Open Access Article
Open Access ArticleSERS-based cascade amplification bioassay protocol of miRNA-21 by using sandwich structure with biotin–streptavidin system†
Zhaoheng
Liang
a,
Jun
Zhou
 *a,
Lucia
Petti
*a,
Lucia
Petti
 b,
Liyang
Shao
c,
Tao
Jiang
b,
Liyang
Shao
c,
Tao
Jiang
 a,
Yanping
Qing
d,
Shusen
Xie
e,
Guanyi
Wu
a and
Pasquale
Mormile
b
a,
Yanping
Qing
d,
Shusen
Xie
e,
Guanyi
Wu
a and
Pasquale
Mormile
b
aInstitute of Photonics, Faculty of Science, Ningbo University, Ningbo 315211, Zhejiang, China. E-mail: zhoujun@nbu.edu.cn
bInstitute of Applied Sciences & Intelligent Systems-ISASI, CNR, Via Campi Flegrei 34, 80078 Pozzuoli, Napoli, Italy
cDepartment of Electrical and Electronic Engineering, Southern University of Science and Technology, Shenzhen 518055, Guangdong, China
dAffiliated Hospital, School of Medicine, Ningbo University, Ningbo 315211, Zhejiang, China
eKey Laboratory of Optoelectronic Science & Technology for Medicine of Ministry of Education, Provincial key Laboratory for Photonics Technology, Fujian Normal University, Fuzhou 350007, Fujian, China
First published on 21st January 2019
Abstract
In our bioassay protocol, the Ag@4MBA@DNA-biotin probes were synthesized by linking biotin-modified DNA and 4-mercaptobenzoic acid-covered Ag nanoparticles, and the Si@Ag@anti-digoxin/digoxin-DNA substrate was fabricated by immune linking of digoxin-DNA and anti-digoxin immobilized on a Ag-coated wafer. Then, the probes, miRNA-21 and the substrate were constructed into a “sandwich structure” to detect the variation in the SERS signals with respect to miRNA-21 concentrations. Next, streptavidin and extra probes were alternately introduced to implement the cascade amplification of the SERS signal to increase the detection sensitivity. The results show that the limit of detection (LOD) with cascade amplification is remarkably improved from 97.81 pM to 38.02 fM, which is three orders of magnitude higher than the original data without cascade amplification. Furthermore, the SERS-based cascade amplification mechanism was analyzed and is attributed to the “hot spots effect” of the noble metal nanostructure. The biotin-streptavidin (B–S) system was introduced into the SERS detection platform, and the novel SERS-based cascade amplification bioassay protocol has significant creativity for the detection of nucleic acids.
1 Introduction
A nucleic acid, also called a polynucleotide, is a biological macromolecule formed by polymerizing a plurality of nucleotides, which is not only an indispensable substance for protein biosynthesis, but also a material basis for biological inheritance.1 Over the decades, nucleic acid research has made great progress and has involved various fields of life sciences.2 It is well known that there are two kinds of nucleic acid types, namely, ribonucleic acid (RNA) and deoxyribonucleic acid (DNA), which both play a major role in the growth, development, reproduction, inheritance and variation of organisms.3 Typically, in terms of the growth and gene regulation of cancer cells, circulating tumor DNA (ctDNA) and microRNA (miRNA) have been revealed to have cancer-specific biological features and have emerged as potential biomarkers for cancer detection.4–7 Moreover, some highly sensitive analytical techniques have been developed to accurately detect the amount of nucleic acid as a tumor marker.8 For example, Zhang et al. proposed a gold nanoparticle-based chronocoulometric DNA sensor to conveniently analyze breast cancer-associated BRCA-1 mutant DNA and achieved a limit of detection (LOD) of as low as 10 fM.9 Kilic et al. designed an enzyme-based electrochemical miRNA sensor that can rapidly and selectively detect miRNA-21 in real total RNA samples without any need for isolation or pre-concentration of miRNA-21.10 Although these techniques have been used to perform qualitative and quantitative analysis of nucleic acids, the urgent need for a “point of care test” in clinical cancer diagnosis still requires exploration of rapid and ultrasensitive detection methods for cancer-specific genes. Due to the advantages of non-destructive and highly-sensitive fingerprint spectroscopy analysis, surface-enhanced Raman scattering (SERS) technology can satisfy the requirement of trace detection of analytes and has great potential for application in clinical medicine.11Currently, the SERS-based bioassay has attracted considerable interest in detecting tumor markers of proteins and nucleic acid species.12 For instance, a SERS-based multiplex immunoassay was proposed for simultaneously detecting tumor markers and achieved a lower limit of detection (LOD) of 3.38 and 4.87 fg mL−1 for PSA and AFP, respectively.13 A silicon-based reproducible and active surface-enhanced Raman scattering substrate was prepared by growing silver nanoparticles in situ on a silicon wafer and was employed to construct high-performance sensor for multiplex DNA detection.14 Nevertheless, there are still many challenges for the development of the SERS-based bioassay for tumor markers, especially, increasing the detection sensitivity, which is an important issue in practical bioassays.15 In fact, the SERS-based bioassay often relies on a bio-modified metal substrate to capture target molecules and form a sandwich structure with Raman-tag nanoprobes for increasing the Raman signals drastically.16–18 In recent years, an ultra-sensitive sensor based on the hairpin structure molecular beacon and high-performance SERS substrate has also achieved great success in improving the sensitivity.19 As a new development, some signal amplification solutions were proposed and successfully applied to the SERS-based bioassay. Typically, Li et al. have achieved the ultrasensitive SERS detection of nucleic acids by an Exonuclease III-boosted cascade signal amplification strategy20 and the triple signal amplification of the bimetallic cavity, gold islands and the target-catalyzed hairpin assembly.21 Indeed, besides the sandwich SERS structure and molecular beacon-based SERS sensor, the above signal amplification strategies offer beneficial insight for further improving the detection sensitivity of the SERS-based bioassay. Therefore, it is desirable to design more signal amplification solutions to implement the ultrasensitive SERS detection of nucleic acids.
