Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Single cell analysis of proliferation and movement of cancer and normal-like cells on nanowire array substrates†
Zhen
Li‡
ab,
Sofia
Kamlund‡
 cd,
Till
Ryser
abe,
Mercy
Lard
cd,
Till
Ryser
abe,
Mercy
Lard
 ab,
Stina
Oredsson
*c and
Christelle N.
Prinz
ab,
Stina
Oredsson
*c and
Christelle N.
Prinz
 *ab
*ab
aDivision of Solid State Physics, Lund University, 221 00 Lund, Sweden. E-mail: christelle.prinz@ftf.lth.se
bNanoLund, Lund University, 221 00 Lund, Sweden
cDepartment of Biology, Lund University, 223 62 Lund, Sweden. E-mail: stina.oredsson@biol.lu.se
dPhase Holographic Imaging AB, 223 63 Lund, Sweden
eDepartment of Biomedicine, Basel University, 4058 Basel, Switzerland
First published on 10th October 2018
Abstract
Nanowires are presently investigated in the context of various biological and medical applications. In general, these studies are population-based, which results in sub-populations being overlooked. Here, we present a single cell analysis of cell cycle and cell movement parameters of cells seeded on nanowires using digital holographic microscopy for time-lapse imaging. MCF10A normal-like human breast epithelial cells and JIMT-1 breast cancer cells were seeded on glass, flat gallium phosphide (GaP), and on vertical GaP nanowire arrays. The cells were monitored individually using digital holographic microscopy for 48 h. The data show that cell division is affected in cells seeded on flat GaP and nanowires compared to glass, with much fewer cells dividing on the former two substrates compared to the latter. However, MCF10 cells that are dividing on glass and flat GaP substrates have similar cell cycle time, suggesting that distinct cell subpopulations are affected differently by the substrates. Altogether, the data highlight the importance of performing single cell analysis to increase our understanding of the versatility of cell behavior on different substrates, which is relevant in the design of nanowire applications.
Introduction
Nanowire arrays have been used extensively as cell culture substrates, for various applications such as mechanosensing,1–3 neural interfacing,4–8 and cell transfection.9–11 Nanowire arrays have been shown to be biocompatible with limited detrimental effects on cell viability, although other parameters such as cell migration and cell proliferation may be affected.12 However, like standard culture substrates, nanowire arrays are different from the in vivo cell environment. Indeed, the cell behavior differs on nanowire substrates, depending on the cell type and nanowire array geometry. For instance, L929 fibroblast cells seeded on low density (0.1 nanowires per μm2) arrays exhibit a lower motility compared to flat substrates, whereas their motility on high density arrays (4 nanowires per μm2) is similar to the one of cells seeded on flat substrates.13 The rate of proliferation of L929 cells is also lower on nanowire substrates compared to cells seeded on flat substrates.14 We have recently proposed the use of nanowire arrays to investigate cellular mechanosensing in the field of cancer research.15 We have used hexagonal arrays of vertical nanowires to measure the traction forces exerted by MCF7 breast cancer cells and MCF10A breast epithelial cells when growing on top of the array.3 In that study, we suggest that monitoring cellular traction forces is useful for evaluating the mechanisms of action of anticancer drugs. However, in order to use these arrays routinely, it is necessary to closely examine how nanowires affect the cell proliferation and migration of both cancer and normal cells. Moreover, in order to avoid cell sub-populations being overlooked, which is of special importance in cancer research, it is crucial to perform single cell analysis.Here, we assess how the proliferation and movement of human JIMT-1 breast cancer cells and normal-like MCF10A cells are affected when seeded on gallium phosphide (GaP) nanowire array substrates, compared to flat GaP and glass. To gain an increased understanding of the variability in cell behavior, and detect cell sub-populations, we have performed single cell analysis using digital holographic microscopy, in addition to bulk analysis.
