs in the ECL system of CdS nanocrystals (NCs)/K2S2O8 on account of the dual ECL quenching effects. The ECL emission of CdS NCs served as the energy acceptor of the CdS NCs ECL and matched perfectly with the absorption band of OND-encapsulated AgNCs to trigger an effective ERET. Many atoms with sizes below 2 nm that have size-dependent discrete energy levels formed the nanoclusters, which was the reason why the AgNPs also showed “molecular” properties, such as catalytic activity. Based on these, the ECL emissions from CdS NCs by ECL RET was quenched by the Ag NCs as well as catalyzing the electro-reduction reaction of K2S2O8, leading to an obvious decrease in ECL intensity. A wide linear range and acceptable selectivity were achieved.
Recently, a strategy was constructed for the determination of miRNA using target-assisted isothermal exponential amplification coupled with fluorescent DNA-scaffolded AgNCs.105 Unimolecular DNA containing three functional domains served as the amplification template, namely as polymerases. Nicking enzymes were utilized as mechanical activators and target miRNA represented the trigger to enable the conversion of miRNA to abundant reporter ONDs within minutes. The method established the superior selectivity to quantify miRNA concentration and was able to distinguish the differences among miRNA family members.
3.5. Cu nanoparticle-based optical sensing
Functional copper NPs (CuNPs) were also reported for miRNA sensors. For example, Mokhir et al. developed a sensor based on random double-stranded DNA-templated/CuNPs (dsDNA–CuNPs).106 Xu et al. prepared poly (thymine)-templated CuNPs, and used dsDNA–CuNPs as the model to fabricate the RCR-mediated concatemeric CuNPs strategy.107 A primer induced the RCR process with a continual replication of the circular template, leading to a short OND primer, which was extended to long concatemeric ssDNA with periodically repeated complementary parts of regions R and H. The fluorescence signal was detected as reserved at ∼60% at 2.5 h after formation, revealing a ∼2 times enhanced stability. In addition, the sensitivity of the concatemeric dsDNA–CuNPs sensor was improved with a ∼10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold amplification. Despite these studies showing good results, the exploration of CuNPs in biochemical applications is still at its early stage.
000-fold amplification. Despite these studies showing good results, the exploration of CuNPs in biochemical applications is still at its early stage.
3.6. Carbon nanoparticle-based optical sensing
Carbon nanoparticles (CNPs) have been intensively explored and applied to the field of biosensing due to their remarkable characteristics, such as great chemical activity and biocompatibility, high stability and water dispersibility, and convenient and green preparation.108,109 Such carbon atoms usually have a diameter of 25–40 nm. CNPs can closely absorb ssDNA but not dsDNA, which generates the fluorescence quenching. Wang et al. described an assay through the introduction of microRNA complementary to the DNA probe. A double-stranded DNA/miRNA hybrid could be formed and released from the surface of CNPs, leading to the fluorescence recovery.110 Ju's team rationally applied CNPs to the construction of an intracellular miRNA sensor, whereby CNPs were used as a nanocarrier for the introduction of gene probes into cells.111 CNPs came into play through a fluorescence quenching effect, leading to a dye-labeled ssDNA assembled on the surface of them.112 A functionalized carbon nanosphere probe was designed to quantitatively detect special target cells and to monitor the miRNA change intracellularly.ISDPR: isothermal strand-displacement polymerase reaction; LOD: limit of detection; GNRs: gold nanorods.
4. Conclusion and perspective
The biological role of miRNAs as cancer-prevention genes has raised good prospects in cancer diagnosis, prognosis, and treatment, which has led to the sustained increasing demand for miRNA profiling. In the last few years for the detection of miRNAs, a wide variety of strategies have been applied to detect miRNA. Among these, nanomaterial-based biosensors continue to be the engine for the development of electrochemical and optical strategies on account of their inherent sensitivity, selectivity, capacity, simplicity, speed, and cost benefits compared to conventional detection strategies.High sensitivity, high selectivity, and the capacity of in situ measurement can be afforded by the use of optical methods. However, there are some limitations of such systems too, such as the need for costly equipment and their non-portability. On the other hand, electrochemical strategies also exhibit high sensitivity, high specificity, and a low detection limit for miRNA determination without enzymatic amplification. Usually nanomaterials serve as the sensing matrix, probe carriers, the catalyst for the reaction, tracer, magnetic separators, and collectors and have supported the construction of a series of miRNA amplification sensing strategies in electrochemical assays. In the area of optical sensing, fluorescence quenching, fluorescence enhancement, and other properties of nanomaterials are well utilized in the construction of optical sensors. What is worth mentioning is that two or more types of nanomaterials are usually used to fabricate multiple amplification sensors as this can vastly improve the analytical performance as well as address the problems of single nanomaterial-based sensing amplification unable to handle certain materials properly.
