Modelling DNA extension and fragmentation in contractive microfluidic devices: a Brownian dynamics and computational fluid dynamics approach†
Abstract
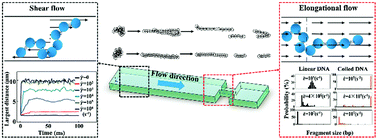
Fragmenting DNA into short pieces is an essential manipulation in many biological studies, ranging from genome sequencing to molecular diagnosis. Among various DNA fragmentation methods, microfluidic hydrodynamic DNA fragmentation has huge advantages especially in terms of handling small-volume samples and being integrated into automatic and all-in-one DNA analysis equipment. Despite the fast progress in experimental studies and applications, a systematic understanding of how DNA molecules are distributed, stretched and fragmented in a confined microfluidic field is still lacking. In this work, we investigate the extension and fragmentation of DNA in a typical contractive microfluidic field, which consists of a shear flow-dominated area and an elongational flow-dominated area, using the Brownian dynamics-computational fluid dynamics method. Our results show that the shear flow at the straight part of the microfluidic channel and the elongational flow at the contractive bottleneck together determine the performance of DNA fragmentation. The average fragment size of DNA decreases with the increase of the strain rate of the elongational flow, and the upstream shear flow can significantly precondition the conformation of DNA to produce shorter and more uniform fragments. A systematic study of the dynamics of DNA fragmentation shows that DNA tends to break at the mid-point when the strain rate of elongational flow is small, and the breakage point largely deviates from the midpoint as the strain rate increases. Our simulation of the thorough DNA fragmentation process in a realistic microfluidic field agrees well with experimental results. We expect that our study can shed new light on the development of future microfluidic devices for DNA fragmentation and integrated DNA analysis devices.


 Please wait while we load your content...
Please wait while we load your content...