Dual role of BSA for synthesis of MnO2 nanoparticles and their mediated fluorescent turn-on probe for glutathione determination and cancer cell recognition†
Qi
Wang
 ab,
Yuan
Zhang
a,
Xiaodong
Wang
a,
Yuehuan
Wu
b,
Chuan
Dong
ab,
Yuan
Zhang
a,
Xiaodong
Wang
a,
Yuehuan
Wu
b,
Chuan
Dong
 a and
Shaomin
Shuang
a and
Shaomin
Shuang
 *a
*a
aCollege of Chemistry and Chemical Engineering, Institute of Environmental Science, Shanxi University, Taiyuan, 030006, PR China. E-mail: smshuang@sxu.edu.cn
bChemistry & Chemical Engineering Department, Taiyuan Institute of Technology, Taiyuan, 030008, PR China
First published on 23rd January 2019
Abstract
A MnO2 nanoparticle (MnO2 NP)-mediated fluorescent turn-on probe for sensitively and selectively detecting glutathione (GSH) and recognizing cancer cells was established in this work. MnO2 NPs were synthesized simply and quickly through an in situ redox reaction by mixing bovine serum albumin (BSA) and KMnO4, in which BSA served the dual roles of template and reductant. It was found that the MnO2 NPs served as an effective energy acceptor and quenched the fluorescence intensity of carbon dots (CDs), owing to the fluorescence resonance energy transfer (FRET) process. Further, the addition of GSH triggered the decomposition of MnO2, breaking the FRET between MnO2 NPs and CDs and thus restoring the fluorescence intensity of CDs. Based on this mechanism, quantitative determination of GSH was performed. Under optimal conditions, a satisfactory linear range of 0.05–90 μM and limit of detection of 39 nM were obtained, and GSH content in human serum samples was detected. Moreover, taking advantage of the higher levels of GSH in cancer cells than in normal cells, the MnO2 NP-CD probe was applied to distinguish SMMC-7721 cancer cells from L02 normal cells. The FRET was interrupted by GSH in cancer cells, and strong fluorescence was observed. This work provides a facile approach for synthesizing MnO2 NPs, and this rapid, low-cost method with no need for reductants makes synthesis green and convenient. The MnO2 NP-mediated fluorescent turn-on response to GSH could improve the MnO2 nanomaterial-based biochemical analysis applications.
1. Introduction
It is universally recognized that glutathione, a tripeptide (Glu-Cys-Gly, GSH), plays an irreplaceable role in human health. Glutathione is directly involved in physiological processes and metabolism as antioxidant and antidote by maintaining the redox equilibrium, removing heavy metals and protecting against free radicals in cells.1–3 Disordered levels of GSH in human tissues or cells can be caused by some diseases, such as diabetes, neurodegenerative diseases, AIDS, heart diseases, and liver damage,4–7 making it a marker for disease surveillance. Particularly, abnormally high amounts of GSH are discovered in cancer cells, and the overexpressed GSH can be used to differentiate cancer cells from normal ones.8–12 Therefore, it is meaningful to detect and monitor the GSH content in biological samples as well as cells.To date, a variety of methods have been developed by researchers for GSH determination, ranging from mass spectrometry,13 high-performance liquid chromatography (HPLC),14 electrochemistry,15,16 surface enhanced Raman scattering,17,18 nuclear magnetic resonance (NMR),19 chemiluminescence,20,21 to colorimetry22–25 and fluorescence. Fluorescence methods have raised our interest because their quick response, high sensitivity and diverse sensing models facilitate the analysis. Various fluorescence methods with different principles have been established for the sensitive determination of GSH.26–29 Particularly, carbon nanodots (CDs) draw much attention because of their excellent bioimaging property and low toxicity,30–32 and consequently, they have been widely applied for GSH determination.33–35 Interestingly, a promising trend focuses on the MnO2 nanomaterial-mediated fluorescent turn-on sensing model, taking advantage of the unique reaction between MnO2 and GSH. MnO2 nanosheet-modified upconversion nanoparticles were applied for intracellular glutathione detection through GSH stimulated energy transformation.36 Cai et al.37 fabricated MnO2 nanosheets by ultrasound directly in carbon dot solutions to form nanocomposites for fluorescent detection of GSH in human serum samples. Similarly, MnO2 nanosheets were combined with fluorescent carbon quantum dots through electrostatic interaction and applied to detect GSH.38 A MnO2 nanoflower was synthesized and connected chemically with carbon dots through amidization for fluorescent turn-on sensing of GSH.39 Xu et al.40 established a carbon dot-MnO2 nanoplatform for GSH detection and further achieved a magnetic resonance image using MR-active Mn2+. An iridium(III)-based fluorescent complex was also constructed with MnO2 nanosheets to detect GSH through fluorescence and magnetic resonance image dual model.41
In this work, MnO2 nanoparticles were synthesized through a facile and quick method using bovine serum albumin (BSA) as both template and reductant. These MnO2 nanoparticles quenched the fluorescence of carbon nanodots (CDs) through fluorescence resonance energy transfer (FRET). GSH interrupted this process through decomposition of the quencher. Based on this mechanism, a sensitive and selective fluorescent turn-on method for GSH detection was established, and the MnO2 NP-CD probe was successfully applied in human serum sample determination and cancer cell recognition.