Some nanomaterial-based miRNA detection methods are superior to the more traditional methods in terms of their simplicity, speed, cost, and option for automation. However, there are significant challenges in the applications of nanomaterials in miRNA-based electrochemical sensing related to a number of factors: (1) due to the high surface area of nanomaterials, methods are needed for eliminating the non-specific adsorption of co-existing species on the nanomaterials in the complex matrix; (2) the controllable synthesis of nanomaterials in different batches; (3) the practical application of nanomaterials. Clinical samples are complex mixtures containing large amounts of components and biological species. The ultimate goal of all biosensing devices should be oriented toward the end user. For clinical applications, research on nanomaterial-based miRNA biosensor development should also focus on their portability, long-term stability, and compatibility. Nevertheless, we expect the research on the nanomaterial-based miRNA sensing will remain a hot topic. These sensors will be further improved to solve problems related to real sample applications and to meet society's needs and the market demands.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21475115), Henan Provincial Science and Technology Innovation Team (C20150026), Graduate Scientific Research Innovation Fund of Xinyang Normal University (No. 2017KYJJ10) and Nanhu Scholars Program of XYNU.References
- M. Zouari, S. Campuzano, J. M. Pingarrón and N. Raouafi, Competitive RNA-RNA hybridization-based integrated nanostructured-disposable electrode for highly sensitive determination of miRNAs in cancer cells, Biosens. Bioelectron., 2017, 91, 40–45 CrossRef CAS.
- Y. X. Chen, K. J. Huang and K. X. Niu, Recent advances in signal amplification strategy based on oligonucleotide and nanomaterials for microRNA detection-a review, Biosens. Bioelectron., 2018, 99, 612–624 CrossRef CAS.
- H. L. Shuai, K. J. Huang, W. J. Zhang, X. Cao and M. P. Jia, Sandwich-type microRNA biosensor based on magnesium oxide nanoflower and graphene oxide-gold nanoparticles hybrids coupling with enzyme signal amplification, Sens. Actuators, B, 2017, 243, 403–411 CrossRef CAS.
- Y. H. Wang, K. J. Huang and X. Wu, Recent advances in transition-metal dichalcogenides based electrochemical biosensors: A review, Biosens. Bioelectron., 2017, 97, 305–316 CrossRef CAS; Y. H. Wang, K. J. Huang and X. Wu, Recent advances in transition-metal dichalcogenides based electrochemical biosensors: A review, Biosens. Bioelectron., 2017, 97, 305–316 CrossRef.
- M. Pu, J. Chen, Z. Tao, L. Miao, X. Qi, Y. Wang and J. Ren, Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression, Cell. Mol. Life Sci., 2019, 76, 1–11 CrossRef.
- W. Yang, Z. Lu, Z. Zhi, L. Liu, L. Deng, X. Jiang and L. Pang, High-throughput transcriptome-Seq and small RNA-Seq reveal novel functional genes and microRNAs for early embryonic arrest in humans, Gene, 2019, 697, 19–25 CrossRef CAS.
- R. Liu, Q. Wang, Q. Li, X. Yang, K. Wang and W. Nie, Surface plasmon resonance biosensor for sensitive detection of microRNA and cancer cell using multiple signal amplification strategy, Biosens. Bioelectron., 2017, 87, 433–438 CrossRef CAS.
- K. A. Pillman, G. J. Goodall, C. P. Bracken and M. P. Gantier, miRNA length variation during macrophage stimulation confounds the interpretation of results: implications for miRNA quantification by RT-qPCR, RNA, 2019, 25, 232–238 CrossRef CAS.
- A. A. Jamali, M. Pourhassan-Moghaddam, J. E. N. Dolatabadi and Y. Omidi, Nanomaterials on the road to microRNA detection with optical and electrochemical nanobiosensors, TrAC, Trends Anal. Chem., 2014, 55, 24–42 CrossRef CAS.
- H. L. Shuai, K. J. Huang and Y. X. Chen, A layered tungsten disulfide/acetylene black composite based DNA biosensing platform coupled with hybridization chain reaction for signal amplification, J. Mater. Chem. B, 2016, 4, 1186–1196 RSC.
- W. Wang, T. Kong, D. Zhang, J. Zhang and G. Cheng, Label-free microRNA detection based on fluorescence quenching of gold nanoparticles with a competitive hybridization, Anal. Chem., 2015, 87, 10822–10829 CrossRef CAS.
- K. J. Huang, H. L. Shuai and Y. X. Chen, Layered molybdenum selenide stacking flower-like nanostructure coupled with guanine-rich DNA sequence for ultrasensitive ochratoxin A aptasensor application, Sens. Actuators, B, 2016, 225, 391–397 CrossRef CAS.
