Recent advances in single cell manipulation and biochemical analysis on microfluidics
Abstract
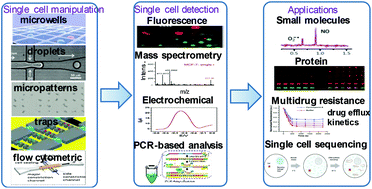
Single cell analysis has become of great interest with unprecedented capabilities for the systematic investigation of cell-to-cell variation in large populations. Rapid and multi-parametric analysis of intercellular biomolecules at the single-cell level is imperative for the improvement of early disease diagnosis and personalized medicine. However, the small size of cells and the low concentration levels of target biomolecules are critical challenges for single cell analysis. In recent years, microfluidic platforms capable of handling small-volume fluid have been demonstrated to be powerful tools for single cell analysis. In addition, microfluidic techniques allow for precise control of the localized microenvironment, which yield more accurate outcomes. Many different microfluidic techniques have been greatly improved for highly efficient single-cell manipulation and highly sensitive detection over the past few decades. To date, microfluidics-based single cell analysis has become the hot research topic in this field. In this review, we particularly highlight the advances in this field during the past three years in the following three aspects: (1) microfluidic single cell manipulation based on microwells, micropatterns, droplets, traps and flow cytometric methods; (2) detection methods based on fluorescence, mass spectrometry, electrochemical, and polymerase chain reaction-based analysis; (3) applications in the fields of small molecule detection, protein analysis, multidrug resistance analysis, and single cell sequencing with droplet microfluidics. We also discuss future research opportunities by focusing on key performances of throughput, multiparametric target detection and data processing.
- This article is part of the themed collections: Recent Review Articles, In celebration of Chinese New Year 2020 and Next wave advances in single cell analyses


 Please wait while we load your content...
Please wait while we load your content...