Molecular profiling of single axons and dendrites in living neurons using electrosyringe-assisted electrospray mass spectrometry†
Abstract
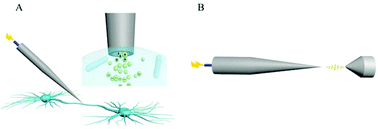
The molecular profiling of single axons and dendrites in living neurons could provide important information for the better understanding of neuron function. Here, electrosyringe-assisted electrospray mass spectrometry (MS) is established for the first time to achieve intracellular sampling from one axon or dendrite in living neurons for mass spectrometric analysis. The key is the insertion of a ∼130 nm capillary tip into one axon or dendrite to load the cytosol through electro-osmotic flow. The ionization efficiency from the nano-capillary is enhanced to guarantee mass spectrometric analysis of multiple components from the axon and dendrite. Higher levels of pyroglutamic acid and glutamic acid are revealed in the axon compared to in the body and dendrite. This uneven distribution is in accordance with the accumulation of neurotransmitters in the axon for information delivery. The achievement of electrosyringe-assisted electrospray MS is to unveil the molecular distribution in the whole living neuron, which offers the feasibility to deeply investigate molecular communication between the axon/dendrite and the body inside neurons.
- This article is part of the themed collections: Celebrating 100 years of Chemistry at Nanjing University, Recent analytical chemistry science from China and Next wave advances in single cell analyses


 Please wait while we load your content...
Please wait while we load your content...