Ultrastructural and SINS analysis of the cell wall integrity response of Aspergillus nidulans to the absence of galactofuranose†
Abstract
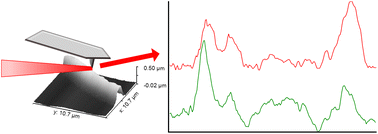
With lethal opportunistic fungal infections on the rise, it is imperative to explore new methods to examine virulence mechanisms. The fungal cell wall is crucial for both the virulence and viability of Aspergillus nidulans. One wall component, Galf, has been shown to contribute to important fungal processes, integrity of the cell wall and pathogenesis. Here, we explore gene deletion strains lacking the penultimate enzyme in Galf biosynthesis (ugmAΔ) and the protein that transports Galf for incorporation into the cell wall (ugtAΔ). In applying gene deletion technology to the problem of cell wall integrity, we have employed multiple micro- and nano-scale imaging tools, including confocal fluorescence microscopy, electron microscopy, X-Ray fluorescence and atomic force microscopy. Atomic force microscopy allows quantification of ultrastructural cell wall architecture while near-field infrared spectroscopy provides spatially resolved chemical signatures, both at the nanoscale. Here, for the first time, we demonstrate correlative data collection with these two emerging modalities for the multiplexed in situ study of the nanoscale architecture and chemical composition of fungal cell walls.
- This article is part of the themed collection: Next wave advances in single cell analyses


 Please wait while we load your content...
Please wait while we load your content...