Experimental
Nanowire array fabrication
Hexagonal nanowire arrays were defined by patterning a GaP(111)B substrate with gold seeds using electron beam lithography (EBL). The substrate was coated with the EBL resist SX AR-P 6200/6 (4000 rpm, 60 s) and subsequently baked on a hot plate at 180 °C for 5 min. A hexagonal array of holes at a density of 2 μm−2 was defined in the resist using EBL (VOYAGER, Raith GmbH). The holes were made using single pixel dot exposure mode (single pixel dose of 30 fAs), followed by development in n-amyl acetate for 60 s, followed by 15 s rinsing in isopropanol (IPA) and drying under nitrogen. A 20 nm thick gold layer was deposited on the substrate using thermal evaporation followed by a lift-off process in 1165 Remover at 60 °C. The sample was subsequently rinsed in IPA and dried under nitrogen. GaP nanowires were synthesized using metal organic vapor phase epitaxy (MOVPE) based on a previously reported method.16 Briefly, GaP substrates with Au nanoparticle arrays (prepared as described above) were placed in a MOVPE reactor (Aixtron 200/4) and annealed at 650 °C in a H2/phosphine atmosphere for 10 min in order to remove native oxide from the surface. The temperature was then gradually lowered to 440 °C. A 3.3 ± 0.2 μm long and 96 ± 3 nm thick GaP segment (Fig. S1, ESI†) was grown from trimethylgallium and phosphine precursors at a molar fraction of 8.9 × 10−6 and 6.9 × 10−3, respectively. The nanowire array geometry was characterized using scanning electron microscopy.Cell culture
The human breast cancer cell line JIMT-1 (ACC-589) was purchased from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany) and the human normal-like breast epithelial cell line MCF10A (CRL-10317) was purchased from the American Type Culture Collection (Manassas, VA, USA).JIMT-1 cells were routinely cultured in DMEM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ham's F-12 (1
Ham's F-12 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) medium supplemented with 10% fetal calf serum (FCS) (VWR, Sweden), 1 mM non-essential amino acids (VWR), 100 U ml−1 penicillin (VWR), 100 μg ml−1 streptomycin (VWR), 2 mM L-glutamine (VWR), and 10 μg ml−1 insulin (Sigma-Aldrich, Stockholm, Sweden). The MCF10A cells were routinely cultured in RPMI 1640 medium (VWR) containing 10% FCS (VWR), 1 mM non-essential amino acids (VWR), 100 U ml−1 penicillin (VWR), 100 μg ml−1 streptomycin (VWR), 10 μg ml−1 insulin (Sigma-Aldrich), 20 ng ml−1 epidermal growth factor (Sigma-Aldrich), 50 ng ml−1 cholera toxin (Sigma-Aldrich), and 250 ng ml−1 hydrocortisol (Sigma-Aldrich). Both cell lines were routinely passaged twice a week. The cells were kept in an incubator with 5% CO2 in air at 37 °C. For time-lapse imaging, cells were seeded on a glass, flat GaP, or vertical nanowire array substrates placed in Petri dishes (35 mm diameter, Nunc, Roskile, Denmark) at a density of 15
1) medium supplemented with 10% fetal calf serum (FCS) (VWR, Sweden), 1 mM non-essential amino acids (VWR), 100 U ml−1 penicillin (VWR), 100 μg ml−1 streptomycin (VWR), 2 mM L-glutamine (VWR), and 10 μg ml−1 insulin (Sigma-Aldrich, Stockholm, Sweden). The MCF10A cells were routinely cultured in RPMI 1640 medium (VWR) containing 10% FCS (VWR), 1 mM non-essential amino acids (VWR), 100 U ml−1 penicillin (VWR), 100 μg ml−1 streptomycin (VWR), 10 μg ml−1 insulin (Sigma-Aldrich), 20 ng ml−1 epidermal growth factor (Sigma-Aldrich), 50 ng ml−1 cholera toxin (Sigma-Aldrich), and 250 ng ml−1 hydrocortisol (Sigma-Aldrich). Both cell lines were routinely passaged twice a week. The cells were kept in an incubator with 5% CO2 in air at 37 °C. For time-lapse imaging, cells were seeded on a glass, flat GaP, or vertical nanowire array substrates placed in Petri dishes (35 mm diameter, Nunc, Roskile, Denmark) at a density of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2 for JIMT-1 cells and 5000 cells per cm2 for MCF10A cells, in 3 ml of regular growth medium.
000 cells per cm2 for JIMT-1 cells and 5000 cells per cm2 for MCF10A cells, in 3 ml of regular growth medium.