2. Experimental
2.1 Reagents and apparatus
Bovine serum albumin (BSA, 96%) and glutathione (GSH, 98%) were purchased from Aladdin Reagent Company (Shanghai, China). Potassium permanganate (KMnO4, Mw = 158.03) and other reagents were at least of analytical grade and were purchased from Kemiou Chemical Co. Ltd (Tianjin, China). HAc–NaAc buffer (0.2 M) of different pH levels were prepared by titrating NaAc solutions (0.2 M) with sodium hydroxide solution or acetic acid solution (both 0.2 M). Deionized water with conductivity of 18.2 MΩ cm−1 was used throughout this experiment. SMMC-7721 cancer cells, L02 normal cells and human serum samples were kindly provided by Shanxi Medical University (Taiyuan, China).Transmission electron microscopy (TEM) of MnO2 NPs and CDs was carried out on a G2 F20 S-TWIN transmission electron microscope (FEI, USA) for the morphology study. The dynamic light scattering (DLS) and zeta potential test were performed using a laser particle sizer (Malvern Nano-ZS90) to measure the size distribution of as-prepared MnO2 NPs and the surface charge of CDs and MnO2 NPs. The surface composition and chemical state test of products were determined by X-ray photoelectron spectroscopy (XPS) on an AXIS ULTRA DLD electron spectrometer (Kratos) with monochromatic Al Kα radiation. The content of Mn element in MnO2 NPs was measured on an atomic absorption spectrometer (900T, PerkinElmer, USA). The photoluminescence spectra were recorded on an F-7000 fluorescence spectrophotometer (Hitachi, Tokyo, Japan). The fluorescence lifetimes of CDs before and after adding MnO2 NPs were measured on an Edinburgh FLS 1000 lifetime and steady state spectrometer (Edinburgh, UK). The UV-vis absorption spectra were recorded on a TU-1901 UV-vis spectrophotometer (Puxi, China). The cell imaging tests were carried out on a confocal laser scanning microscope (CLSM, Olympus FV1000, Tokyo, Japan).
2.2 Synthesis of MnO2 NPs and CDs
The manganese dioxide nanoparticles were synthesized from KMnO4 by adding the latter to bovine serum albumin solutions. In a typical process, 50 mg bovine serum albumin powder was dissolved in 9 mL deionized water to form a transparent solution. Next, 1 mL KMnO4 solution with concentration of 10 mM was added into BSA solutions dropwise under stirring. The reaction system was performed under room temperature and kept stirring for 3 min, and a final brown colour of the mixed solution was obtained. Then, the resultant solutions were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. Finally, the supernatant was dialyzed with deionized water for 48 h to remove unreacted molecules and stored at 4 °C for future characterization and use. The concentration of as-prepared MnO2 NPs was calculated through mass conservation of Mn element, whose content was measured by atomic absorption spectrometry.
000 rpm for 10 min. Finally, the supernatant was dialyzed with deionized water for 48 h to remove unreacted molecules and stored at 4 °C for future characterization and use. The concentration of as-prepared MnO2 NPs was calculated through mass conservation of Mn element, whose content was measured by atomic absorption spectrometry.
Carbon nanodots were synthesized according to a previous work.42 Typically, 3 g citric acid and 3 g urea were dissolved into 10 mL deionized water, and the mixed solution was heated in a domestic 750 W microwave oven for 5 min. Dark brown solids were obtained and then transferred to a vacuum oven and heated at 60 °C for 1 h to remove the residual small molecules. Finally, CD solutions were obtained after dissolving the solids and centrifuging at 3000 rpm for 20 min. The CD solids were obtained through freeze-drying, and CD solutions were prepared after weighing for further use.