Furthermore, in many efforts to raise the detection sensitivity, the biotin–streptavidin (B–S) system composed of a biotin-linked reactant and a tag-labeled streptavidin, as an effective signal amplification model, has been widely used to quantitatively detect various analytes.22,23 For example, the B–S-amplified enzyme-linked immunosorbent assay (ELISA) was proposed for the determination of chloramphenicol residues in milk and the LOD of 42 pg mL−1 was obtained, which is 8-fold more sensitive than the traditional ELISA.24 The homogeneous immunoassay has been successfully developed to determine the content of aflatoxin B1 in agricultural products by coupling immunomagnetic beads and the B–S system, with a low LOD of 5.79 pg mL−1 and a wide detection range from 0.0183 to 17.9 ng mL−1.25 A rapid and sensitive miRNA detection was implemented by the sandwich hybridization and the laminar flow-assisted dendritic amplification at a low LOD of 0.5 pM due to the multiple signal-amplification between the fluorescein isothiocyanate-labeled streptavidin and the biotinylated anti-streptavidin.26 Based on the hybridization chain reaction and B–S signal amplification protocol, a sandwich-type electrochemical immunosensor was designed to implement the multiplex immunoassay of biomarkers and high sensitivities with LODs of 62, 48, 77 and 60 fg mL−1 were acquired for a-fetoprotein (AFP), carcinoembryonic antigen (CEA), carbo-hydrate antigen (CA125) and prostate special antigen (PSA), respectively.27 A microfluidic sandwich immunoassay platform was developed to detect free PSA, and the LOD was reduced from 21.4 to 2.7 ng mL−1 by using an additional bio-affinity amplification strategy of the B–S system.28 However, to our knowledge, although many bioassay strategies have adopted the B–S system, a SERS-based bioassay grafted with a B–S system has not yet been explored.
In this study, by introducing the B–S system, we proposed the SERS-based cascade amplification bioassay protocol to achieve highly sensitive detection of miRNA-21. First, a biotin-modified DNA (DNA-biotin) was linked with 4-mercaptobenzoic acid-covered Ag nanoparticles (Ag@4MBA NPs) to synthesize the Ag@4MBA@DNA-biotin probes and the electron-beam physical vapor deposition (EBPVD) technology was used to fabricate the Ag-film substrate. Next, the probe DNA linked to the Ag@4MBA NPs and the capture DNA immobilized on the Ag-film substrate were hybridized with target RNA to construct a sandwich structure. Then, streptavidin and the Ag@4MBA@DNA-biotin probes were alternately added to the above sandwich structure to form the SERS-based cascade amplification bioassay platform for the detection of miRNA-21. Herein, the SERS-based cascade amplification effect is produced by the B–S system composed of streptavidin and Ag@4MBA@DNA-biotin probes and is attributed to the “hot spots effect” of the noble metal nanostructure. That is, when Ag NPs are arranged in the closely spaced configurations of the B–S system, more hot spots are formed at the nanoparticle gaps to generate a high-intensity electromagnetic field that greatly enhances the SERS signal.29,30 The experimental results show that the quantitative detection of miRNA-21 has a wide concentration range from 100 fM to 100 nM and an ultralow LOD of 38.02 fM. The specificity of the bioassay protocol was verified by using non-complementary RNA (miRNA-106a), single-base mismatch miRNA and double-base mismatch miRNA in our control experiment. Moreover, compared with the complex fabrication processes of the SERS substrates in other detection platforms,31–33 the simple fabrication process of the Ag-film substrate is an attractive merit due to the direct deposition of Ag film onto the polished surface of the silicon wafer by EBPVD. Therefore, the SERS-based cascade amplification bioassay protocol will have great significance and potential applications in the detection of nucleic acids.
2 Experimental
2.1 Materials and reagents
Silver nitrate (AgNO3, 99.5%), 4-mercaptobenzoic acid (4MBA), sodium citrate tribasic dihydrate, N-hydroxysuccinimide (NHS), and N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) were acquired from Sigma-Aldrich (Milwaukee, WI, USA). Bovine serum albumin (BSA) was purchased from Zeke Biotech Co., Ltd (Suzhou, China). TBS/0.05% Tween 20 buffer solution (0.05 M Tris, 0.138 M NaCl, 0.0027 M KCl, 0.05% Tween 20, pH 8) and streptavidin (SA) were obtained from Solarbio Science & Technology Co., Ltd (Beijing, China). Rabbit anti-digoxin was purchased from Biosynthesis Biotechnology Co., Ltd (Beijing, China). Diethyl pyrocarbonate-treated water (DEPC-H2O) was acquired from Generay Biotech Co., Ltd (Shanghai, China). Phosphate buffer solution (PBS, pH 7.4) was bought from Sangon Biotech Co., Ltd (Shanghai, China). All oligonucleotides were purchased from Generay Biotech Co., Ltd (Shanghai, China), and the sequences of the oligonucleotides are shown in Table 1. All the chemicals used were of analytical grade, and the solutions were prepared with Milli-Q quality water (resistivity of 18.2 MΩ cm−1) or absolute ethanol.| Oligonucleotide | Sequence |