Digital holographic imaging and tracking
HoloMonitor® M4 (Phase Holographic Imaging AB (PHI), Lund, Sweden) with a motorized stage was used for time-lapse imaging. Imaging was started 24 h after seeding. Images were acquired using the software Hstudio™ (PHI) at the same position on the substrate (glass, flat GaP, or nanowires) every five minutes for 48 h. To increase the image quality, the standard lid of the Petri dish was replaced with HoloLid™ 71 110 (PHI) prior to the start of imaging. The experiments were repeated three times with each substrate and two time-lapse movies in different areas were acquired per replicate.The HoloMonitor® M4 is a quantitative imaging system based on digital holographic microscopy.17,18 In digital holography, the image is a computer reconstruction of a hologram. The hologram is acquired by the interference of two laser beams, of which one is phase-shifted due to passing through the sample and one is the original laser beam. The hologram is imprinted on a CCD-camera19 coupled to a computer. The hologram contains information about the three-dimensional (3D) sample it is imaging, in this case a 3D-reconstructed cell image. The HoloMonitor® M4 uses a low power laser (635 nm wavelength, 0.2 mW cm−2) with no associated phototoxicity, making it suitable for extended time imaging.
After image acquisition, the time-lapses were analyzed by individual cell tracking using Hstudio™. The tracking is semi-automated, using a frame-by-frame algorithm attempting to find each tracked cell in the next frame based on the centroid position. The software allows for manual changes when the algorithm fails to predict the correct position. From Hstudio™, data about cell cycle time and cell movement can be extracted.
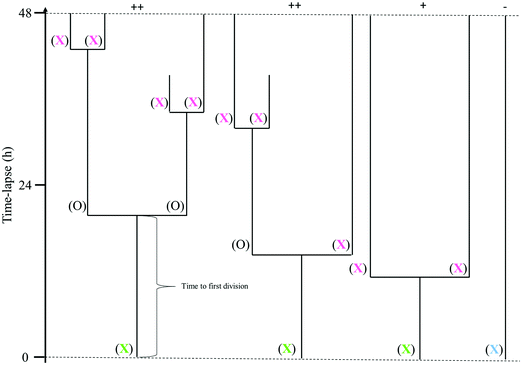
The data acquired from the tracking of cells in the digital holographic images was used to create cell family trees (Fig. 1). The tracking of each cell family started in the first image and the tracked cells were characterized by their fate. A cell tracked from the first image of the time-lapse until its division is called a mother cell and is marked with a green X in Fig. 1. For a mother cell, the start of the cell cycle is unknown. After division, the individual daughter cells were also tracked. If it was possible to track them throughout a full cell cycle, i.e. until the next division, they are marked with a full circle in Fig. 1. If it was not possible to track a daughter cell throughout the entire cell cycle, it is marked with a pink X in Fig. 1. A cell that does not divide at all throughout the tracking period is marked with a blue X in Fig. 1. Different factors contribute to interrupted tracking before the end of the time-lapse, the most common ones being that cells migrate out of the field of view, or that cells clump together and can no longer be distinguished.
Statistics
The computer language R was used for drawing figures and statistical analysis (R Core Team, 2015). For comparison of the effect of the substrates, a post hoc pairwise comparison of two-way ANOVA was performed. For all samples, six time-lapse movies from three different samples were analyzed.Results and discussion
In the following, we refer to time as the elapsed time in the time-lapse image acquisition, i.e. t = 0 corresponds to the beginning of time-lapse imaging and t = 48 h to the end of the imaging period. Representative videos of both cell lines on all substrates can be found in the ESI† (Fig. S2–S7).Bulk analysis of cell density and proliferation
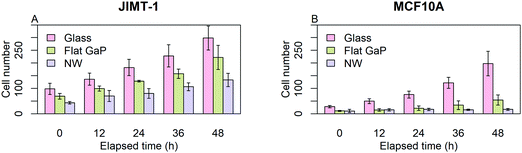
Before initiating single cell analysis, we used the data to get an overview of cell proliferation. Fig. 2 shows the number of cells obtained from individual frames of the time-lapse images at 0, 12, 24, 36, and 48 h. Each data point is the mean of the cell numbers obtained from six frames, each with an area of 0.0025 cm2. This gives only an approximate estimate of the bulk cell proliferation, as the field of view is small.For both cell lines, the number of cells counted on flat GaP and nanowire substrates is lower than on glass (Fig. 2A and B). The MCF10A cells were more affected by seeding on flat GaP and nanowires compared to glass than were the JIMT-1 cells i.e. the difference in cell number between glass and the two GaP substrates was larger in the former cell line. The lower cell density on flat GaP and nanowires suggests that, cells seeded on these substrates, either have a poor attachment, are slower at initiating proliferation, and/or have a lower rate of proliferation. It has been shown that the density of possible binding points for integrin receptors on the cell substrate is critical for efficient cell attachment and spreading.20 Glass and GaP substrates are both planar with potentially no physical restraint for cell attachment. However, the surface of GaP wafers used in this study is atomically flat, therefore cells may have difficulties attaching to it, as topography is another important factor in cell attachment.21,22 In contrast, on the nanowire substrates used here, the distance between nanowires is around 680 nm, which could be too large to allow for optimal cell attachment.20,23 That could result in a delayed onset of cell proliferation after seeding, which would be manifested in the lower cell number 24 h after seeding as shown in Fig. 2A and B (time 0 in the time-lapse).