2.3 GSH detection procedure
The detection of GSH was performed at room temperature in HAc–NaAc buffer (0.2 M, pH 5.0). In a typical run, 50 μL of CD solution was added into 650 μL buffer, followed by the addition of 200 μL of MnO2 NPs. Then, 100 μL of different concentrations of GSH were introduced to the MnO2 NP-CD probe (final concentrations: CDs: 3 μg mL−1, MnO2 NPs: 15 μg mL−1). The fluorescence emission spectra were recorded after reaction for 120 min. The sensitivity and selectivity measurements were conducted in triplicate. All the fluorescence measurements were performed under the same conditions: excitation wavelength was 360 nm; the slit widths for excitation and emission were 5 nm. Human serum samples were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min before use. 100 μL of supernatant was added into the buffer containing CDs (3 μg mL−1), MnO2 NPs (15 μg mL−1) and GSH (1, 50, 90 μM), with a final volume of 1 mL. After incubation for 120 min, the fluorescence intensity was measured at the above-mentioned conditions, and the content and recovery of GSH in human serum samples were calculated.
000 rpm for 10 min before use. 100 μL of supernatant was added into the buffer containing CDs (3 μg mL−1), MnO2 NPs (15 μg mL−1) and GSH (1, 50, 90 μM), with a final volume of 1 mL. After incubation for 120 min, the fluorescence intensity was measured at the above-mentioned conditions, and the content and recovery of GSH in human serum samples were calculated.
2.4 In vitro cytotoxicity assay
The cytotoxicity of MnO2 NP-CD probe was tested by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay using SMMC-7721 cells. Cells were seeded and cultured in a 96-well plate in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), 50 mg mL−1 penicillin, and 50 mg mL−1 streptomycin at 37 °C and 5% CO2. After incubation for 24 h, MnO2 NPs-CDs with different concentrations (0–50 μg mL−1) in culture medium were added into each well and then incubated for 24 h. MTT solution (0.5 mg mL−1) was introduced into each well, and cells were incubated for another 4 h. Then, an enzyme-linked immunosorbent assay (ELISA) reader was used to measure the absorbance of the suspension at 490 nm.2.5 Cell imaging
To further investigate the capacity of the probe to distinguish the normal cells and cancer cells, SMMC-7721 cancer cells and L02 normal cells were seeded into 35 mm dishes and cultured for 12 h. Before imaging, the cells were pretreated with fresh medium and washed three times with PBS solution and then incubated with MnO2 NPs-CDs (32 μg mL−1) for 3 h. To decrease GSH concentration in living cells, both cells were treated with N-ethylmaleimide (NEM, 400 μM) for 20 min before incubation with MnO2 NPs-CDs. The cell lines were further washed with PBS solution (0.01 M, pH = 7.4) and subsequently imaged at ambient temperature.3. Results and discussion
3.1 Synthesis and characterization of MnO2 NPs and CDs
MnO2 NPs were synthesized through an in situ redox reaction by mixing KMnO4 and BSA solutions. BSA performed the dual roles of template and reductant in this procedure. It was reported that amino acid residues with reducibility (such as –NH2, –SH, –OH, etc.) exist in the BSA molecule.43,44 KMnO4 molecules were reduced into MnO2 around these active sites and grew gradually on the protein, so that BSA-templated MnO2 NPs were obtained. The reaction equation for this process is listed below:| BSA(Red) + KMnO4 + H2O → MnO2/BSA(Ox) + K+ + OH− |
