Integration of a 3D-printed read-out platform with a quantum dot-based immunoassay for detection of the avian influenza A (H7N9) virus‡
Abstract
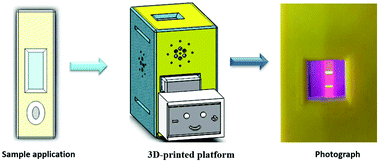
Outbreaks and potential epidemics of the highly pathogenic avian influenza virus pose serious threats to human health and the global economy. As such, its timely and accurate detection is critically important. In the present study, positive hybridoma cells (6B3) were obtained, which were used to secrete high-titer avian influenza virus (AIV) H7N9 monoclonal antibodies (H7N9 mAb). Based on these mAbs, quantum dot-based lateral flow immunochromatographic strips (QD-LFICS) were developed for AIV H7N9 detection. Under optimized conditions, results from a commercial fluorescent strip reader indicated that the limit of detection of QD-LFICS was 0.0268 HAU. To achieve rapid on-site testing, a mini 3D-printed read-out platform was fabricated to allow observation of QD-LFICS by the naked eye. More importantly, QD-LFICS were found to be practical and specific for the detection of actual samples compared with a real-time polymerase chain reaction.
- This article is part of the themed collection: Coronavirus articles - free to access collection


 Please wait while we load your content...
Please wait while we load your content...