In order to quantify the effect of underlying substrates on cells, the data of Fig. 2 was used to calculate the number of generations and the population-doubling time (PDT) using the mean cell count values of time-lapse images at time 0 and 48 (Table 1). Whereas for JIMT-1 cells, the PDT is constant irrespective of the substrate, the PDT of MCF10A cells is slightly longer on flat GaP compared to glass and more than 3 times longer on nanowires compared to when seeded on flat GaP substrates.
| Glass | Flat GaP | Nanowires | |
|---|---|---|---|
| a Number of generations calculated using the formula N = N0 × 2n where N is the cell number in the frame at time 48 and N0 the cell number in the first frame i.e. t = 0. N is the number of generations. b Population-doubling time (PDT) calculated from the number of generations during 48 h. | |||
| JIMT-1 | |||
| Generationsa | 1.6 | 1.7 | 1.6 |
| PDTb (h) | 29.8 | 28.5 | 29.9 |
| MCF-10A | |||
| Generations | 2.8 | 2.2 | 0.7 |
| PDT (h) | 17.0 | 21.9 | 67.6 |
Single cell analysis of cell division
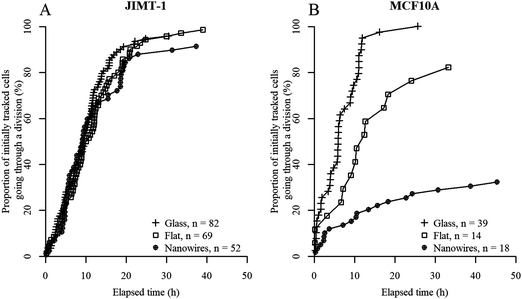
In order to get a deeper understanding of cell division on the different substrates, individual cells were tracked in each frame of the time-lapse images and the data were analyzed longitudinally. We began by investigating the time to first division of all cells that are visible in the first time-lapse frame, and which also divided during this measurement window (i.e. cells labeled with green X in Fig. 1). Cells that divided immediately when starting the measurement have proceeded through interphase during the first 24 h after seeding and are in mitosis at the starting point. If all initially observed cells are in active cell cycle progression, the plot should reach 100% when a time equivalent to the cell cycle time has passed.Fig. 3 shows the percentage of the initially visible cells (i.e. cells described as blue X and green X in Fig. 1) that have divided as a function of elapsed time for JIMT-1 and MCF10A cells seeded on the different substrates. Note that although the data are compiled from 6 time lapse movies and 3 different samples, the data points are aligned on the same curve without discontinuities for a given cell line and substrate type. This shows the low variability in cell cycle time between cells in different movies and in different replica experiments and it illustrates the reproducibility of the experiments.
From Fig. 3, it is apparent that the two cell lines react differently to the three substrates. JIMT-1 cells are moderately affected by the nanowire and flat GaP substrates while the MCF10A cells are highly sensitive to the substrate they are seeded on. These differences in behavior may be due to genetic alterations present in the cancer cells, giving them a higher probability to adapt to various environmental stresses, which promotes cancer cell survival.24
After analyzing the time to first division, cell family trees were constructed from cells that were observed during the measurement window from start to end. For each tree, the number of cell divisions in the branch exhibiting most cell divisions was quantified. The number and proportion in percentage of all cell trees that could be tracked during the entire measurement is shown in Table 2. For JIMT-1 cells, this proportion varies between 65% and 78% without any marked difference between substrates. For MCF10A, however, fewer cell trees could be tracked during the entire measurement window, compared to JIMT-1 cells, except for those on nanowire substrates. This could possibly be explained by differences in cell migration between the two cell types, with more mobile cells of MCF10A leaving the field of view at earlier time points (see section below on cell migration).