- H. L. Shuai, X. Wu and K. J. Huang, Molybdenum disulfide sphere-based electrochemical aptasensors for protein detection, J. Mater. Chem. B, 2017, 5, 5362–5372 RSC.
- Y. X. Chen, K. J. Huang, F. Lin and L. X. Fang, Ultrasensitive electrochemical sensing platform based on graphene wrapping SnO2 nanocorals and autonomous cascade DNA duplication strategy, Talanta, 2017, 175, 168–176 CrossRef CAS.
- A. E. Vilian, B. Dinesh, S. M. Kang, U. M. Krishnan, Y. S. Huh and Y. S. Han, Recent advances in molybdenum disulfide-based electrode materials for electroanalytical applications, Microchim. Acta, 2019, 186, 203 CrossRef.
- J. Mandli, H. Mohammadi and A. Amine, Electrochemical DNA sandwich biosensor based on enzyme amplified microRNA-21 detection and gold nanoparticles, Bioelectrochemistry, 2017, 116, 17–23 CrossRef CAS.
- M. Azimzadeh, M. Rahaie, N. Nasirizadeh, K. Ashtari and H. Naderi-Manesh, An electrochemical nanobiosensor for plasma miRNA-155, based on graphene oxide and gold nanorod, for early detection of breast cancer, Biosens. Bioelectron., 2016, 77, 99–106 CrossRef CAS.
- M. Wang, H. Yin, Y. Zhou, X. Meng, G. I. Waterhouse and S. Ai, A novel photoelectrochemical biosensor for the sensitive detection of dual microRNAs using molybdenum carbide nanotubes as nanocarriers and energy transfer between CQDs and AuNPs, Chem. Eng. J., 2019, 365, 351–357 CrossRef CAS.
- X. Wu, Y. Chai, R. Yuan, Y. Zhuo and Y. Chen, Dual signal amplification strategy for enzyme-free electrochemical detection of microRNAs, Sens. Actuators, B, 2014, 203, 296–302 CrossRef CAS.
- Y. X. Chen, K. J. Huang, L. L. He and Y. H. Wang, Tetrahedral DNA probe coupling with hybridization chain reaction for competitive thrombin aptasensor, Biosens. Bioelectron., 2018, 100, 274–281 CrossRef CAS.
- L. L. Xing, X. Wu and K. J. Huang, High-performance supercapacitor based on three-dimensional flower-shaped Li4Ti5O12-graphene hybrid and pine needles derived honeycomb carbon, J. Colloid Interface Sci., 2018, 529, 171–179 CrossRef CAS.
- L. L. Xing, K. J. Huang and K. J. Fang, Preparation of layered graphene and tungsten oxide hybrids for enhanced performance supercapacitors, Dalton Trans., 2016, 45, 17439–17446 RSC.
- K. J. Huang, J. Z. Zhang, Y. Liu and Y. M. Liu, Synthesis of reduced graphene oxide wrapped-copper sulfide hollow spheres as electrode material for supercapacitor, Int. J. Hydrogen Energy, 2015, 40, 10158–10167 CrossRef CAS.
- K. J. Huang, J. Z. Zhang and J. L. Cai, Preparation of porous layered molybdenum selenide-graphene composites on Ni foam for high-performance supercapacitor and electrochemical sensing, Electrochim. Acta, 2015, 180, 770–777 CrossRef CAS.
- E. Sun, L. Wang, X. Zhou, C. Ma, Y. Sun, M. Lei and R. Han, Graphene oxide/DNA-decorated electrode for the fabrication of microRNA biosensor, RSC Adv., 2015, 5, 69334–69338 RSC.
- H. V. Tran, B. Piro, S. Reisberg, H. T. Duc and M. C. Pham, Antibodies directed to RNA/DNA hybrids: an electrochemical immunosensor for microRNAs detection using graphene-composite electrodes, Anal. Chem., 2013, 85, 8469–8474 CrossRef CAS.
- H. L. Shuai, K. J. Huang, L. L. Xing and Y. X. Chen, Ultrasensitive electrochemical sensing platform for microRNA based on tungsten oxide-graphene composites coupling with catalyzed hairpin assembly target recycling and enzyme signal amplification, Biosens. Bioelectron., 2016, 86, 337–345 CrossRef CAS.
- Q. Tian, Y. Wang, R. Deng, L. Lin, Y. Liu and J. Li, Carbon nanotube enhanced label-free detection of microRNAs based on hairpin probe triggered solid-phase rolling-circle amplification, Nanoscale, 2015, 7, 987–993 RSC.
- F. Li, J. Peng, Q. Zheng, X. Guo, H. Tang and S. Yao, Carbon nanotube-polyamidoamine dendrimer hybrid-modified electrodes for highly sensitive electrochemical detection of microRNA24, Anal. Chem., 2015, 87, 4806–4813 CrossRef CAS.
