A cascade toehold-mediated strand displacement strategy for label-free and sensitive non-enzymatic recycling amplification detection of the HIV-1 gene†
Qiong
Li‡
ab,
Zhi
Liu‡
a,
Danhua
Zhou
bc,
Jiafeng
Pan
b,
Chengshuai
Liu
d and
Junhua
Chen
 *b
*b
aCollege of Bioscience and Biotechnology, Hunan Agricultural University, Changsha 410128, China
bGuangdong Key Laboratory of Integrated Agro-environmental Pollution Control and Management, Guangdong Institute of Eco-Environmental and Science &Technology, Guangzhou 510650, China. E-mail: 222chenjunhua@163.com
cCollege of Natural Resources and Environment, South China Agricultural University, Guangzhou 510462, China
dState Key Laboratory of Environmental Geochemistry, Institute of Geochemistry, Chinese Academy of Sciences, Guiyang 550081, China
First published on 31st January 2019
Abstract
In this work, a label-free fluorescence biosensor for simple detection of the HIV-1 gene was proposed by using toehold-mediated strand displacement reactions (TMSDRs) combined with a non-enzymatic target recycling amplification strategy. In this system, two TMSDRs were used. In the presence of the HIV-1 gene, an autocatalytic DNA machine can be activated. This leads to the generation of numerous free G-rich sequences, which can associate with a fluorescent dye N-methylmesoporphyrin IX (NMM) to yield an amplified fluorescence signal for the target detection. This sensing platform showed a high sensitivity towards the HIV-1 gene with a detection limit as low as 1.9 pM without any labelling, immobilization, or washing steps. The designed sensing system also exhibits an excellent selectivity for the HIV-1 gene compared with other interference DNA sequences. Furthermore, the presented biosensor is robust and has been successfully applied for the detection of the HIV-1 gene in a real biological sample with satisfactory results, suggesting that this method is promising for simple and early clinical diagnosis of HIV infection. Thanks to its simplicity, cost-effectiveness and ultrasensitivity, our proposed sensing strategy provides a universal platform for the detection of other genes by substituting the target-recognition element.
Introduction
Acquired immunodeficiency syndrome (AIDS), firstly reported by the US Centers for Disease Control and Prevention in 1981, is a severe communicable immune deficiency disease caused by the human immune deficiency virus (HIV).1,2 The World Health Organization (WHO)/Joint United Nations Programme on HIV/AIDS (UNAIDS) estimates that, up to 2014, about 36.9 million people were living with HIV in the world. Almost 46% of them didn't know their HIV status, and almost 60% were not accessing reverse transcription treatment.3 Therefore, the development of a simple and highly sensitive biosensor for HIV detection in the early infection stage is urgently needed.Conventional methods for the detection of HIV infection in clinical diagnosis are primarily based on virus isolation, and include polymerase chain reaction (PCR),4–6 enzyme-linked immune-absorbent assay (ELISA),7,8 and western blotting (WB).9 Unfortunately, they are expensive and time-consuming, and require fussy pre-treatment. Thus, it is of great significance to design a convenient, rapid, and low-cost detection approach for HIV infection analysis. As alternatives to conventional methods, biosensors have been widely used for virus detection,10–12 such as colorimetric biosensors,13 electrochemical biosensors14 and fluorescence biosensors.15 However, most of them exhibit relativity poor sensitivity and require labelling and immobilization, or washing steps, which inhibit the application of these technologies. To overcome these limitations, a label-free and non-enzymatic toehold-mediated strand displacement reaction (TMSDR) strategy has been applied.16–18