Fig. 1A displays the schematic synthesis of BSA-based manganese dioxide nanoparticles, in which the purple solution of KMnO4 turned brown, indicating the formation of MnO2 NPs. Fig. S1† shows photos of the reaction product. Based on the abovementioned reaction equation, the generation of OH− caused the increase of pH, and this is shown in Fig. S2.† TEM image of as-prepared MnO2 NPs is shown in Fig. 1B; uniformly dispersed nanoparticles were observed. The size of MnO2 NPs was measured in Fig. S3,† and it was found to be ∼3.6 nm. The HRTEM image in Fig. 1C indicates a lattice spacing of 3.5 Å that is well coincident with the (220) lattice plane of α-MnO2.45
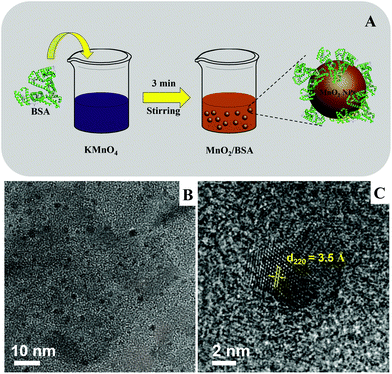 | ||
| Fig. 1 (A) The synthetic scheme of BSA-templated MnO2 NPs. (B) The TEM and (C) HRTEM images of as-prepared MnO2 NPs; scale bars are 10 nm and 2 nm, respectively. | ||
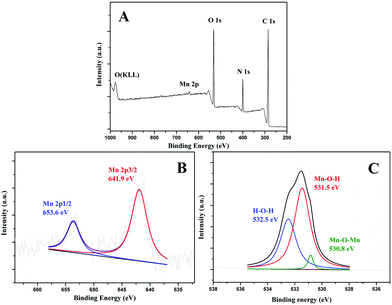
XPS technique was performed to test the elemental composition of the obtained materials, as shown in Fig. 2. The full scan spectrum demonstrated that this material is composed of elements Mn 2p, O 1s, N 1s and C 1s (Fig. 2A). C 1s, N 1s and a portion of O 1s come from BSA. The partial spectra of Mn 2p (Fig. 2B) can be divided into two peaks at 653.6 and 641.9 eV, which were assigned to Mn 2p1/2 and Mn 2p3/2, respectively. In O 1s spectrum (Fig. 2C), three component peaks at 532.5, 531.5 and 530.8 eV were obtained corresponding to H–O–H, Mn–O–H and Mn–O–Mn, respectively. Therefore, the above data demonstrate that the as-prepared materials were MnO2 NPs.
Fig. S4† shows the TEM image of CDs, and the zeta potentials of CDs and as-prepared MnO2 NPs were tested and are shown in Fig. S5.† CDs showed a zeta potential of +0.95 mV, which was attributed to the protonation of nitrogen atoms, which were doped in CDs through the addition of urea.38 The zeta potential of MnO2 NPs was found to be −12.87 mV. This was caused by the presence of Mn–O− groups on the surface of MnO2.46 The positive and negative charges endowed CDs and MnO2 NPs a mutual attraction through electrostatic force.
3.2 Fluorescence response of CDs to MnO2 NPs and GSH
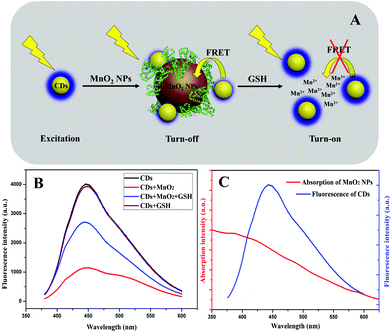
Based on the above results, we designed a fluorescent turn-off-on probe for GSH, taking advantage of fluorescent CDs and BSA-templated MnO2 NPs. Fig. 3A displays the schematic model of the fluorescent probe. The CDs showed a maximal fluorescent emission around 450 nm under excitation of 360 nm, and the fluorescence intensity was efficiently quenched once MnO2 NPs were introduced. Furthermore, the addition of GSH triggered a unique redox reaction between GSH and MnO2 NPs, which caused the decomposition of MnO2 NPs so that the fluorescence intensity of CDs was restored. Fig. 3B indicates the fluorescence spectra of CDs, CDs + MnO2 NPs, CDs + MnO2 NPs + GSH, and CDs + GSH, respectively, and the fluorescent turn-off-on process is demonstrated clearly. To study the fluorescence quenching mechanism, the fluorescence spectrum of CDs and UV-vis absorption spectrum of MnO2 NPs were measured. As recorded in Fig. 3C, there was an effective overlap between these two spectra. Furthermore, Fig. S6† shows the fluorescence lifetime measurement of CDs in the absence and presence of MnO2 NPs, and the results are listed in Table S1.