- S. Fortunati, A. Rozzi, F. Curti, M. Giannetto, R. Corradini and M. Careri, Single-Walled Carbon Nanotubes as Enhancing Substrates for PNA-Based Amperometric Genosensors, Sensors, 2019, 19, 588 CrossRef.
- H. V. Tran, B. Piro, S. Reisberg, L. H. Nguyen, T. D. Nguyen, H. T. Duc and M. C. Pham, An electrochemical ELISA-like immunosensor for miRNAs detection based on screen-printed gold electrodes modified with reduced graphene oxide and carbon nanotubes, Biosens. Bioelectron., 2014, 62, 25–30 CrossRef CAS.
- F. Li, J. Peng, J. Wang, H. Tang, L. Tan, Q. Xie and S. Yao, Carbon nanotube-based label-free electrochemical biosensor for sensitive detection of miRNA-24, Biosens. Bioelectron., 2014, 54, 158–164 CrossRef CAS.
- Y. H. Wang, K. J. Huang, X. Wu, Y. Y. Ma, D. L. Song, C. Y. Du and S. H. Chang, Ultrasensitive supersandwich-type biosensor for enzyme-free amplified microRNA detection based on N-doped graphene/Au nanoparticles and hemin/G-quadruplexes, J. Mater. Chem. B, 2018, 6, 2134–2142 RSC.
- Y. X. Chen, W. J. Zhang, K. J. Huang, M. Zheng and Y. C. Mao, An electrochemical microRNA sensing platform based on tunsten diselenide nanosheets and competitive RNA-RNA hybridization, Analyst, 2017, 142, 4843–4851 RSC.
- Y. X. Chen, X. Wu and K. J. Huang, A sandwich-type electrochemical biosensing platform for microRNA-21 detection using carbon sphere-MoS2 and catalyzed hairpin assembly for signal amplification, Sens. Actuators, B, 2018, 270, 179–186 CrossRef CAS.
- H. L. Shuai, K. J. Huang, Y. X. Chen, L. X. Fang and M. P. Jia, Au nanoparticles/hollow molybdenum disulfide microcubes based biosensor for microRNA-21 detection coupled with duplex-specific nuclease and enzyme signal amplification, Biosens. Bioelectron., 2017, 89, 989–997 CrossRef CAS.
- J. Lei and H. Ju, Signal amplification using functional nanomaterials for biosensing, Chem. Soc. Rev., 2012, 41, 2122–2134 RSC.
- K. J. Huang, Y. J. Liu, J. Z. Zhang and Y. M. Liu, A sequence-specific DNA electrochemical sensor based on acetylene black incorporated two-dimensional CuS nanosheets and gold nanoparticles, Sens. Actuators, B, 2015, 209, 570–578 CrossRef CAS.
- J. Li, S. Tan, R. Kooger, C. Zhang and Y. Zhang, MicroRNAs as novel biological targets for detection and regulation, Chem. Soc. Rev., 2014, 43, 506–517 RSC.
- K. J. Huang, Y. J. Liu, J. Z. Zhang and Y. M. Liu, A novel aptamer sensor based on layered tungsten disulfide nanosheets and Au nanoparticles amplification for 17β-estradiol detection, Anal. Methods, 2014, 6, 8011–8017 RSC.
- K. J. Huang, Y. J. Liu, H. B. Wang, Y. Y. Wang and Y. M. Liu, Sub-femtomolar DNA detection based on layered molybdenum disulfide/multi-walled carbon nanotube composites, Au nanoparticle and enzyme multiple signal amplification, Biosens. Bioelectron., 2014, 55, 195–202 CrossRef CAS.
- K. J. Huang, Y. J. Liu, J. T. Cao and H. B. Wang, An aptamer electrochemical assay for sensitive detection of immunoglobulin E based on tungsten disulfide-graphene composites and gold nanoparticles, RSC Adv., 2014, 4, 36742–36748 RSC.
- K. J. Huang, Y. J. Liu, G. W. Shi, X. R. Yang and Y. M. Liu, Label-free aptamer sensor for 17β-estradiol based on vanadium disulfide nanoflowers and Au nanoparticles, Sens. Actuators, B, 2014, 201, 579–585 CrossRef CAS.
- K. J. Huang, Y. J. Liu, H. B. Wang, T. Gan, Y. M. Liu and L. L. Wang, Signal amplification for electrochemical DNA biosensor based on two-dimensional graphene analogue tungsten sulfide-graphene composites and gold nanoparticles, Sens. Actuators, B, 2014, 191, 828–836 CrossRef CAS.
- K. J. Huang, J. Z. Zhang, Y. J. Liu and L. L. Wang, Novel electrochemical sensing platform based on molybdenum disulfide nanosheets-polyaniline composites and Au nanoparticles, Sens. Actuators, B, 2014, 194, 303–310 CrossRef CAS.
- J. Wan, X. Liu, Y. Zhang, Q. Gao, H. Qi and C. Zhang, Sensitive impedimetric detection of microRNAs using a hairpin probe based on DNAzyme-functionalized gold nanoparticle tag-initiated deposition of an insulating film on gold electrode, Sens. Actuators, B, 2015, 213, 409–416 CrossRef CAS.
- E. Spain, K. Adamson, M. Elshahawy, I. Bray, T. E. Keyes, R. L. Stallings and R. J. Forster, Hemispherical platinum: silver core: shell nanoparticles for miRNA detection, Analyst, 2017, 142, 752–762 RSC.
- H. Cheng, Y. Zhao, Y. Fan, X. Xie, L. Qu and G. Shi, Graphene-quantum-dot assembled nanotubes: a new platform for efficient Raman enhancement, ACS Nano, 2012, 6, 2237–2244 CrossRef CAS.
- Y. Li, Y. Hu, Y. Zhao, G. Shi, L. Deng, Y. Hou and L. Qu, An electrochemical avenue to green-luminescent graphene quantum dots as potential electron-acceptors for photovoltaics, Adv. Mater., 2011, 23, 776–780 CrossRef CAS.
- X. Wang, X. Sun, H. He, H. Yang, J. Lao, Y. Song and F. Huang, A two-component active targeting theranostic agent based on graphene quantum dots, J. Mater. Chem. B, 2015, 3, 3583–3590 RSC.
- S. N. Baker and G. A. Baker, Luminescent carbon nanodots: emergent nanolights, Angew. Chem., Int. Ed., 2010, 49, 6726–6744 CrossRef CAS.
- T. Hu, L. Zhang, W. Wen, X. Zhang and S. Wang, Enzyme catalytic amplification of miRNA-155 detection with graphene quantum dot-based electrochemical biosensor, Biosens. Bioelectron., 2016, 77, 451–456 CrossRef CAS.
- Y. He, X. Wang, J. Sun, S. Jiao, H. Chen, F. Gao and L. Wang, Fluorescent blood glucose monitor by hemin-functionalized graphene quantum dots based sensing system, Anal. Chim. Acta, 2014, 810, 71–78 CrossRef CAS.
- J. Das, H. Kim, K. Jo, K. H. Park, S. Jon, K. Lee and H. Yang, Fast catalytic and electrocatalytic oxidation of sodium borohydride on palladium nanoparticles and its application to ultrasensitive DNA detection, Chem. Commun., 2009, 42, 6394–6396 RSC.
- H. Dong, S. Jin, H. Ju, K. Hao, L. P. Xu, H. Lu and X. Zhang, Trace and label-free microRNA detection using oligonucleotide encapsulated silver nanoclusters as probes, Anal. Chem., 2012, 84, 8670–8674 CrossRef CAS.
- Z. Wang, L. Si, J. Bao and Z. Dai, A reusable microRNA sensor based on the electrocatalytic property of heteroduplex-templated copper nanoclusters, Chem. Commun., 2015, 51, 6305–6307 RSC.
- Y. Peng and Z. Gao, Amplified detection of microRNA based on ruthenium oxide nanoparticle-initiated deposition of an insulating film, Anal. Chem., 2011, 83, 820–827 CrossRef CAS.
- N. Yu, Z. Wang, C. Wang, J. Han and H. Bu, Combining padlock exponential rolling circle amplification with CoFe2O4 magnetic nanoparticles for microRNA detection by nanoelectrocatalysis without a substrate, Anal. Chim. Acta, 2017, 962, 24–31 CrossRef CAS.
- Y. H. Wang, H. Xia, K. J. Huang, X. Wu, Y. Y. Ma, R. Deng and Z. W. Han, Ultrasensitive determination of thrombin by using an electrode modified with WSe2 and gold nanoparticles, aptamer-thrombin-aptamer sandwiching, redox cycling, and signal enhancement by alkaline phosphatase, Microchim. Acta, 2018, 185, 502 CrossRef.
- L. Liu, N. Xia, H. Liu, X. Kang, X. Liu, C. Xue and X. He, Highly sensitive and label-free electrochemical detection of microRNAs based on triple signal amplification of multifunctional gold nanoparticles, enzymes and redox-cycling reaction, Biosens. Bioelectron., 2014, 53, 399–405 CrossRef CAS.
- N. Xia, L. Zhang, G. Wang, Q. Feng and L. Liu, Labelfree and sensitive strategy for microRNAs detection based on the formation of boronate ester bonds and the dual-amplification of gold nanoparticles, Biosens. Bioelectron., 2013, 47, 461–466 CrossRef CAS.
- K. Zhang, H. Dong, W. Dai, X. Meng, H. Lu, T. Wu and X. Zhang, Fabricating Pt/Sn-In2O3 nanoflower with advanced oxygen reduction reaction performance for high-sensitivity microRNA electrochemical detection, Anal. Chem., 2016, 89, 648–655 CrossRef.
- Y. Wu, K. Sheng, Y. Liu, Q. Yu and B. Ye, Enzyme spheres as novel tracing tags coupled with target-induced DNAzyme assembly for ultrasensitive electrochemical microRNA assay, Anal. Chim. Acta, 2016, 948, 1–8 CrossRef CAS.
- Z. Wu, W. Li, P. A. Webley and D. Zhao, General and controllable synthesis of novel mesoporous magnetic iron oxide@carbon encapsulates for efficient arsenic removal, Adv. Mater., 2012, 24, 485–491 CrossRef CAS.
- A. Abbaspour and A. Noori, Electrochemical detection of individual single nucleotide polymorphisms using monobase-modified apoferritin-encapsulated nanoparticles, Biosens. Bioelectron., 2012, 37, 11–18 CrossRef CAS.
- O. Mykhaylyk, Y. S. Antequera, D. Vlaskou and C. Plank, Generation of magnetic nonviral gene transfer agents and magnetofection in vitro, Nat. Protoc., 2007, 2, 2391–2411 CrossRef CAS.
- W. Lu, Y. Chen, Z. Liu, W. Tang, Q. Feng, J. Sun and X. Jiang, Quantitative detection of microRNA in one step via next generation magnetic relaxation switch sensing, ACS Nano, 2016, 10, 6685–6692 CrossRef CAS.
- Z. Lu, H. Tang, D. Wu, Y. Xia, M. Wu, X. Yi and J. Wang, Amplified voltammetric detection of miRNA from serum samples of glioma patients via combination of conducting magnetic microbeads and ferrocene-capped gold nanoparticle/streptavidin conjugates, Biosens. Bioelectron., 2016, 86, 502–507 CrossRef CAS.
- W. Cai, S. Xie, Y. Tang, Y. Chai, R. Yuan and J. Zhang, A label-free electrochemical biosensor for microRNA detection based on catalytic hairpin assembly and in situ formation of molybdophosphate, Talanta, 2017, 163, 65–71 CrossRef CAS.
- L. Dyadyusha, H. Yin, S. Jaiswal, T. Brown, J. J. Baumberg, F. P. Booy and T. Melvin, Quenching of CdSe quantum dot emission, a new approach for biosensing, Chem. Commun., 2005, 25, 3201–3203 RSC.
- Z. S. Qian, X. Y. Shan, L. J. Chai, J. J. Ma, J. R. Chen and H. Feng, A universal fluorescence sensing strategy based on biocompatible graphene quantum dots and graphene oxide for the detection of DNA, Nanoscale, 2014, 6, 5671–5674 RSC.
- Y. Song, Y. Luo, C. Zhu, H. Li, D. Du and Y. Lin, Recent advances in electrochemical biosensors based on graphene two-dimensional nanomaterials, Biosens. Bioelectron., 2016, 76, 195–212 CrossRef CAS.
- K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Gold nanoparticles in chemical and biological sensing, Chem. Rev., 2012, 112, 2739–2779 CrossRef CAS.
- M. Ren, S. Wang, C. Cai, C. Chen and X. Chen, A simple and sensitive resonance light scattering method based on aggregation of gold nanoparticles for selective detection of microRNA-21, RSC Adv., 2016, 6, 83078–83083 RSC.
- Y. Cheng, J. Lei, Y. Chen and H. Ju, Highly selective detection of microRNA based on distance-dependent electrochemiluminescence resonance energy transfer between CdTe nanocrystals and Au nanoclusters, Biosens. Bioelectron., 2014, 51, 431–436 CrossRef CAS.
- S. Xu, Y. Liu, T. Wang and J. Li, Positive potential operation of a cathodic electrogenerated chemiluminescence immunosensor based on luminol and graphene for cancer biomarker detection, Anal. Chem., 2011, 83, 3817–3823 CrossRef CAS.
- Q. Wang, R. D. Li, B. C. Yin and B. C. Ye, Colorimetric detection of sequence-specific microRNA based on duplex-specific nuclease-assisted nanoparticle amplification, Analyst, 2015, 140, 6306–6312 RSC.
- C. Wang, H. Zhang, D. Zeng, W. Sun, H. Zhang, A. Aldalbahi and X. Mi, Elaborately designed diblock nanoprobes for simultaneous multicolor detection of microRNAs, Nanoscale, 2015, 7, 15822–15829 RSC.
- K. J. Huang, Y. J. Liu, H. B. Wang and Y. Y. Wang, A sensitive electrochemical DNA biosensor based on silver nanoparticles-polydopamine@ graphene composite, Electrochim. Acta, 2014, 118, 130–137 CrossRef CAS.