| Glass | Flat GaP | Nanowires | |
|---|---|---|---|
| a Percentage of the total number of trees undergoing 0, 1, 2, 3, 4, and 5 cell divisions, extracted from trees where at least one branch could be tracked throughout the entire 48 h time-lapse. Data from Fig. S8 and S9 (ESI). b Number of divisions for a branch in each tree. If different number of divisions occurred in one tree, the highest number was registered. c Number of initially tracked cells where at least one branch was followed for 48 h; corresponding percentage of initially tracked cells. d Distribution in percent of the total number of cell trees. | |||
| JIMT-1 | |||
| Number of divisionsb | (57; 65%)c | (57; 77%) | (44; 78%) |
| 0 | 5d | 7 | 13 |
| 1 | 9 | 24 | 17 |
| 2 | 60 | 57 | 49 |
| 3 | 26 | 12 | 21 |
| MCF-10A | |||
| Number of divisions | (20; 43%) | (11; 29%) | (55; 71%) |
| 0 | 0 | 36 | 73 |
| 1 | 0 | 0 | 18 |
| 2 | 0 | 27 | 9 |
| 3 | 55 | 27 | 0 |
| 4 | 35 | 9 | 0 |
| 5 | 10 | 0 | 0 |
On glass, the shorter cell cycle time of MCF10A cells compared to JIMT-1 cells is reflected by the higher number of cell divisions, up to five cell divisions for MCF10A cells. In line with the observed variations in PDT in Table 1, there is a shift towards fewer cell divisions when JIMT-1 and MCF10A cells are seeded on flat GaP and nanowires compared to controls. The decrease in number of cell divisions is more pronounced for MCF10A cells than for JIMT-1 cells.
Next, we analyzed the fates of all cells, including cells that migrated out of the field of view during the time lapse imaging period (Fig. 4). The upper part of each panel shows the distribution of cell cycle times for all cells that progressed from mitosis to mitosis i.e. a whole cell cycle during the observation time. The lower part of each panel shows the tracking time period of cells that did not divide (i.e. the time interval where they are in the field of view). The green crosses are mother cells, visible from the first frame. Therefore, their individual tracking time is equal to the elapsed time. The data in Fig. 4 are gathered from 6 time-lapse movies and 3 different samples. All the different cell types were found in each data set in comparable proportions (mother cells, daughter cells with complete cell cycles, cells that did not divide during the total experiment duration, and cells that could not be tracked during the entire experiment duration).
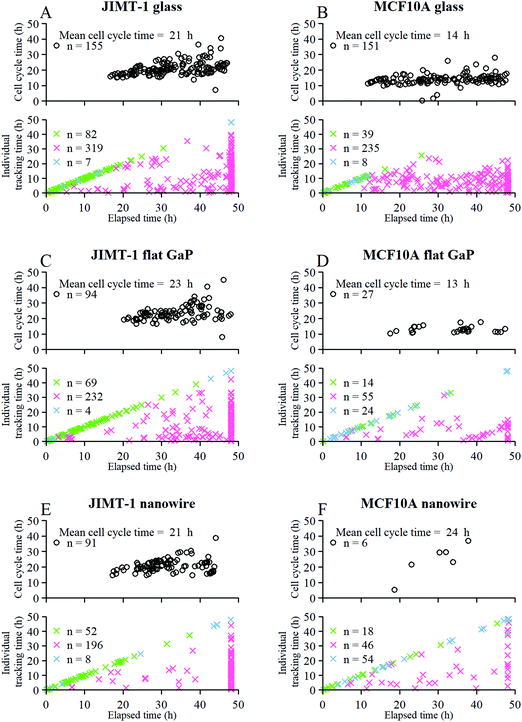 | ||
Fig. 4 Fate of all cells observed during the acquired time-lapses of JIMT-1 and MCF10A cells seeded on different substrates. Cell cycle times for cells with completed cell cycles (upper panel in each sub-figure) and tracking time for cells with non-completed cell cycles (lower part in each sub-figure). n is number of cells. 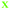 , mother cell. ○, daughter cells with complete cell cycles. , mother cell. ○, daughter cells with complete cell cycles. 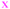 , tracking not complete due to different causes. , tracking not complete due to different causes. 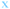 , cell that did not divide during the total time-lapse. See Fig. 1 for clarification. , cell that did not divide during the total time-lapse. See Fig. 1 for clarification. | ||
The mean cell cycle time and number of cells that divide is presented for JIMT-1 and MCF10A (Fig. 4) cells cultured on the different substrates. The mean cell cycle time of JIMT-1 cells that divide is similar on all three substrates (Fig. 4A, C and E).
For MCF10A cells, the mean cell cycle time is similar for cells seeded on glass and flat GaP (Fig. 4B and D), despite much fewer cells completing a full cell cycle on the latter substrate. In contrast, the cell cycle time of the few MCF10A cells that completed a whole cell cycle on nanowires was substantially prolonged compared to glass and flat GaP substrates (Fig. 4F).
In this study we observed that the bulk cell proliferation is hampered when cells are seeded on flat GaP and nanowire substrates. However, individual cell analysis revealed that the cell cycle time of cells that actually divide on these substrates is similar to that of cells seeded on glass. One may speculate that the few cells that continue dividing on flat GaP and nanowire substrates constitute a specific phenotypic population of cells. Thus, further studies of cell behavior on these substrates should include phenotypic identification to further our understanding of individual cell behavior.
Cell movement
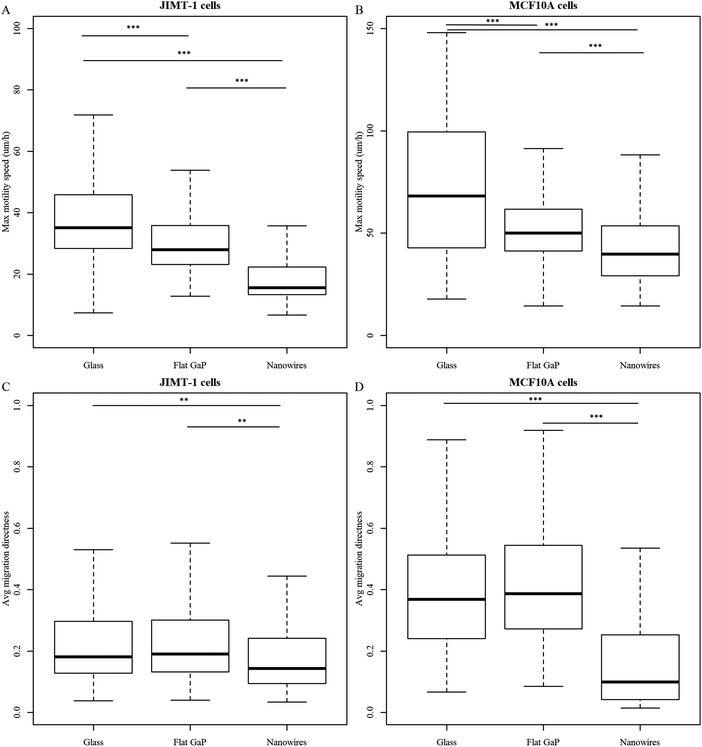
The cell substrate strongly affects the number of cells that can be tracked through the entire time-lapse imaging period (Fig. 4). For instance, fewer cells could be tracked on flat GaP and nanowire substrates compared to glass for both cell types. The reduction in traceable cells was more pronounced for MCF10A cells than for JIMT-1 cells. However, this could be the result of different degrees of cell movement on the three substrates, resulting in a variation in the number of cells leaving the field of view.Using the HoloMonitor® M4 time-lapse data, we have extracted information about cell movement. The total accumulated cell movement over time is defined as motility and the shortest distance between the first cell position and the point where the cell can be found in each image is defined as migration. During the time-lapse acquisition, motility constantly increases, while migration can increase or decrease depending on the cell trajectory. In the cell trees shown in Fig. S8 and S9 (ESI†), the color correlates to motility and the size of the rings correlates to migration. Fig. S10 displays how cell motility and migration are distributed within the cell trees found in Fig. S8 and S9 (ESI†). Motility and migration are dependent on the time of tracking. For cells exhibiting high migration, the risk of moving out of the frame during the tracking period is increased and when they have moved out of the frame, they will not be included in the calculation of migration. Therefore, to be able to include all cells in the analysis of movement parameters, we have analyzed the speed of all tracked cells, which is independent of tracking time (Fig. 5A and B, max motility speed and Fig. S11 (ESI†), max migration speed). For both cell lines, the maximum motility speed is the highest for cells seeded on glass and the lowest for cells seeded on nanowire substrates (Fig. 5A and B). The maximum motility speed (Fig. 5A and B) and the maximum migration speed (Fig. S11, ESI†) are higher for MCF10A cells compared to JIMT-1 cells on all substrates. This can explain the lower number of MCF10A cells that are traceable throughout the entire imaging period, compared to JIMT-1 cells (Table 2).
We have also determined the average migration directness, which is a measure of the linearity in migration (migration/motility) (Fig. 5C and D). When the value is one, the cell has moved away from its starting position in a straight line. MCF10A cells display higher migration directness when seeded on glass and flat GaP compared to JIMT-1 cells on those substrates (Fig. 5C and D). The migration directness is significantly lower for MCF10A cells seeded on nanowires compared to the two other substrates (Fig. 5D). This suggests that the difference in number of MCF10A traceable cells on nanowires compared to flat substrate is due to differences in cell proliferation rather than migration.
Our data shows that cells move differently on glass, flat GaP, and nanowire substrates. We have previously shown that L929 cells move at different rates depending on nanowire density.13 Others have also shown that the composition of the substrate affects cell movement.25
It is important to stress that cell behavior may be different on arrays of different nanowire array geometries. For instance, we and others have shown that cells behave differently on arrays of various densities and nanowire lengths.6,14,26 In contrast, the nanowire diameter and material do not affect the cell behavior significantly.6,27 In the present study, we have tested the same nanowire array topography as the ones used in our force assays in order to gain knowledge on how cells are affected by the underlying nanowire arrays.
Conclusions
We have tracked individual JIMT-1 breast cancer cells and MCF10A breast epithelial cells on glass, flat GaP, and GaP nanowire substrates using digital holographic microscopy. We investigated cell proliferation and cell movement using bulk data analysis and single cell analysis. The two cell lines studied behave differently, both in terms of proliferation and cell movement, on flat GaP and GaP nanowire substrates compared to when seeded on glass. Compared to JIMT-1 cells, MCF10A cells are more severely affected when cultured on flat GaP or nanowires than on glass. One may speculate that this is related to the higher adaptability of cancer cells to a foreign environment driven by genomic instability. Whereas bulk analysis revealed an increase in PDT of MCF-10A cells on flat GaP and nanowire substrates, single cell analysis of MCF-10A cells revealed that this increase in PDT is due to the presence of different sub-populations. Therefore, our data suggest that there are sub-populations of cells that react differently to the substrates, which highlights the importance to perform individual cell analysis. These different populations presumably represent different phenotypes, which are not observed in bulk cell analysis. In addition, our data clearly show the importance of investigating many cells, although this is a time-consuming process at present. Here, we choose to analyze all cells in order to avoid excluding relevant data. Future studies will aim at understanding the molecular mechanisms responsible for the different behaviors on nanowire substrates, which is a requirement for using nanowires in cell biological applications.Conflicts of interest
SK is an industrial PhD student at PHI financed by the Swedish Research Council.Acknowledgements
This study was founded by the Swedish Research Council, NanoLund, the Crafoord Foundation, the European Research Council (ERC-CoG NanoPokers) and by a donation from Carolina LePrince with the “Kalenderflickorna” and associated sponsors. The nanowire array fabrication was performed at Lund Nano Lab.References
- W. Hällström, M. Lexholm, D. B. Suyatin, G. Hammarin, D. Hessman, L. Samuelson, L. Montelius, M. Kanje and C. N. Prinz, Nano Lett., 2010, 10, 782–787 CrossRef PubMed.
- P. K. Sahoo, R. Janissen, M. P. Monteiro, A. Cavalli, D. M. Murillo, M. V. Merfa, C. L. Cesar, H. F. Carvalho, A. A. de Souza, E. P. A. M. A. M. Bakkers and M. A. Cotta, Nano Lett., 2016, 16, 4656–4664 CrossRef CAS PubMed.
- Z. Li, H. Persson, K. Adolfsson, L. Abariute, M. T. Borgström, D. Hessman, K. Åström, S. Oredsson and C. N. Prinz, Nanoscale, 2017, 9, 19039–19044 RSC.
- C. Prinz, W. Hällström, T. Mårtensson, L. Samuelson, L. Montelius and M. Kanje, Nanotechnology, 2008, 19, 345101 CrossRef PubMed.
- G. Piret, M.-T. Perez and C. N. Prinz, ACS Appl. Mater. Interfaces, 2015, 7, 18944–18948 CrossRef CAS PubMed.