† It was demonstrated that the lifetime of CDs decreased from 9.24 ns to 7.44 ns after adding MnO2 NPs. Thirdly, combining the above research on zeta potential, the electrostatic attraction shortened the distance between CDs and MnO2 NPs. As reported, FRET occurs between CDs in the excited state and quencher in the ground state when three conditions are reached: (a) the fluorescence spectra of CDs and the absorption spectra of the quencher overlap; (b) the fluorescence lifetime of CDs decreases; and (c) CD-quencher distance is in the range of 10–100 Å.47 A similar mechanism for fluorescence quenching of CDs by MnO2 was also claimed with FRET.37–39 Therefore, it was concluded that the fluorescence quenching of CDs is due to the FRET between CDs and MnO2 NPs.3.3 Optimization of parameters for GSH detection
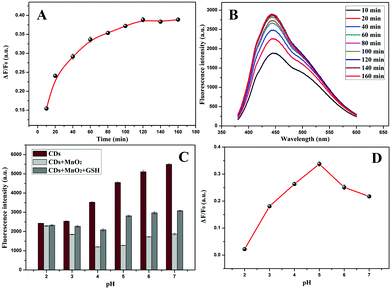
The fluorescence intensity of CDs was quenched by MnO2 NPs rapidly at room temperature and then remained unchanged (Fig. S7†), indicating CDs and MnO2 NPs formed stable composites. Therefore, the detection accuracy would not be affected by the environment. In this work, 200 μL of as-prepared MnO2 NPs (final concentration: 15 μg mL−1) was employed as quencher. The influence of time and pH on fluorescence restoration for GSH was investigated. Fig. 4A shows the relationship between the restoration efficiency (ΔF/F0) and time after adding GSH, and the corresponding FL spectra are displayed in Fig. 4B. It was concluded that the reaction between GSH and MnO2 NPs kept stable after 120 min, and an optimal restoration efficiency was obtained. HAc–NaAc buffers with different pH levels were used to investigate the effect on the recovery efficiency in the range of pH 2.0 to 7.0 in Fig. 4C and D. It was calculated that the fluorescence recovery efficiency of the MnO2 NP-CD probe displayed a maximum at pH = 5.0. This phenomenon was because of the pH-dependent fluorescent emission of CDs and the strong oxidation capacity of MnO2 at pH = 5.0.3.4 Calibration and selectivity
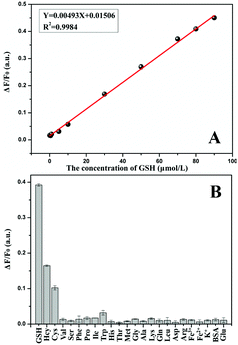
Under optimal conditions, the calibration curve of GSH detection was obtained between ΔF/F0 and the concentration of GSH. Fig. 5A indicates that the restoration efficiency of the fluorescence was proportional to the concentration of GSH in the range from 0.05–90 μM and followed the equation ΔF/F0 = 0.00493C + 0.01506 (R2 = 0.9984), where C is the GSH concentration (μM), ΔF is the recovered fluorescence intensity of the MnO2 NP-CD probe, and F0 is the fluorescence intensity of CDs. The limit of detection was 39 nM (3σ).To study the selectivity of MnO2 NP-CD probe for detecting GSH, equal concentrations of amino acids and some possible coexisting substances (80 μM) were used to investigate the effects. As shown in Fig. 5B, the MnO2 NP-CD probe demonstrated significant fluorescence recovery in the presence of GSH, and others did not show the same phenomenon. Hcy and Cys also showed a certain degree of fluorescence recovery because of their thiol content. However, the content of Cys and Hcy in cells is much lower than that of GSH,48,49 and it was reported that the concentration of GSH is about one thousand times more than that of Cys or Hcy.37,50 Further, we conducted the contrast experiment on the fluorescence recovery efficiency of Cys and Hcy, in which concentration was one-thousandth that of GSH. Fig. S8† demonstrates that Cys and Hcy, at low concentrations, did not affect the detection of GSH, indicating the influence of Cys and Hcy can be ignored during GSH detection. Therefore, the MnO2 NP-CD probe shows good selectivity to GSH response.
3.5 Determination of GSH in human serum
Because of the satisfactory sensitivity and selectivity of this fluorescent turn-on probe, MnO2 NPs-CDs were applied to the determination of GSH in human serum. The resultant concentration of GSH accorded with the reported literature.51 To investigate the accuracy of this method, three known concentrations of standard GSH solutions were spiked, and recoveries were calculated. The results are listed in Table 1, which shows recoveries in the range of 94.28% to 102.77%. The good recovery demonstrates that the MnO2 NP-mediated fluorescent turn-on probe is feasible in real sample analysis.| Found in sample (μM) | Added (μM) | Total found (μM) | Recovery (%, n = 6) | RSD (%, n = 6) |
|---|---|---|---|---|
| 1 | 6.29 | 102.77 | 2.33 | |
| 5.26 | 50 | 52.40 | 94.28 | 1.38 |
| 90 | 96.78 | 101.69 | 1.08 |
3.6 Recognition of cancer cell
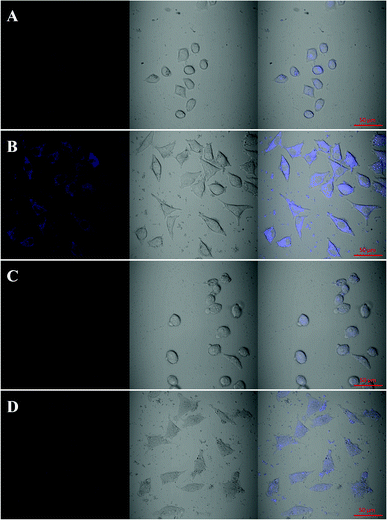
The cell toxicity of MnO2 NPs-CDs was studied in Fig. S9† by measuring SMMC-7721 cell viability after incubation with different concentrations of MnO2 NPs-CDs. The results demonstrate that the cytotoxicity of the probe is very low. According to the distinct difference of GSH levels in cancer cells against normal cells, the MnO2 NP-mediated fluorescent turn-on sensing model was applied in cell imaging for the recognition of cancer cells. The confocal laser scanning microscope (CLSM) images of L02 normal cells and SMMC-7721 cancer cells (Fig. 6) were recorded for comparison. Fig. 6A and B show images of the L02 and SMMC-7721 cells after treatment with MnO2 NPs-CDs, in which a strong blue fluorescence emission was observed in SMMC-7721 cells compared with L02 cells. The results indicate that a high concentration of GSH in cancer cells turns on the fluorescence of MnO2 NPs-CDs. To further investigate the effect of intracellular GSH level, N-ethylmaleimide (NEM), a GSH scavenger, was added to both cell samples before treating with MnO2 NPs-CDs. As shown in Fig. 6C and D, the fluorescence signal weakened significantly in cancer cells after NEM treatment, and it was not obvious in normal cells. This was attributed to the decline of GSH level after NEM treatment, decreasing the fluorescence restoration. The above results imply that the MnO2 NP-mediated fluorescence turn-on model can identify cancer cells from normal cells.4. Conclusions
MnO2 nanoparticles were synthesized from BSA and KMnO4 through an in situ redox reaction in this work. The nanoparticles were combined with CDs by electrostatic attraction, leading to the fluorescence quenching of CDs through FRET. According to this mechanism, a MnO2-mediated fluorescent turn-on sensing model was established for GSH determination and cancer cell recognition. The sensitive and selective determination of GSH in human serum samples was performed, and satisfactory linear range and good recovery were obtained. The bright fluorescence observed in cancer cells demonstrates the obvious distinction of cancer cells from normal cells. The MnO2 nanoparticle-mediated fluorescent turn-on sensing model has a promising prospect in bioanalytical chemistry.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21874087 and 21575084) and Shanxi Scholarship Council of China (2017-Key1). We thank the Scientific Instrument Center of Shanxi University and its technician for helping us to identify samples and for technical support.References
- S. Y. Zhang, C. N. Ong and H. M. Shen, Cancer Lett., 2004, 208, 143–153 CrossRef CAS PubMed.
- S. Kanzok, R. H. Schirmer, I. Turbachova, R. Lozek and K. Becker, J. Biol. Chem., 2000, 275, 40180–40186 CrossRef CAS PubMed.
- J. H. Pan, Z. Y. Zheng, J. Y. Yang, Y. Y. Wu, F. S. Lu, Y. W. Chen and W. H. Gao, Talanta, 2017, 166, 1–7 CrossRef CAS PubMed.
- P. K. Mandal, S. Saharan, M. Tripathi and G. Murari, Biol. Psychiatry, 2015, 78, 702–710 CrossRef CAS PubMed.
- J. M. Estrela, A. Ortega and E. Obrador, Crit. Rev. Clin. Lab. Sci., 2006, 43, 143–181 CrossRef CAS PubMed.
- A. Chauhan and V. Chauhan, Pathophysiology, 2006, 13, 171–181 CrossRef CAS PubMed.
- X. B. Zhang, R. M. Kong, Q. Q. Tan, F. Qu and F. L. Qu, Talanta, 2017, 169, 1–7 CrossRef CAS PubMed.
- L. Y. Niu, Y. S. Guan, Y. Z. Chen, L. Z. Wu, C. H. Tung and Q. Z. Yang, J. Am. Chem. Soc., 2012, 134, 18928–18931 CrossRef CAS PubMed.
- S. Kim, S. M. Ahn, J.-S. Lee, T. S. Kim and D.-H. Min, 2D Mater., 2017, 4, 025069 CrossRef.
- C. Liu, D. P. Wang, Y. Zhan, L. Y. Yan, Q. Lu, M. Y. Z. Chang, J. W. Luo, L. D. Wang, D. Du, Y. H. Lin, J. Xia and Y. Wu, ACS Appl. Mater. Interfaces, 2018, 10, 44231–44239 CrossRef CAS PubMed.
- G. B. Yang, L. G. Xu, Y. Chao, J. Xu, X. Q. Sun, Y. F. Wu, R. Peng and Z. Liu, Nat. Commun., 2017, 8, 902 CrossRef PubMed.
- H. T. Bi, Y. L. Dai, P. P. Yang, J. T. Xu, D. Yang, S. L. Gai, F. He, G. H. An, C. N. Zhong and J. Lin, Chem. Eng. J., 2019, 356, 543–553 CrossRef CAS.
- T. Fahrenholz, M. M. Wolle, H. M. S. Kingston, S. Faber, J. C. KernII, M. Pamuku, L. Miller, H. Chatragadda and A. Kogelnik, Anal. Chem., 2015, 87, 1232–1240 CrossRef CAS PubMed.
- B. Baytam, G. Rimbach, J. Frank and T. Esatbeyoglu, Food Chem., 2014, 62, 402–408 CrossRef PubMed.
- J. C. Harfield, C. Batchelor-McAuleya and R. G. Compton, Analyst, 2012, 137, 2285–2296 RSC.
- M. C. C. Areias, K. Shimizu and R. G. Compton, Analyst, 2016, 141, 2904–2910 RSC.
- A. Saha and N. R. Jana, Anal. Chem., 2013, 85, 9221–9228 CrossRef CAS PubMed.
- C. H. Wei, X. Liu, Y. Gao, Y. P. Wu, X. Y. Guo, Y. Ying, Y. Wen and H. F. Yang, Anal. Chem., 2018, 90, 11333–11339 CrossRef CAS PubMed.
- V. G. Kontogianni, C. G. Tsiafoulis, L. G. Roussis and I. P. Gerothanassis, Anal. Methods, 2017, 9, 4464–4470 RSC.
- Y. Wang, J. Lu, L. Tang, H. Chang and J. Li, Anal. Chem., 2009, 81, 9710–9715 CrossRef CAS PubMed.
- Y. R. Tang, H. J. Song, Y. Y. Su and Y. Lv, Anal. Chem., 2013, 85, 11876–11884 CrossRef CAS PubMed.
- Y. Xianyu, Y. Xie, N. X. Wang, Z. Wang and X. Y. Jiang, Small, 2015, 11, 5510–5514 CrossRef CAS PubMed.
- W. Y. Li, J. Y. Wang, J. C. Zhu and Y. Q. Zheng, J. Mater. Chem. B, 2018, 6, 6858–6864 RSC.
- Y. Yuan, J. Zhang, M. J. Wang, B. Mei, Y. F. Guan and G. L. Liang, Anal. Chem., 2013, 85, 1280–1284 CrossRef CAS PubMed.
- X. Liu, Q. Wang, Y. Zhang, L. C. Zhang, Y. Y. Su and Y. Lv, New J. Chem., 2013, 37, 2174–2178 RSC.
- J. Qin, L. M. Zhang and R. Yang, Spectrochim. Acta, Part A, 2019, 207, 54–60 CrossRef CAS PubMed.
- P. J. Ni, D. F. Jiang, C. X. Chen, Y. Y. Jiang, Y. Z. Lu and Z. L. Zhao, Analyst, 2018, 143, 4442–4447 RSC.
- Y. X. Guo, X. D. Zhang and F. G. Wu, J. Colloid Interface Sci., 2018, 530, 511–520 CrossRef CAS PubMed.
- Y. Zhang, Y. R. Tang, X. Liu, L. C. Zhang and Y. Lv, Sens. Actuators, B, 2013, 185, 363–369 CrossRef CAS.
- Z. H. Li, S. Guo, Z. Q. Yuan and C. Lu, Sens. Actuators, B, 2017, 241, 821–827 CrossRef CAS.
- S. Q. Dong, Z. Q. Yuan, L. J. Zhang, Y. J. Lin and C. Lu, Anal. Chem., 2017, 89, 12520–12526 CrossRef CAS PubMed.
- S. Y. Lim, W. Shen and Z. Gao, Chem. Soc. Rev., 2015, 44, 362–381 RSC.
- J. H. Pan, Z. Y. Zheng, J. Y. Yang, Y. Y. Wu, F. S. Lu, Y. W. Chen and W. H. Gao, Talanta, 2017, 166, 1–7 CrossRef CAS PubMed.
- Z. J. Bai, F. Y. Yan, J. X. Xu, J. Zhang, J. F. Wei, Y. M. Luo and L. Chen, Spectrochim. Acta, Part A, 2018, 205, 29–39 CrossRef CAS PubMed.
- Q. Wang, X. Liu, L. C. Zhang and Y. Lv, Analyst, 2012, 137, 5392–5397 RSC.
- R. Deng, X. Xie, M. Vendrell, Y.-T. Chang and X. Liu, J. Am. Chem. Soc., 2011, 133, 20168–20171 CrossRef CAS PubMed.
- Q.-Y. Cai, J. Li, J. Ge, L. Zhang, Y.-L. Hu, Z.-H. Li and L.-B. Qu, Biosens. Bioelectron., 2015, 72, 31–36 CrossRef CAS PubMed.
- D. G. He, X. X. Yang, X. X. He, K. M. Wang, X. Yang, X. He and Z. Zou, Chem. Commun., 2015, 51, 14764–14767 RSC.
- C. L. Yang, W. P. Deng, H. Y. Liu, S. G. Ge and M. Yan, Sens. Actuators, B, 2015, 216, 286–292 CrossRef CAS.
- Y. Xu, X. Chen, R. Chai, C. F. Xing, H. R. Li and X.-B. Yin, Nanoscale, 2016, 8, 13414–13421 RSC.
- Z.-Z. Dong, L. H. Lu, C.-N. Ko, C. Yang, S. N. Li, M.-Y. Lee, C.-H. Leung and D.-L. Ma, Nanoscale, 2017, 9, 4677–4682 RSC.
- S. N. Qu, X. Y. Wang, Q. P. Lu, X. Y. Liu and L. J. Wang, Angew. Chem., Int. Ed., 2012, 51, 12215–12218 CrossRef CAS PubMed.
- C. Y. Hu, D.-P. Yang, Z. H. Wang, P. Huang, X. S. Wang, D. Chen, D. X. Cui, M. Yang and N. Q. Jia, Biosens. Bioelectron., 2013, 41, 656–662 CrossRef CAS PubMed.
- Q. W. Liu, T. Zhang, L. L. Yu, N. Q. Jia and D.-P. Yang, Analyst, 2013, 138, 5559–5562 RSC.
- X. Q. Tan, Y. F. Wan, Y. J. Huang, C. He, Z. L. Zhang, Z. Y. He, L. L. Hu, J. W. Zeng and D. Shu, J. Hazard. Mater., 2017, 321, 162–172 CrossRef CAS PubMed.
- H. Tamura, T. Oda, M. Nagayama and R. Furuichi, J. Electrochem. Soc., 1989, 136, 2782–2786 CrossRef CAS.
- F. L. Zu, F. Y. Yan, Z. J. Bai, J. X. Xu, Y. Y. Wang, Y. C. Huang and X. G. Zhou, Microchim. Acta, 2017, 184, 1899–1914 CrossRef CAS.
- F. Yu, P. Li, B. Wang and K. Han, J. Am. Chem. Soc., 2013, 135, 7674–7680 CrossRef CAS PubMed.
- A. Saha and N. R. Jana, Anal. Chem., 2013, 85, 9221–9228 CrossRef CAS PubMed.
- J. H. Pan, Z. Y. Zheng, J. Y. Yang, Y. Y. Wu, F. S. Lu, Y. W. Chen and W. H. Gao, Talanta, 2017, 166, 1–7 CrossRef CAS PubMed.
- F. Michelet, R. Gueguen, P. Leroy, M. Wellman, A. Nicolas and G. Siest, Clin. Chem., 1995, 41, 1509–1517 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8an02501k |
| This journal is © The Royal Society of Chemistry 2019 |