- K. J. Huang, L. Wang, J. Li, M. Yu and Y. M. Liu, Electrochemical sensing of catechol using a glassy carbon electrode modified with a composite made from silver nanoparticles, polydopamine, and graphene, Microchim. Acta, 2013, 180, 751–757 CrossRef CAS.
- K. J. Huang, L. Wang, J. Li and Y. M. Liu, Electrochemical sensing based on layered MoS2-graphene composites, Sens. Actuators, B, 2013, 178, 671–677 CrossRef CAS.
- S. Guo, F. Yang, Y. Zhang, Y. Ning, Q. Yao and G. J. Zhang, Amplified fluorescence sensing of miRNA by combination of graphene oxide with duplex-specific nuclease, Anal. Methods, 2014, 6, 3598–3603 RSC.
- Y. Tu, W. Li, P. Wu, H. Zhang and C. Cai, Fluorescence quenching of graphene oxide integrating with the site-specific cleavage of the endonuclease for sensitive and selective microRNA detection, Anal. Chem., 2013, 85, 2536–2542 CrossRef CAS.
- H. Dong, J. Zhang, H. Ju, H. Lu, S. Wang, S. Jin and X. Zhang, Highly sensitive multiple microRNA detection based on fluorescence quenching of graphene oxide and isothermal strand-displacement polymerase reaction, Anal. Chem., 2012, 84, 4587–4593 CrossRef CAS.
- A. Zunger, Semiconductor quantum dots, MRS Bull., 1998, 23, 15–17 CrossRef.
- F. X. Redl, K. S. Cho, C. B. Murray and S. O'Brien, Three-dimensional binary superlattices of magnetic nanocrystals and semiconductor quantum dots, Nature, 2003, 423, 968 CrossRef CAS.
- R. Yuan, X. Yu, Y. Zhang, L. Xu, W. Cheng, Z. Tu and S. Ding, Target-triggered DNA nanoassembly on quantum dots and DNAzyme-modulated double quenching for ultrasensitive microRNA biosensing, Biosens. Bioelectron., 2017, 92, 342–348 CrossRef CAS.
- P. Miao, Y. Tang, B. Wang and F. Meng, Near-infrared Ag2S quantum dots-based DNA logic gate platform for miRNA diagnostics, Anal. Chem., 2016, 88, 7567–7573 CrossRef CAS.
- S. Lv, F. Chen, C. Chen, X. Chen, H. Gong and C. Cai, A novel CdTe quantum dots probe amplified resonance light scattering signals to detect microRNA-122, Talanta, 2017, 165, 659–663 CrossRef CAS.
- F. Khakbaz and M. Mahani, Micro-RNA detection based on fluorescence resonance energy transfer of DNA-carbon quantum dots probes, Anal. Biochem., 2017, 523, 32–38 CrossRef CAS.
- X. Pang, J. Qi, Y. Zhang, Y. Ren, M. Su, B. Jia and B. Du, Ultrasensitive photoelectrochemical aptasensing of miR-155 using efficient and stable CH3NH3PbI3 quantum dots sensitized ZnO nanosheets as light harvester, Biosens. Bioelectron., 2016, 85, 142–150 CrossRef CAS.
- B. Dong, C. Li, G. Chen, Y. Zhang, Y. Zhang, M. Deng and Q. Wang, Facile synthesis of highly photoluminescent Ag2Se quantum dots as a new fluorescent probe in the second near-infrared window for in vivo imaging, Chem. Mater., 2013, 25, 2503–2509 CrossRef CAS.
- J. M. Frost, K. T. Butler, F. Brivio, C. H. Hendon, M. Van Schilfgaarde and A. Walsh, Atomistic origins of high-performance in hybrid halide perovskite solar cells, Nano Lett., 2014, 14, 2584–2590 CrossRef CAS.
- G. Xing, N. Mathews, S. Sun, S. S. Lim, Y. M. Lam, M. Grätzel and T. C. Sum, Long-range balanced electron-and hole-transport lengths in organic-inorganic CH3NH3PbI3, Science, 2013, 342, 344–347 CrossRef CAS.
- B. Yang, S. Lv, F. Chen, C. Liu, C. Cai, C. Chen and X. Chen, A resonance light scattering sensor based on bioinspired molecularly imprinted polymers for selective detection of papain at trace levels, Anal. Chim. Acta, 2016, 912, 125–132 CrossRef CAS.
- L. P. Wu, Y. F. Li, C. Z. Huang and Q. Zhang, Visual detection of Sudan dyes based on the plasmon resonance light scattering signals of silver nanoparticles, Anal. Chem., 2006, 78, 5570–5577 CrossRef CAS.
- J. Shen, Y. Zhu, X. Yang and C. Li, Graphene quantum dots: emergent nanolights for bioimaging, sensors, catalysis and photovoltaic devices, Chem. Commun., 2012, 48, 3686–3699 RSC.
- O. S. Wolfbeis, An overview of nanoparticles commonly used in fluorescent bioimaging, Chem. Soc. Rev., 2015, 44, 4743–4768 RSC.
- Q. Liu, C. Ma, X. P. Liu, Y. P. Wei, C. J. Mao and J. J. Zhu, A novel electrochemiluminescence biosensor for the detection of microRNAs based on a DNA functionalized nitrogen doped carbon quantum dots as signal enhancers, Biosens. Bioelectron., 2017, 92, 273–279 CrossRef CAS.
- W. Y. Chen, G. Y. Lan and H. T. Chang, Use of fluorescent DNA-templated gold/silver nanoclusters for the detection of sulfide ions, Anal. Chem., 2011, 83, 9450–9455 CrossRef CAS.
- E. Braun, Y. Eichen, U. Sivan and G. Ben-Yoseph, DNA-templated assembly and electrode attachment of a conducting silver wire, Nature, 1998, 391, 775–778 CrossRef CAS.
- P. Shah, S. W. Choi, H. J. Kim, S. K. Cho, P. W. Thulstrup, M. J. Bjerrum and S. W. Yang, DNA/RNA chimera templates improve the emission intensity and target the accessibility of silver nanocluster-based sensors for human microRNA detection, Analyst, 2015, 140, 3422–3430 RSC.
- P. Shah, A. Rørvig-Lund, S. B. Chaabane, P. W. Thulstrup, H. G. Kjaergaard, E. Fron and T. Vosch, Design aspects of bright red emissive silver nanoclusters/DNA probes for microRNA detection, ACS Nano, 2012, 6, 8803–8814 CrossRef CAS.
- Y. Y. Zhang, Q. M. Feng, J. J. Xu and H. Y. Chen, Silver nanoclusters for high-efficiency quenching of CdS nanocrystal electrochemiluminescence and sensitive detection of microRNA, ACS Appl. Mater. Interfaces, 2015, 7, 26307–26314 CrossRef CAS.
- Y. Q. Liu, M. Zhang, B. C. Yin and B. C. Ye, Attomolar ultrasensitive microRNA detection by DNA-scaffolded silver-nanocluster probe based on isothermal amplification, Anal. Chem., 2012, 84, 5165–5169 CrossRef CAS.
- A. Rotaru, S. Dutta, E. Jentzsch, K. Gothelf and A. Mokhir, Selective dsDNA-Templated Formation of Copper Nanoparticles in Solution, Angew. Chem., Int. Ed., 2010, 49, 5665–5667 CrossRef CAS.
- F. Xu, H. Shi, X. He, K. Wang, D. He, Q. Guo and J. Tang, Concatemeric dsDNA-templated copper nanoparticles strategy with improved sensitivity and stability based on rolling circle replication and its application in microRNA detection, Anal. Chem., 2014, 86, 6976–6982 CrossRef CAS.
- X. Sun and Y. Li, Colloidal carbon spheres and their core/shell structures with noble-metal nanoparticles, Angew. Chem., 2004, 116, 607–611 CrossRef.
- L. Kong, X. Lu, X. Bian, W. Zhang and C. Wang, Accurately tuning the dispersity and size of palladium particles on carbon spheres and using carbon spheres/palladium composite as support for polyaniline in H2O2 electrochemical sensing, Langmuir, 2010, 26, 5985–5990 CrossRef CAS.
- L. Wang, Y. Cheng, H. Wang and Z. Li, A homogeneous fluorescence sensing platform with water-soluble carbon nanoparticles for detection of microRNA and nuclease activity, Analyst, 2012, 137, 3667–3672 RSC.
- X. Liao and H. Ju, In situ quantitation of intracellular microRNA in the whole cell cycle with a functionalized carbon nanosphere probe, Chem. Commun., 2015, 51, 2141–2144 RSC.
- H. Li, Y. Zhang, T. Wu, S. Liu, L. Wang and X. Sun, Carbon nanospheres for fluorescent biomolecular detection, J. Mater. Chem., 2011, 21, 4663–4668 RSC.
- H. Zhao, Y. Qu, F. Yuan and X. Quan, A visible and label-free colorimetric sensor for miRNA-21 detection based on peroxidase-like activity of graphene/gold-nanoparticle hybrids, Anal. Methods, 2016, 8, 2005–2012 RSC.
- K. Hao, Y. He, H. Lu, S. Pu, Y. Zhang, H. Dong and X. Zhang, High-sensitive surface plasmon resonance microRNA biosensor based on streptavidin functionalized gold nanorods-assisted signal amplification, Anal. Chim. Acta, 2017, 954, 114–120 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2019 |