- G. Piret, M. T. Perez and C. N. Prinz, Biomaterials, 2013, 34, 875–887 CrossRef CAS PubMed.
- J. T. Robinson, M. Jorgolli, A. K. Shalek, M.-H. H. Yoon, R. S. Gertner and H. Park, Nat. Nanotechnol., 2012, 7, 180–184 CrossRef CAS PubMed.
- C. Xie, Z. L. Lin, L. Hanson, Y. Cui and B. X. Cui, Nat. Nanotechnol., 2012, 7, 185–190 CrossRef CAS PubMed.
- N. Sköld, W. Hällström, H. Persson, L. Montelius, M. Kanje, L. Samuelson, C. N. Prinz and J. O. Tegenfeldt, Nanotechnology, 2010, 21, 155301 CrossRef PubMed.
- H. Persson, J. P. Beech, L. Samuelson, S. Oredsson, C. N. Prinz and J. O. Tegenfeldt, Nano Res., 2012, 5, 190–198 CrossRef CAS.
- A. K. Shalek, J. T. Robinson, E. S. Karp, J. S. Lee, D.-R. Ahn, M.-H. Yoon, A. Sutton, M. Jorgolli, R. S. Gertner, T. S. Gujral, G. MacBeath, E. G. Yang and H. Park, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 1870–1875 CrossRef CAS PubMed.
- C. N. Prinz, J. Phys.: Condens. Matter, 2015, 27, 233103 CrossRef PubMed.
- H. Persson, Z. Li, J. O. Tegenfeldt, S. Oredsson and C. N. Prinz, Sci. Rep., 2015, 5, 18535 CrossRef CAS PubMed.
- H. Persson, C. Købler, K. Mølhave, L. Samuelson, J. O. Tegenfeldt, S. Oredsson and C. N. Prinz, Small, 2013, 9, 4006–4016 CrossRef CAS PubMed.
- Z. Li, H. Persson, K. Adolfsson, L. Abariute, M. T. Borgström, D. Hessman, K. Åström, S. Oredsson and C. N. Prinz, Nanoscale, 2017, 9, 19039–19044 RSC.
- K. Adolfsson, H. Persson, J. Wallentin, S. Oredsson, L. Samuelson, J. O. Tegenfeldt, M. T. Borgstrom and C. N. Prinz, Nano Lett., 2013, 13, 4728–4732 CrossRef CAS PubMed.
- S. Kamlund, D. Strand, B. Janicke, K. Alm and S. Oredsson, Cell Cycle, 2017, 4101, 1–11 Search PubMed.
- E. Cuche, F. Bevilacqua and C. Depeursinge, Opt. Lett., 1999, 24, 291 CrossRef CAS PubMed.
- U. Schnars and W. Juptner, Inst. Phys. Publ., 2002, 13, 17 Search PubMed.
- E. A. Cavalcanti-Adam, T. Volberg, A. Micoulet, H. Kessler, B. Geiger and J. P. Spatz, Biophys. J., 2007, 92, 2964–2974 CrossRef CAS PubMed.
- M. J. Dalby, C. C. Berry, M. O. Riehle, D. S. Sutherland, H. Agheli and A. S. Curtis, Exp. Cell Res., 2004, 295, 387–394 CrossRef CAS PubMed.
- A. S. Curtis, B. Casey, J. Gallagher, D. Pasqui, M. Wood and C. D. Wilkinson, Biophys. Chem., 2001, 94, 275–283 CrossRef CAS PubMed.
- C. Selhuber-Unkel, T. Erdmann, M. López-García, H. Kessler, U. S. Schwarz and J. P. Spatz, Biophys. J., 2010, 98, 543–551 CrossRef CAS PubMed.
- L. K. Boroughs and R. J. Deberardinis, Nat. Cell Biol., 2016, 17, 351–359 CrossRef PubMed.
- K. M. Hakkinen, J. S. Harunaga, A. D. Doyle and K. M. Yamada, Tissue Eng., Part A, 2011, 17, 713–724 CrossRef CAS PubMed.
- N. Buch-Månson, D.-H. Kang, D. Kim, K. E. Lee, M.-H. Yoon and K. L. Martinez, Nanoscale, 2017, 9, 5517–5527 RSC.
- Z. Li, H. Persson, K. Adolfsson, S. Oredsson and C. N. Prinz, Sci. China: Life Sci., 2018, 61, 427–435 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8tb02049c |
| ‡ These authors corresponded equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |