Advancing single-cell proteomics and metabolomics with microfluidic technologies
Abstract
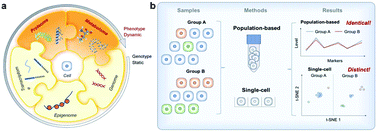
Recent advances in single-cell analysis have unraveled substantial heterogeneity among seemingly identical cells at genomic and transcriptomic levels. These discoveries have urged scientists to develop new tools that are capable of investigating single cells from a broader set of “omics”. Proteomics and metabolomics, for instance, are of particular interest as they are closely correlated with a dynamic picture of cellular behaviors and phenotypic identities. The development of such tools requires highly efficient isolation and processing of a large number of individual cells, where techniques such as microfluidics are extremely useful. Here, we review the recent advances in single-cell proteomics and metabolomics, with a focus on microfluidics-based platforms. We highlight a vast array of emerging microfluidic formats for single-cell isolation and manipulation, and how the state-of-the-art analytical tools are coupled with such platforms for proteomic and metabolomic profiling.
- This article is part of the themed collections: Recent Review Articles, In celebration of Chinese New Year 2020 and Next wave advances in single cell analyses


 Please wait while we load your content...
Please wait while we load your content...