The TMSDR is an enzyme-free DNA strand displacement reaction based on the principle of the toehold exchange,19–21 which is a spontaneous phenomenon driven forward by entropy without any enzymes.22 Due to the programmability and versatility, the TMSDR-based signal amplification technique has been used for the construction of various biosensing systems.23–27
G-quadruplexes are structures that are formed by stacked G-tetrads, a planar association of four guanines by Hoogsteen hydrogen bonding.28 Owing to their structure polymorphism and tunable conformation, G-quadruplexes have been widely used as building blocks for directing the assembly of nanoscale components into sophisticated structures or as a functional tool for constructing nanoscale devices and biosensors.29–32N-Methyl mesoporphyrin IX (NMM) is a commercially available unsymmetrical anionic porphyrin characterized by a pronounced structural selectivity for G-quadruplex but not for duplex (DNA, RNA and RNA–DNA hybrid), triplex or single-stranded forms.33,34 Generally, NMM only shows a weak fluorescence in the free state, but exhibits a 20-fold enhancement in its fluorescence upon interacting with the G-quadruplex.35
Herein, a simple, facile, highly sensitive and selective detection method for the HIV-1 gene was developed by using cascaded TMSDRs and non-enzymatic target recycling amplification assay. In this system, the HIV-1 gene could bind to the terminal toehold region of the triple-stranded DNA substrate probe (SP) through a base pair and initiate the TMSDRs, resulting in the reuse of the target DNA and the production of plentiful G-quadruplexes. Ultimately, the increasing G-quadruplex/NMM complex leads to a dramatic enhancement in the fluorescence intensity. Taking advantage of the signal amplification method, the designed biosensor could achieve a high sensitivity for the HIV-1 gene detection. Owing to its cost-effectiveness (the detecting probe used in this research does not require any modification, and the entire reaction can be carried out in an isothermal manner without the involvement of a higher-precision thermal cycler), easy operation, and flexibility, the sensing system could be a promising platform for the early point-of-care diagnosis of the HIV-1 infection.
Experimental
Materials and reagents
Dimethyl sulfoxide (DMSO) and tris-(hydroxymethyl) aminomethane (Tris) were purchased from Sigma-Aldrich (St Louis, Mo). Human serums were provided by the fourth Affiliated Hospital of Guangzhou Medical University (Guangzhou, China). NMM was purchased from Frontier Scientific Inc. (Logan, Utah, USA). A 5 mM NMM stock solution was prepared in DMSO and stored at −20 °C before use. All other reagents and materials were of analytical grade and used without purification. All of the solutions were prepared with ultrapure water (18.2 MΩ cm−1) from a Millipore Milli-Q water purification system (Billerica MA).All of the DNA oligonucleotides were ULTRAPAGE-purified and were purchased from Shanghai Sangon Biotechnology Co., Ltd (Shanghai, China) and their sequences are listed as follows:
Signal strand (SS): 5′-GCACAGATTGGGTAGGGCGGGTTGGG-3′
Helper strand (HS): 5′-TCTCTTTACTATTTTATTT-3′
Long strand (LS): 5′-CTGGGATTAAATAAAATAGTAAAGAGAGCCCTACCCAATCTGTGC-3′
Fuel strand (FS): 5′-GCACAGATTGGGTAGGGCTCTCTTTACTATTTTATTT-3′
HIV-1 gene: 5′-GCTATACATTCTTACTATTTTATTTAATCCCAG-3′
M1: 5′-GCTATACATTCTTACTATTTTATTTAATC![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) CAG-3′
CAG-3′
M2: 5′-GCTATACATTCTTACTATTTTATTTAAT![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) CAG-3′
CAG-3′
M3: 5′-GCTATACATTCTTACTATTTTATT![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) AAT
AAT![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) CAG-3′
CAG-3′
NC: 5′-![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) -3′
-3′
Underlined letters represent the mismatched bases.