|---|---|
| Probe DNA (amino and biotin conjugated DNA) | 5′-Amino-TCAACATCAGT-Biotin-3′ |
| Capture DNA (digoxin conjugated DNA) | 5′-CTGATAAGCTA-Digoxin-3′ |
| Target RNA (miRNA-21) | 5′-UAGCUUAUCAGACUGAUGUUGA-3′ |
| Control RNA (miRNA-106a) | 5′-AAAAGUGCUUACAGUGCAGGUAG-3′ |
| Single-base mismatch miRNA | 5′-UAGCUUAACAGACUGAUGUUGA-3′ |
| Double-base mismatch miRNA | 5′-UAGCUUAACAGACUGAGGUUGA-3′ |
2.2 Synthesis of Ag NPs
Ag NPs were synthesized by the hydrothermal reduction of silver nitrate using Lee and Meisel's method with some modifications.34 First, 9 mg of AgNO3 was dissolved in 50 mL of ultrapure water in a flask, and the prepared aqueous solution was heated to 90 °C under magnetic stirring with a 2.5 cm × 0.5 cm stir bar at a rotation speed of 300 rpm. Next, 1 mL of trisodium citrate solution (1%, w/v) was gradually added into the flask and the solution color rapidly changed from colorless to gray. Immediately, the mixture was heated until boiling and continuously reacted for 2 h. Lastly, the Ag NPs were obtained when the solution became grayish green. Subsequently, the Ag colloid was cooled to room temperature and washed more than twice by centrifugation at 8000 rpm for 15 min to remove the anionic surfactant. Finally, the centrifuged sediment was dispersed in 20 mL absolute ethanol for further use.2.3 Preparation of Ag@4-MBA@DNA-biotin probes
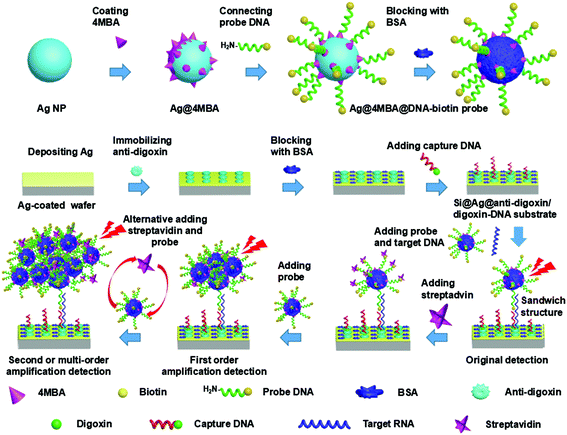
The preparation process of the Ag@4MBA@DNA-biotin probes is illustrated in Fig. 1. The specific steps are as follows. (1) Coating of 4MBA onto Ag NPs: 10 μL of 4MBA solution (10 mM, dissolved in absolute ethanol) was first added to 3 mL of the Ag colloid with ultrasonic treatment for 5 min; this solution was then kept for 2 h at room temperature. Next, the prepared Ag@4MBA NPs were washed by centrifugation and the sediment was resuspended in 3 mL of ultrapure water with an ultrasonic treatment of 30 min. Then, the monodispersed Ag@4MBA NP solution was obtained. (2) Linking of the probe DNA and Ag@4MBA NPs: 10 μL of EDC (4 mg mL−1) and 10 μL of NHS (2 mg mL−1) were added to the prepared Ag@4MBA NPs solution (∼0.3 mg mL−1) with gentle shaking at 37 °C for 30 min to activate the carboxyl group of 4MBA. The reactants were then washed by centrifugation and dispersed in 3 mL of ultrapure water with an ultrasonic treatment for 20 min. Afterwards, probe DNA (0.5 OD, diluted in 30 μL of DEPC-H2O) was dispersed in 200 μL of TBS/0.05% Tween@20 buffer solution and then added to the above mixture solution and incubated at 37 °C for 3 h. The products were then washed twice by centrifugation to remove the excess reagents. Lastly, the sediment was ultrasonically dispersed into 3 mL of ultrapure water to obtain the monodisperse Ag@4MBA@DNA-biotin NPs solution. It is worth noting that the concentration of the probe DNA (0.5 OD) is sufficient to fully link with Ag@4MBA NPs as described in part 1 of ESI.† (3) Blocking the remnant active sites and the nonspecific adsorption sites: 10 μL BSA (3% w/v) was added to the Ag@4MBA@DNA-biotin NP solution at 37 °C for 3 h; then, the mixture was washed at least three times by centrifugation to remove the excess BSA, and the precipitate was resuspended in 3 mL of ultrapure water. Finally, the Ag@4MBA@DNA-biotin probe was obtained by an ultrasonic treatment for 30 min. The probes were then stored at 4 °C for later use. The centrifugation parameters (8000 rpm, 15 min) were the same in all of the washing steps listed above.2.4 Preparation of the Si@Ag@anti-digoxin/digoxin-DNA substrate
The fabrication process of the Si@Ag@anti-digoxin/digoxin-DNA substrate is also shown in Fig. 1. First, a cleaned wafer was modified via plasma treatment (15 Pa, 35 W of radio power) for 15 min, and then 100 nm-thick Ag film was thermally deposited onto the surfaces of the wafer using the EBPVD system. Subsequently, the saturation concentrations of the anti-digoxin and capture DNA were estimated by the steps described in part 2 of ESI† and were set as 1 μg mL−1 and 1.5 μM, respectively. Then, the Ag-coated wafer was immersed in 700 μL of rabbit anti-digoxin solution (1 μg mL−1, diluted by PBS), and immediately immobilized at 4 °C and 60% relative humidity overnight.35 Subsequently, the substrate was washed successively with a TBS/0.05% Tween@20 buffer solution, PBS, and ultrapure water to remove any redundant proteins, followed by drying under a gentle flow of nitrogen gas. Third, the nonspecific adsorption sites on the surface of the substrate were blocked via treatment with a blocking solution (3% BSA, diluted with PBS) at room temperature for more than 3 h. Subsequently, the substrate was successively washed with TBS/0.05% Tween@20 buffer solution, PBS (pH 7.4), and ultrapure water to remove residual BSA and dried again with nitrogen gas. Fourth, the substrate was immersed in the capture DNA solution (1.5 μM, diluted by DEPC-H2O) and allowed to stand for 3 h to ensure completion of the reaction between digoxin and anti-digoxin. After washing the substrate with PBS (pH 7.4) and ultrapure water, the Si@Ag@anti-digoxin/digoxin-DNA substrate was finally obtained and stored at 4 °C for later use.2.5 Detection protocol
As schematically depicted in Fig. 1, the SERS-based cascade amplification bioassay protocol was performed by imitating ELISA and grafting the B–S system. First, the Si@Ag@anti-digoxin/digoxin-DNA substrate was immersed in the target RNA DEPC-H2O solution and placed in a constant temperature shaker at 15 °C for 20 min (the hybridization time and temperature are related to the chain length of the oligonucleotide and the hybridization environment).36,37 The products were quickly washed twice with ultrapure water. Next, the cleaned Si@Ag@anti-digoxin/digoxin-DNA substrate was immersed in the Ag@4MBA@DNA-biotin probe solutions, and the mixture was similarly placed in the constant temperature shaker at 15 °C for 20 min. Subsequently, the substrate was washed successively with PBS and ultrapure water to remove redundant probes. At this point, the sandwich complex composed of the Ag@4MBA@DNA-biotin probes, target RNA and the Si@Ag@anti-digoxin/digoxin-DNA substrate was used for the original detection of the SERS signal. Furthermore, by the alternate addition of streptavidin and the Ag@4MBA@DNA-biotin probes to the sandwich structure, the B–S system was grafted onto the sandwich complex to enable ultrasensitive detection with SERS cascade amplifications.2.6 Apparatus
Scanning electron microscopy (SEM) images were obtained by a field-emission scanning electron microscope (SU-70, Hitachi, Japan) at an accelerating voltage of 5 kV. The UV–visible absorption spectra were measured by a spectrometer (TU1901, Pgeneral, China). ζ-Potential measurements were performed with a nanoparticle size analyzer (Zetasizer Nano S90, Malvern, United Kingdom). The surface of the silicon wafer was modified using a low pressure plasma system (Zepto, Diener electronic, Germany) at a vacuum pressure of 15 Pa and a radio power of 35 W. Ag film was deposited by the electron-beam physical vapor deposition (EBPVD) system (Auto306, HHV Ltd, United Kingdom) at a rate of 0.4 nm s−1 and a vacuum pressure of 5 × 10−4 Pa. Atomic force microscopy (AFM) images were measured using an atomic force microscope (Cypher-HV, Oxford Instruments Asylum Research, Inc., USA) in contact mode. The SERS signals were measured by using a Raman spectrometer (BWS415, B&W Tek Inc., USA) equipped with a semiconductor laser (785 nm, 499.95 mW), a grating rate of 1200 lines per mm, and a charge coupled device (CCD, 2048 pixels) detector. All the Raman signals were collected under 25 mW laser power, 20× objective lens (numerical aperture = 0.4), and 10 s accumulation time. A baseline correction of the spectra was performed using BWS software to remove the fluorescence background. Each SERS spectrum was obtained by averaging tested data at 30 random sites on the substrate to eliminate the influence of outliers.3 Results and discussion
3.1 Characterizations of Ag NPs and Ag@4MBA@DNA-biotin probes
The SEM image of the synthesized Ag NPs is shown in Fig. 2(a). From the SEM image, we can see that most of the Ag NPs are spherical and have a particle size of about 40 nm. The formation process of the Ag@4MBA@DNA-biotin probes was investigated by UV-vis absorption spectroscopy and ζ-potential measurements. As shown in Fig. 2(b), the absorption peaks of Ag NPs, Ag@4MBA NPs and Ag@4MBA@DNA-biotin probes are located at 408, 416 and 421 nm, respectively. Clearly, the absorption peaks of the Ag@4MBA NPs and Ag@4MBA@DNA-biotin probes have significant red-shifts with respect to the absorption peak of the Ag NPs, which suggests that the size of the nanoparticles gradually increases during the synthesis of the probe and confirms that 4MBA molecules and probe DNA are successively linked to the Ag NPs.38 Furthermore, as listed in Table 2, during the immobilization of 4MBA molecules, the zeta-potential of the Ag NPs changed from −20.7 to −10.9 mV as many weakly electrolyzed 4MBA molecules bonded to the surface of the Ag NPs to replace the negatively charged citrates. After linking the probe DNA to the Ag@4MBA NPs, the surface potential of the Ag@4MBA@DNA-biotin probe was −21.0 mV due to the electronegativity of DNA molecules.39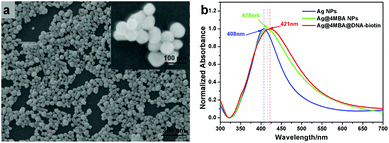 | ||
| Fig. 2 (a) SEM image of Ag NPs. (b) UV-vis absorption spectra of Ag NPs (blue), Ag@4MBA NPs (green) and Ag@4MBA@DNA-biotin probes (red). | ||
| Samples | ζ-Potential (mV) |
|---|---|
| Ag NPs | −20.7 ± 2.15 |
| Ag@4MBA NPs | −10.9 ± 2.69 |
| Ag@4-MBA@DNA-biotin probe | −21.0 ± 1.54 |
3.2 Characterizations of the Si@Ag@anti-digoxin/digoxin-DNA substrate
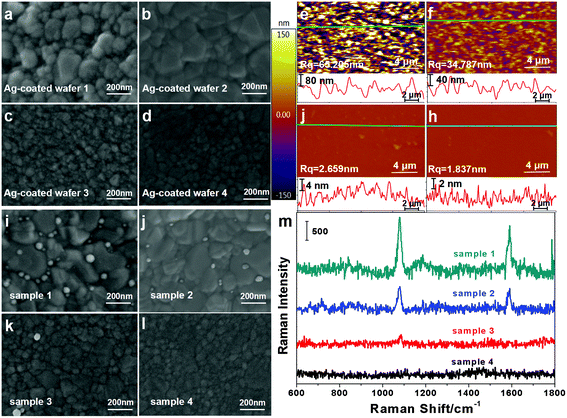
As described in section 2.4, the Si@Ag@anti-digoxin/digoxin-DNA substrates were successfully fabricated using the EBPVD technique, protein binding and immune reactions. In order to prevent the non-specific adsorption of probes from interfering with the detection results, a set of control experiments were performed as follows. First, the Ag-coated wafers with 100 nm-thick Ag film were fabricated at different deposition rates, and their SEM images are shown in Fig. 3(a–d), respectively. The roughness of these Ag-coated wafers was measured by AFM, and the AFM images are shown in Fig. 3(e–h). The roughness of the Ag-coated wafers decreases from 65.205 nm to 1.837 nm as the deposition rate of Ag increases from 0.05 to 0.4 nm s−1. The Ag-coated wafers were then used to prepare the Si@Ag@anti-digoxin/digoxin-DNA substrates. After immersion in the Ag@4MBA@DNA-biotin probe solution for 20 min, the substrates were filtered and washed carefully to function as control samples. Fig. 3(i–l) show the SEM images of the control samples, which shows that the number of probes adsorbed onto the substrates decreases as the roughness of the Ag-coated wafers decreases. Additionally, Fig. 3(m) shows the Raman spectra of the control samples and shows that the SERS signal of the samples gradually declines (samples 1–3) and ultimately disappears (sample 4) as the number of absorbed probes decreases. This result demonstrates that the substrate fabricated by the smooth Ag-coated wafers exhibits an excellent characteristic of low non-specific adsorption to the probes. Thus, the Ag-coated wafer fabricated at a deposition rate of 0.4 nm s−1 was used in our subsequent experiments.3.3 Cascade SERS amplification-based bioassay of miRNA-21
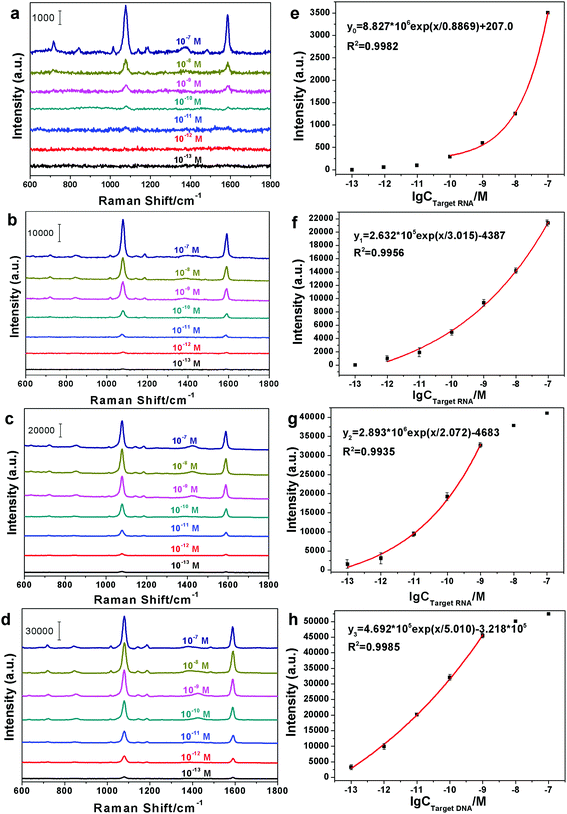
According to the SERS-based cascade amplification bioassay protocol described in section 2.5, the miRNA-21 was detected by measuring the SERS signal of 4MBA in the sandwich structure during the original detection, first order amplification detection and multi-order amplification detection. In our experiments, the 100 nM miRNA-21 solution was diluted with DEPC-H2O at room temperature to prepare 7 standard samples with a 10-fold dilution ratio from 1 to 106. Corresponding to the initial detection and multi-order amplification, the SERS spectra of the standard samples were recorded and are shown in Fig. 4(a)–(d), respectively. It can be clearly seen from Fig. 4(a) that the SERS signals decrease with a decrease in the miRNA-21 concentrations and show a negligible weak background signal at concentrations of 10−11 M and below. Compared with Fig. 4(a), the SERS spectra in Fig. 4(b) exhibit stronger SERS signals for various concentrations of samples, except for the sample with a concentration of 10−13 M. These SERS signals can be attributed to the amplification role of the B–S system because the extra probes added to the original sandwich structure by specific binding between biotin and streptavidin results in more “hotspots” to produce the distinct Raman enhancement. Similarly, Fig. 4(c) and (d) present stronger SERS signals for all of the samples due to multiple amplifications from the B–S system. Thus, the sandwich complexes grafted to the B–S system provide superior SERS performance than that of the original sandwich structure.Subsequently, the peaks at 1078 cm−1 in Fig. 4(a)–(d) were selected for generating the standard dose–response curve for miRNA-21, and their peak intensities as a function of the logarithm of the concentrations are presented in Fig. 4(e)–(h), respectively. As shown in Fig. 4(e), within the concentration range from 100 pM to 100 nM, the SERS intensity of the designated peak exponentially changes with the concentration of miRNA-21 according to the curve fitting equation y0 = 8.827 × 106![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) exp(x/0.8869) + 207.0, and the LOD of 97.81 pM can be estimated using the blank determination method.40 Similarly, the change in the SERS peak intensity in Fig. 4(f) is also exponential within the concentration range of 1 pM–100 nM, and the LOD = 0.7825 pM. Comparing Fig. 4(f) with (e), the sandwich structure with the B–S system clearly has a wider detection range and a lower LOD than those of the sandwich structure without the B–S system. From Fig. 4(g) and (h), it is clearly seen that the standard dose–response curves still display exponential variation between 100 fM and 1 nM and their LOD are 65.77 fM and 38.02 fM, respectively. It is worth noting that although the values of peak intensities at 10 nM and 100 nM have large deviations with the exponential fitting curves in Fig. 4(g) and (h), there are no effects on the practical applications due to the lack of multiple amplifications for the high concentrations of miRNA-21. In other words, as shown in Fig. 4(e), the original sandwich structure is sufficient for the testing requirements above 100 pM. Also, as described in part 3 of ESI†,† the LOD (42.95 fM) of the SERS-based bioassay with fourth-order amplification is higher than that of the third-order amplification (38.02 fM). Thus, the third order amplification was selected as an optimal amplification order in our detection scheme. Therefore, the appropriate multiple amplifications provided by the B–S system can further reduce the LOD for highly sensitive trace detection of miRNA-21.
exp(x/0.8869) + 207.0, and the LOD of 97.81 pM can be estimated using the blank determination method.40 Similarly, the change in the SERS peak intensity in Fig. 4(f) is also exponential within the concentration range of 1 pM–100 nM, and the LOD = 0.7825 pM. Comparing Fig. 4(f) with (e), the sandwich structure with the B–S system clearly has a wider detection range and a lower LOD than those of the sandwich structure without the B–S system. From Fig. 4(g) and (h), it is clearly seen that the standard dose–response curves still display exponential variation between 100 fM and 1 nM and their LOD are 65.77 fM and 38.02 fM, respectively. It is worth noting that although the values of peak intensities at 10 nM and 100 nM have large deviations with the exponential fitting curves in Fig. 4(g) and (h), there are no effects on the practical applications due to the lack of multiple amplifications for the high concentrations of miRNA-21. In other words, as shown in Fig. 4(e), the original sandwich structure is sufficient for the testing requirements above 100 pM. Also, as described in part 3 of ESI†,† the LOD (42.95 fM) of the SERS-based bioassay with fourth-order amplification is higher than that of the third-order amplification (38.02 fM). Thus, the third order amplification was selected as an optimal amplification order in our detection scheme. Therefore, the appropriate multiple amplifications provided by the B–S system can further reduce the LOD for highly sensitive trace detection of miRNA-21.
Considering the false positive signal induced by non-complementary RNA, the selectivity of the cascade SERS amplification-based bioassay protocol was examined. In our control experiments, the SERS signals of the miRNA-21, single-base mismatch miRNA, double-base mismatch miRNA, miRNA-106a and blank samples were detected and are shown in Fig. 5(a). The results demonstrate that the strong characteristic SERS signal of the Ag@4MBA@DNA-biotin probes was only generated in the bioassay of the miRNA-21 sample, while very weak signals and even background noises were obtained for the blank samples, non-complementary RNA (miRNA-106a), double-base mismatch miRNA and single-base mismatch miRNA. Therefore, the constructed SERS-based sandwich structure grafted onto the B–S system is a highly specific platform for detecting miRNA-21.
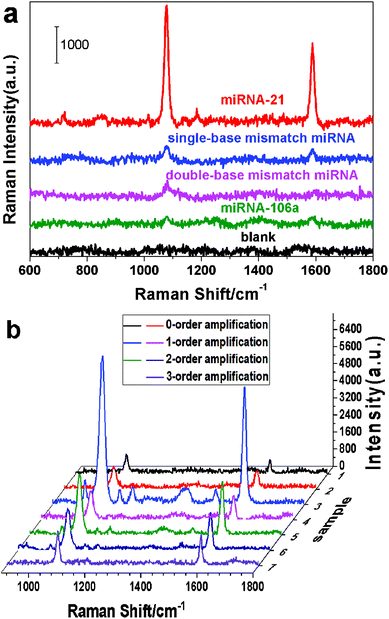 | ||
| Fig. 5 (a) SERS spectra of Ag@4MBA@DNA-biotin probes in the specificity control experiments; (b) the SERS spectra of the seven samples recorded by the SERS-based bioassay platform. | ||
Finally, to check the reliability of the above SERS-based bioassay platform, different concentrations of miRNA-21 samples were detected using both the SERS-based cascade amplification bioassay protocol and the RT-qPCR technology. Fig. 5(b) shows the multi-order amplified SERS spectra of the seven samples. Combining the results in Fig. 5(b) with the standard dose–response curves (Fig. 4(e–h)), the concentration of miRNA-21 in each sample was obtained and are listed in the second column of Table 3. At the same time, the sample concentrations detected by the RT-qPCR technology are listed in the fourth column of Table 3. The results obtained by the SERS-based cascade amplification bioassay protocol are consistent with those of RT-qPCR and display higher detection sensitivities. Furthermore, the amplification orders in Table 3 show that the samples with high concentrations of miRNA-21 (samples 1–2) can be directly detected by the sandwich structure without the B–S system. However, the miRNA-21 concentrations in samples 3–7 were obtained using the multiple-order amplification of the BS system due to their low miRNA-21 content. Additionally, the data listed in Table S3 of the ESI† also demonstrate that the SERS-based sandwich platform with the B–S system provides improved detection of miRNA-21. Therefore, the proposed SERS-based cascade amplification bioassay is a valid strategy for detecting miRNA-21.
| Samples | SERS-based bioassay | RT-qPCR | Relative deviation | ||
|---|---|---|---|---|---|
| Concentrations | Amplification orders | Concentrations | Cycle threshold | ||
| Notes: Symbol “—” denotes no valid data. | |||||
| 1 | 4.21 nM | 0 | 4.33 nM | 13.64739 | −2.77% |
| 2 | 6.53 nM | 0 | 5.97 nM | 13.12042 | 9.38% |
| 3 | 329.43 pM | 1 | 309.79 pM | 17.95382 | 6.34% |
| 4 | 2.95 pM | 1 | 2.71 pM | 25.69209 | 8.86% |
| 5 | 493.57 fM | 2 | 466.57 fM | 28.56744 | 5.79% |
| 6 | 257.42 fM | 2 | — | — | — |
| 7 | 66.29 fM | 3 | — | — | — |
4. Conclusions
In summary, a novel oligonucleotide detection approach is proposed that involves the coupling of SERS technology with the cascade amplification of B–S system. This approach was applied for the detection of miRNA-21. The SERS-based sandwich structure was composed of Si@Ag@anti-digoxin/digoxin-DNA substrates, miRNA-21 and a Ag@4MBA@DNA-biotin probe, and the cascade amplification of the B–S system was implemented by binding the Ag@4MBA@DNA-biotin probe and streptavidin to improve the detection sensitivity. Also, with the multi-order amplification detection, the LOD decreased by three orders of magnitude from 97.81 pM to 38.02 fM and exhibited a wider detection range from 100 fM to 100 nM after the original sandwich structure was grafted onto the B–S system. The control experiments exhibited superior detection selectivity for miRNA-21, and the reliability of this SERS-based cascade amplification bioassay protocol was examined by RT-qPCR. In our experiments, the smooth Ag-coated wafer was selected for preparing the Si@Ag@anti-digoxin/digoxin-DNA substrate as it reduced the non-specific adsorption between the probe and the substrate. However, the sensitivity of our platform was sacrificed to some extent due to the weak SERS effect of the smooth Ag-coated wafer. In addition, by adjusting the sequences of the probe DNA and the capture DNA, our detection protocol may be extended to the detection of various genes with many potential applications in clinical diagnostics.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant No. 61320106014, 61335011, 61475128 and 61675104), Natural Science Foundation of Zhejiang Province (Grant No. LY19F050002), and the K. C. Wong Magna Foundation of Ningbo University, China.Notes and references
- B. Alberts, A. Johnson, J. Lewis, D. Morgan, M. Raff, K. Roberts and P. Walter, Molecular Biology of the Cell, Garland Science, New York, 6th edn, 2014 Search PubMed.
- K. C. Smith, Aging, carcinogenesis, and radiation biology: The role of nucleic acid addition reactions, Springer Science & Business Media, New York, 2013 Search PubMed.
- D. Frias-Lasserre and C. A. Villagra, The Importance of ncRNAs as Epigenetic Mechanisms in Phenotypic Variation and Organic Evolution, Front. Microbiol., 2017, 8, 02483 CrossRef PubMed.
- P. Anker, H. Mulcahy, X. Q. Chen and M. Stroun, Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients, Cancer Metastasis Rev., 1999, 18, 65–73 CrossRef CAS PubMed.
- S. L. Ameres and P. D. Zamore, Diversifying microRNA sequence and function, Nat. Rev. Mol. Cell Biol., 2013, 14, 475–488 CrossRef CAS PubMed.
- G. Siravegna, S. Marsoni, S. Siena and A. Bardelli, Integrating liquid biopsies into the management of cancer, Nat. Rev. Clin. Oncol., 2017, 14, 531–548 CrossRef CAS PubMed.
- J. Gasparello, M. Allegretti, E. Tremante, E. Fabbri, C. Azzurra Amoreo, P. Romania, E. Melucci, K. Messana, M. Borgatti, P. Giacomini, R. Gambari and A. Finotti, Liquid biopsy in mice bearing colorectal carcinoma xenografts: gateways regulating the levels of circulating tumor DNA (ctDNA) and miRNA (ctmiRNA), J. Exp. Clin. Cancer Res., 2018, 37, 124–134 CrossRef PubMed.
- H. Zhou, J. Liu, J. Xu, S. Zhang and H. Chen, Optical nano-biosensing interface via nucleic acid amplification strategy: construction and application, Chem. Soc. Rev., 2018, 47, 1996–2019 RSC.
- J. Zhang, S.-P. Song, Li.-H. Wang, D. Pan and C.-H. Fan, A gold nanoparticle-based chronocoulometric DNA sensor for amplified detection of DNA, Nat. Protoc., 2007, 2, 2888–2895 CrossRef CAS PubMed.
- T. Kilic, S. Nur Topkaya, D. Ozkan Ariksoysal, M. Ozsoz, P. Ballar, Y. Erac and O. Gozen, Electrochemical based detection of microRNA, mir21 in breast cancer cells, Biosens. Bioelectron., 2012, 38, 195–201 CrossRef CAS PubMed.
- W. E. Smith, Practical understanding and use of surface enhanced Raman scattering/surface enhanced resonance Raman scattering in chemical and biological analysis, Chem. Soc. Rev., 2008, 37, 955–964 RSC.
- Y. Wang, B. Yan and L. Chen, SERS Tags: Novel Optical Nanoprobes for Bioanalysis, Chem. Rev., 2013, 113, 1391–1428 CrossRef CAS PubMed.
- Z. Wang, H. Yang, M. Wang, L. Petti, T. Jiang, Z. Jia, S. Xie and J. Zhou, SERS-based multiplex immunoassay of tumor markers using double SiO2@Ag immune probes and gold-film hemisphere array immune substrate, Colloids Surf., A, 2018, 546, 48–58 CrossRef CAS.
- Z. Y. Jiang, X. X. Jiang, S. Su, X. P. Wei, S. T. Lee and Y. He, Silicon-based reproducible and active surface-enhanced Raman scattering substrates for sensitive, specific, and multiplex DNA detection, Appl. Phys. Lett., 2012, 100, 203104 CrossRef.
- C. Song, Y. Yang, B. Yang, Y. Sun, Y. Zhao and L. Wang, Ultrasensitive SERS sensor for simultaneous detection of multiple cancer-related miRNAs, Nanoscale, 2016, 8, 17365–17373 RSC.
- C. Zong, M. Xu, L. Xu, T. Wei, X. Ma, X. Zheng, R. Hu and B. Ren, Surface-Enhanced Raman Spectroscopy for Bioanalysis: Reliability and Challenges, Chem. Rev., 2018, 118, 4946–4980 CrossRef CAS PubMed.
- S.-C. Luo, K. Sivashanmugan, J.-D. Liao, C.-K. Yao and H.-C. Peng, Nanofabricated SERS-active substrates for single-molecule to virus detection in vitro: A review, Biosens. Bioelectron., 2014, 61, 232–240 CrossRef CAS PubMed.
- B. Guven, F. Ceyda Dudak, I. Hakki Boyaci, U. Tamer and M. Ozsoz, SERS-based direct and sandwich assay methods for mir-21 detection, Analyst, 2014, 139, 1141–1147 RSC.
- M. J. Banholzer, J. E. Millstone, L. Qin and C. A. Mirkin, Rationally designed nanostructures for surface-enhanced Raman spectroscopy, Chem. Soc. Rev., 2008, 37, 885–897 RSC.
- Y. Sun, P. Peng, R. Guo, H. Wang and T. Li, Exonuclease III-boosted cascade reactions for ultrasensitive SERS detection of nucleic acids, Biosens. Bioelectron., 2018, 104, 32–38 CrossRef CAS PubMed.
- Y. Sun and T. Li, Composition-Tunable Hollow Au/Ag SERS Nanoprobes Coupled with Target-Catalyzed Hairpin Assembly for Triple-AmplificationDetection of miRNA, Anal. Chem., 2018, 90, 11614–11621 CrossRef CAS PubMed.
- O. H. Laitinen, V. P. Hytönen, H. R. Nordlund and M. S. Kuloma, Genetically engineered avidins and streptavidins, Cell. Mol. Life Sci., 2006, 63, 2992–3017 CrossRef CAS PubMed.
- E. P. Diamandis and T. K. Christopoulos, The Biotin-(Strept)Avidin System: Principles and Applicationsin Biotechnology, Clin. Chem., 1991, 37, 625–636 CAS.
- Li Wang, Y. Zhang, X. Gao, Z. Duan and S. Wang, Determination of Chloramphenicol Residues in Milk by Enzyme-Linked Immunosorbent Assay: Improvement by Biotin-Streptavidin-Amplified System, J. Agric. Food Chem., 2010, 58, 3265–3270 CrossRef CAS PubMed.
- M. Li, Y. Zhang, R. Zhao, Z. Liu, X. Hong, Y. Cui, Y. Xue and D. Du, Immunomagnetic bead-based biotin-streptavidin system for highly efficient detection of aflatoxin B1 in agricultural products, RSC Adv., 2018, 8, 26029–26035 RSC.
- H. Arata, H. Komatsu, K. Hosokawa and M. Maeda, Rapid and Sensitive MicroRNA Detection with Laminar Flow-Assisted Dendritic Amplification on Power-Free Microfluidic Chip, PLoS One, 2012, 7, e48329 CrossRef CAS PubMed.
- Q. Zhu, Y. Chai, Y. Zhuo and R. Yuan, Ultrasensitive simultaneous detection of four biomarkers based on hybridization chain reaction and biotin–streptavidin signal ampli-fication strategy, Biosens. Bioelectron., 2015, 68, 42–48 CrossRef CAS PubMed.
- N. Madaboosi, R. R. G. Soares, V. Chu and J. Pedro Conde, A microfluidic immunoassay platform for the detection of free prostate specific antigen: a systematic and quantitative approach, Analyst, 2015, 140, 4423–4433 RSC.
- N. Pazos-Perez, C. Simone Wagner, J. M. Romo-Herrera, L. M. Liz-Marzán, F. Javier García de Abajo, A. Wittemann, A. Fery and R. A. Alvarez-Puebla, Organized Plasmonic Clusters with High Coordination Number and Extraordinary Enhancement in Surface-Enhanced Raman Scattering (SERS), Angew. Chem., Int. Ed., 2012, 51, 12688–12693 CrossRef CAS PubMed.
- N. J. Halas, S. Lal, W.-S. Chang, S. Link and P. Nordlander, Plasmons in Strongly Coupled Metallic Nanostructures, Chem. Rev., 2011, 111, 3913–3961 CrossRef CAS.
- W. Ahn, Y. Qiu and B. M. Reinhard, Generation of scalable quasi-3D metallo-dielectric SERS substrates through orthogonal reactive ion etching, J. Mater. Chem. C, 2013, 1, 3110–3118 RSC.
- K. Sivashanmugan, J. Der Liao, B. Haochih Liu and C.-K. Yao, Focused-ion-beam-fabricated Au nanorods coupled with Ag nanoparticles used as surface-enhanced Raman scattering-active substrate for analyzing trace melamine constituents in solution, Anal. Chim. Acta, 2013, 800, 56–64 CrossRef CAS PubMed.
- M. Rippa, R. Castagna, J. Zhou, R. Paradiso, G. Borriello, E. Bobeico and L. Petti, Dodecagonal plasmonic quasicrystals for phagebased biosensing, Nanotechnology, 2018, 29, 405501 CrossRef CAS PubMed.
- P. C. Lee and D. Meise, Adsorption and Surface-Enhanced Raman of Dyes on Silver and Gold Sols, J. Phys. Chem., 1982, 86, 3391–3395 CrossRef CAS.
- Lu Zhou, J. Zhou, Z. Feng, F. Wang, S. Xie and S. Bu, Immunoassay for tumor markers in human serum based on Si nanoparticles and SiC@Ag SERS-active substrate, Analyst, 2016, 141, 2534–2541 RSC.
- A. Takashima and M. Oishi, Kinetic study of DNA hybridization on DNA-modified gold nanoparticles with engineered nano-interfaces, RSC Adv., 2015, 5, 76014–76018 RSC.
- J. Sun, Y. Yan, S. Sun, X. Shu, C. Zhu and J. Zhu, DNA binding and reactivity assays based on in-frame protein expression, Chem. Sci., 2013, 4, 633–641 RSC.
- F. Zhang, M. W. A. Skoda, R. M. J. Jacobs, D. G. Dressen, R. A. Martin, C. M. Martin, G. F. Clark, T. Lamkemeyer and F. Schreiber, Gold Nanoparticles Decorated with Oligo (ethylene glycol) Thiols: Enhanced Hofmeister Effects in Colloid-Protein Mixtures, J. Phys. Chem. C, 2009, 113, 4839–4847 CrossRef CAS.
- P. L. Felgner, T. R. Gadek, M. Holm, R. Roman, H. W. Chan, M. Wenz, J. P. Northrop, G. M. Ringold and M. Danielsen, Lipofection: A highly efficient, lipid-mediated DNA-transfection procedure, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 7413–7417 CrossRef CAS.
- A. Shrivastava and V. B. Gupta, Methods for the determination of limit of detection and limit of quantitation of the analytical methods, Chron. Young Sci., 2011, 2, 21–25 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8an02259c |
| This journal is © The Royal Society of Chemistry 2019 |