Assay procedure
The triple-stranded DNA substrate probes (SPs) were first prepared by mixing the LS, HS and SS at a final concentration of 300 nM in 20 mM Tris-HCl buffer (pH = 7.4, 100 mM NaCl, 10 mM MgCl2, 15 mM KCl), heating the mixture to 95 °C for 7 min and then gradually cooling to room temperature. Subsequently, 300 nM FS was added to the mixture. Next, various concentrations of the HIV-1 gene were incubated with the above solution with a reaction volume of 50 μL for 120 min at room temperature. This was followed by the introduction of the NMM (1 μM) and further incubation for another 30 min. Finally, the resulting solutions were diluted to 200 μL. The fluorescence spectra were recorded using a SpectraMax i3x (Molecular Devices, the U. S.) at room temperature. The scanning wavelength ranged from 580 to 640 nm (λex = 399 nm, λem = 610 nm). The fluorescence intensity at 610 nm was used to estimate the performance of the proposed biosensor. The slit widths of both the excitation and emission peaks were 10 nm; the PMT detector voltage was 700 V.Gel electrophoresis
To prepare the hydrogel, 5 mL of 30% gel solution (29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), 1 mL TBE buffer (10×), 100 μL APS, 4 μL TEMED, and 3.8 mL deionized water were mixed together. This mixture contained a final gel percentage of 15%. The gel was polymerized for 1.5 h at room temperature and then soaked in a 1× TBE buffer prior to use. 5 μL of each reacted sample was mixed with 1 μL of 6× loading buffer and was subjected to 15% non-denaturing polyacrylamide gel electrophoresis (PAGE). The PAGE was carried out in 1× TBE at a constant voltage of 60 V for about 70 min at room temperature. After staining in diluted SYBR gold solution, the gels were scanned using a Gel Doc XR documentation system (Bio-RAD Laboratories Inc., USA).
1), 1 mL TBE buffer (10×), 100 μL APS, 4 μL TEMED, and 3.8 mL deionized water were mixed together. This mixture contained a final gel percentage of 15%. The gel was polymerized for 1.5 h at room temperature and then soaked in a 1× TBE buffer prior to use. 5 μL of each reacted sample was mixed with 1 μL of 6× loading buffer and was subjected to 15% non-denaturing polyacrylamide gel electrophoresis (PAGE). The PAGE was carried out in 1× TBE at a constant voltage of 60 V for about 70 min at room temperature. After staining in diluted SYBR gold solution, the gels were scanned using a Gel Doc XR documentation system (Bio-RAD Laboratories Inc., USA).
Results and discussion
Strategy for the HIV-1 gene monitoring
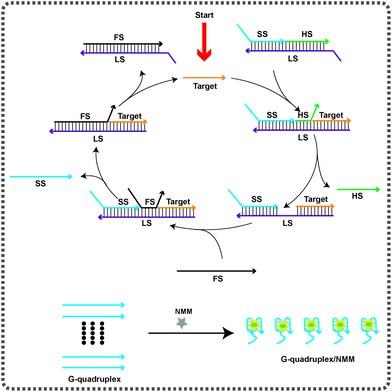
The strategy for the amplified detection of the HIV-1 gene on the basis of the TMSDR cascade signal amplification strategy is schematically demonstrated in Scheme 1. The biosensor consisted of a triple-stranded DNA substrate probe (SP) and a fuel strand (FS). A G-quadruplex forming signal strand (SS), a helper strand (HS) and a long strand (LS) are primarily denatured together to obtain the stable SP, which can prevent the FS from triggering the strand displacement reaction in the absence of the target DNA. The target DNA can specially bind to the toehold region at the 5′-terminus of the LS and displace the HS via an entropy-driven strand displacement reaction. The dissociation of the HS causes the exposure of the initially obstructed toehold in the middle of the LS. Subsequently, the newly exposed toehold can hybridize with the FS, resulting in the liberation of the target DNA and the SS at the same time. Consequently, the released target DNA can activate another round of the strand displacement reaction. Finally, the free G-rich sequence produced can form a G-quadruplex structure, which can be “lit up” by NMM, exhibiting a dramatically enhanced fluorescence intensity. In the absence of the target DNA, the TMSDRs can't be activated. Therefore, the G-rich sequence is still blocked in the LS. Thus, the system only yields a relatively low fluorescence background. Most importantly, the sensing system using G-quadruplex/NMM as the signal reporter avoids the use of any labels or modification procedures.Feasibility study
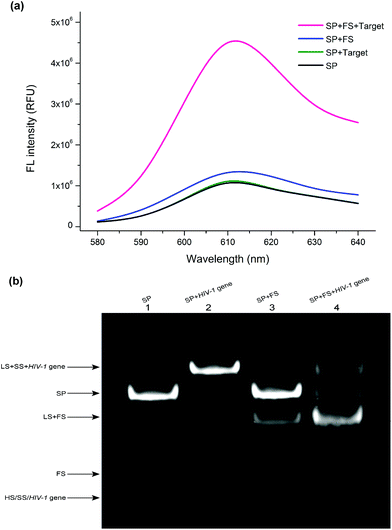
To verify the ability of the designed autocatalytic DNA machine for the target detection, the fluorescence spectra were investigated under different conditions. As shown in Fig. 1a, when the SP was alone in the solution, it exhibited a weak fluorescence emission peak at 610 nm (black curve), implying that the caged G-rich sequence can't interact with NMM. When the HIV-1 gene was added into the SP solution without the FS, the SS was still blocked by the LS. In this state, almost no obvious signal change can be observed compared with the background signal (green curve). When only the SP and the FS were present, the fluorescence intensity was slightly enhanced compared with the background fluorescence signal (blue curve). In the presence of the HIV-1 gene, the SP, and the FS, the fluorescence intensity improved sharply (red curve), suggesting that the TMSDRs were completed. The G-rich sequence was released and then interacted with the fluorescent dye NMM. These results indicated that our designed sensing system can feasibly provide an amplified fluorescence signal for the detection of the target HIV-1 gene. To further verify the feasibility of this study, gel electrophoresis experiments were used to confirm the results of the fluorescence assay. As shown in Fig. 1b, the SP can be formed from the hybridization among the HS, SS, and LS (lane 1). After the HIV-1 gene and the SP incubated together, a new band (the LS/SS/HIV-1 complex) was observed (lane 2), suggesting that the TMSDR has been taken place and the target could displace the HS from the SP to form the LS/SS/HIV-1 gene complex. When both the SP and the FS were present, only negligible leakage, which accounted for the system background, was observed during the 120 min reaction time (lane 3). However, after the addition of the HIV-1 gene, nearly most of the SP was converted into the LS/FS complex, and the released HS, SS, and the target HIV-1 gene can also be observed (lane 4). The above results further indicated that our designed detecting system can be utilized for the amplified detection of the target HIV-1 gene.Optimization of this assay
The experimental parameters (such as the temperature, the reaction time, and the concentration of the NMM) that could affect the analytical performance of the designed sensing system were optimized. The optimizations were performed by varying one experimental condition and keeping the conditions of the other parameters constant. In this work, the reaction temperature plays an important role in the sensing process. It not only affects the stability of the SP, but also the binding kinetics between the SP complex and the target HIV-1 gene. The effect of reaction temperature on the response of the fluorescent sensor was investigated by detecting 100 nM of the HIV-1 gene at different temperatures (4 °C, 25 °C, 37 °C, and 45 °C). The blank sample (in the absence of the HIV-1 gene) at each temperature was tested under the same conditions. As shown in Fig. S1 (ESI†), the fluorescence signal increased with the increase in the temperature (4–45 °C) in the presence of the HIV-1 gene (the green histogram in Fig. S1, ESI†). However, a higher temperature may change the stability of the SP, which increases the background signal.36 In order to obtain the best signal-to-background ratio (S/N) (the red line in Fig. S1, ESI†), 25 °C was considered to be the optimum reaction temperature. The corresponding error bars in Fig. S1† represent the standard deviation of three independent measurements obtained at each reaction temperature.The performance of the signal amplification and detection of the HIV-1 gene was deeply influenced by the reaction time of the TMSDRs. In order to optimize the reaction time, we control the fluorescence signal response at 25 °C. As shown in Fig. S2 (ESI†), the fluorescence intensity of the solution containing 100 nM of the HIV-1 gene increased rapidly when the incubation time ranges from 30 to 150 min and remained at almost a constant level after 120 min, which suggested that the G-rich sequence was almost completely released from the SP for the formation of the G-quadruplex/NMM complex. The issue of the reaction time is surely an important point to be considered. After this first demonstration of the use of this biosensor, it will be important to shorten the reaction time in the future with a more extended study.
The effect of the concentration of the NMM was also taken into consideration. As shown in Fig. S3 (ESI†), the S/N level increased gradually with the increasing concentration of the NMM from 0.25 to 1.5 μM, and reached a peak at 1 μM. However, the S/N level decreased when the NMM concentration continued to increase (1–1.25 μM). Thus, 1 μM of the NMM was chosen for further experiments.
Analytical performance of the biosensor
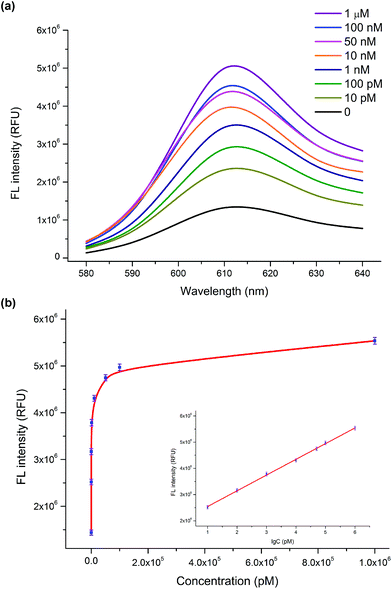
To investigate the dynamic range and sensitivity of the assay, a series of samples containing different concentrations of the HIV-1 gene were tested under optimal experimental conditions. As shown in Fig. 2a, the fluorescence intensity was enhanced as the concentration of the HIV-1 gene increased (0–1 μM). The relationship between the fluorescence intensity at 610 nm and the concentration of the HIV-1 gene is displayed in Fig. 2b. The resulting calibration curve shows that a linear dependence between the fluorescence intensity and the logarithm of the HIV-1 gene concentration was obtained in the range from 10 pM to 1 μM with a correlation coefficient (R2) of 0.99 (Fig. 2b, inset). The linear regression equation is y = 5.9 × 105 lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C + 1.9 × 106 (y and C represent the fluorescence value and the concentration of the HIV-1 gene, respectively). According to the 3δ rule, the calculated limit of detection (LOD) is 1.9 pM. Compared with the previously reported method of the HIV-1 gene detection, our sensing platform proposed herein has an acceptable analytical range and a relatively low detection limit (Table S1, ESI†). Such a high sensitivity can be attributed to the fact that the autocatalytic DNA machine can stimulate the formation of numerous G-quadruplex/NMM complexes even at a low concentration of the target HIV-1 gene. Here, our proposed fluorescence detection strategy holds great potential for monitoring of the HIV-1 gene in real biological samples with high sensitivity.
C + 1.9 × 106 (y and C represent the fluorescence value and the concentration of the HIV-1 gene, respectively). According to the 3δ rule, the calculated limit of detection (LOD) is 1.9 pM. Compared with the previously reported method of the HIV-1 gene detection, our sensing platform proposed herein has an acceptable analytical range and a relatively low detection limit (Table S1, ESI†). Such a high sensitivity can be attributed to the fact that the autocatalytic DNA machine can stimulate the formation of numerous G-quadruplex/NMM complexes even at a low concentration of the target HIV-1 gene. Here, our proposed fluorescence detection strategy holds great potential for monitoring of the HIV-1 gene in real biological samples with high sensitivity.
Selectivity of the biosensor
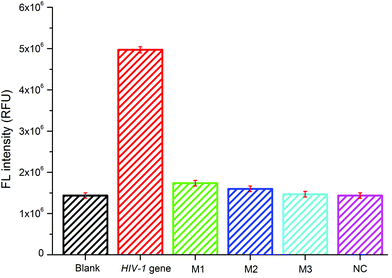
The selectivity of the sensing platform for the HIV-1 gene to other four kinds of DNA sequences, including a single-base mismatched DNA (M1), a two-base mismatched DNA (M2), a three-base mismatched DNA (M3), and the non-complementary DNA (NC) at the same concentration of 100 nM, was also analyzed by the present sensing system. As shown in Fig. 3, the fluorescence intensity varies with the target and the four other kinds of DNA. The relative fluorescence intensity for M1, M2, M3 and NC was about 34%, 32%, 29% and 29% of the HIV-1 gene, respectively. The four kinds of DNA sequences showed almost the same intensity as the blank sample. These results clearly demonstrated the high specificity of our designed sensing system.Real sample analysis
To investigate whether this fabricated monitoring system could be applied to real samples, a recovery experiment was carried out. The collected human serum samples from healthy people were filtered through a 0.2 μM membrane to remove the insoluble particles. Aliquots of the serum samples were spiked with different concentrations, 10, 50, and 100 nM, of the HIV-1 gene. As shown in Table 1, the recovery of all of the measured samples and the relative standard derivations (RSD) were in the range from 87–103% and 4.2–6.5% (n = 5), respectively, which indicated that the possible interference from the human serum sample to the biosensor analysis was negligible. The above data proved that our designed method can be successfully applied for the detection of the HIV-1 gene in real biological samples.Conclusions
In conclusion, a facile, isothermal, and ultrasensitive platform for the detection of the HIV-1 gene was developed by using TMSDRs combined with a label-free and non-enzymatic target recycling amplification strategy. The addition of the HIV-1 gene leads to the displacement of the HS and the exposure of the toehold domains of the SP for the FS to reuse the HIV-1 gene and to generate massive G-quadruplex structures. The increasing G-quadruplex interacts with the NMM to form G-quadruplex/NMM complexes, which result in an enhanced fluorescence intensity of the reaction solution for the highly sensitive detection of the HIV-1 gene at concentrations as low as 1.9 pM. Moreover, the designed sensor also exhibits an excellent selectivity toward the HIV-1 gene compared with other interference DNA samples. Furthermore, this method can be applied for the determination of the HIV-1 gene in spiked real biological samples with good recovery and accuracy. Without the use of any labelling, immobilization, washing, or modification steps, the developed method described herein can be easily applied for routine monitoring of the HIV-1 gene. Importantly, the designed autocatalytic DNA machine could be easily extended to fabricate versatile and robust sensing platforms for the detection of other genes by choosing the corresponding recognition elements. With the characteristics described above, this biosensor has great potential for applications as a platform for the early clinical diagnosis of the HIV infection.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support was provided by the Guangdong National Science Funds for Distinguished Young Scholars (2016A030306012), the NSFC (31671933), and the Local Innovative and Research Teams Project of the Guangdong Pearl River Talents Program (2017BT01Z176).Notes and references
- L. Zheng, J. Li, B. Li, Q. Liu, Q. Wang and N. Gan, Molecules, 2017, 17, 5988–6000 CrossRef PubMed.
- B. Li, Z. Li and Z. Dai, Biosens. Bioelectron., 2014, 52, 330–336 CrossRef CAS PubMed.
- D. Yin, Y. Tao, L. Tang and W. Li, Microchim. Acta, 2017, 184, 3721–3728 CrossRef CAS.
- A. F. Véronique, L. C. Marie, B. Stéphane, B. Marianne, F. Corinne and T. Kadidia, J. Med. Virol., 2009, 81, 217–223 CrossRef PubMed.
- M. Kiselinova, A. O. Pasternak, W. S. De, D. Vogelaers, B. Berkhout and L. Vandekerckhove, PLoS One, 2014, 9, e85999 CrossRef PubMed.
- A. Edelmann, U. Kalus, A. Oltmann, A. Stein, A. Unbehaun, C. Drosten, D. H. Krüger and J. Hofmann, Transfusion, 2010, 50, 685–692 CrossRef CAS PubMed.
- L. R. De and M. M. Stevens, Nat. Nanotechnol., 2012, 7, 821–824 CrossRef PubMed.
- W. Diao, M. Tang and S. Ding, Biosens. Bioelectron., 2018, 100, 228–234 CrossRef CAS PubMed.
- L. Zhou, J. Huang and B. Yu, ACS Appl. Mater. Interfaces, 2015, 7, 24438–24445 CrossRef CAS PubMed.
- Z. Ali, J. Wang, Y. Tang, B. Liu, N. He and Z. Li, Biomater. Sci., 2017, 5, 57–66 RSC.
- T. Le and C. Chang, Anal. Chem., 2017, 89, 6781–6786 CrossRef CAS PubMed.
- R. B. Channon, Y. Yang and K. M. Feibelman, Anal. Chem., 2018, 12, 7777–7783 CrossRef PubMed.
- Y. Long, C. Zhou, C. Wang, H. Cai, C. Yin, Q. Yang and D. Xiao, Sci. Rep., 2016, 6, 23949 CrossRef CAS PubMed.
- X. Wang, A. Jiang, T. Hou and F. Li, Anal. Chim. Acta, 2015, 890, 91–97 CrossRef CAS PubMed.
- S. H. Qaddare and A. Salimi, Biosens. Bioelectron., 2017, 89, 773–780 CrossRef CAS PubMed.
- X. Li, D. Li, W. Zhou, Y. Chai, R. Yuan and Y. Xiang, Chem. Commun., 2015, 51, 11084–11087 RSC.
- Y. Wu, D. Zhang, P. Yin and F. Vollmer, Small, 2014, 10, 2067–2076 CrossRef CAS PubMed.
- N. Zhang, X. Shi and H. Guo, Anal. Chem., 2018, 90, 11892–11898 CrossRef CAS PubMed.
- D. Soloveichik, G. Seelig and E. Winfree, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 5393–5398 CrossRef CAS PubMed.
- G. Seelig, D. Soloveichik, D. Zhang and E. Winfree, Science, 2006, 314, 1585–1588 CrossRef CAS PubMed.
- A. J. Turberfield, J. C. Mitchell, B. Yurke, A. J. Mills, M. I. Blakey and F. C. Simmel, Phys. Rev. Lett., 2003, 90, 118102 CrossRef CAS PubMed.
- D. Zhang, A. J. Turberfield, B. Yurke and E. Winfree, Science, 2007, 318, 1121–1125 CrossRef CAS PubMed.
- W. Engelen, L. H. Meijer, B. Somers and M. Merkx, Nat. Commun., 2017, 8, 14473 CrossRef CAS PubMed.
- P. Peng, L. Shi, H. Wang and T. Li, Nucleic Acids Res., 2016, 45, 541–546 CrossRef PubMed.
- Q. Feng, Y. Guo, J. Xu and H. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 17637–17644 CrossRef CAS PubMed.
- Y. Lv, L. Cui, R. Peng, Z. Zhao, L. Qiu, H. Chen, C. Jin, X. Zhang and W. Tan, Anal. Chem., 2015, 87, 11714–11720 CrossRef CAS PubMed.
- K. Shi, B. Dou, C. Yang, Y. Chai, R. Yuan and Y. Xiang, Anal. Chem., 2015, 87, 8578–8583 CrossRef CAS PubMed.
- D. Hu, Z. Huang, F. Pu, J. Ren and X. Qu, Chem. – Eur. J., 2011, 17, 1635–1164 CrossRef CAS PubMed.
- Y. Peng, X. Wang, Y. Xiao, L. Feng, C. Zhao, J. Ren and X. Qu, J. Am. Chem. Soc., 2009, 131, 13813–13818 CrossRef CAS PubMed.
- F. Pu, Z. Huang, D. Hu, J. Ren, S. Wang and X. Qu, Chem. Commun., 2009, 47, 7357–7359 RSC.
- T. Hou, W. Li, X. Liu and F. Li, Anal. Chem., 2015, 87, 11368–11374 CrossRef CAS PubMed.
- H. Li, J. Chang, P. Gai and F. Li, ACS Appl. Mater. Interfaces, 2018, 10, 4561–4568 CrossRef CAS PubMed.
- J. Ren and J. B. Chaires, Biochemistry, 1999, 38, 16067–16075 CrossRef CAS PubMed.
- H. Arthanari, S. Basu, T. L. Kawano and P. H. Bolton, Nucleic Acids Res., 1998, 26, 3724–3728 CrossRef CAS PubMed.
- J. Zhao, C. Chen, L. Zhang, J. Jiang, G. Shen and R. Yu, Analyst, 2013, 138, 1713–1718 RSC.
- J. Chen and S. Zhou, Biosens. Bioelectron., 2016, 77, 277–283 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. S1–S3 and Table S1. See DOI: 10.1039/c8an02340a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |