Open Access Article
Open Access ArticleProduct-oriented chemical surface modification of a levansucrase (SacB) via an ene-type reaction†
Maria Elena
Ortiz-Soto
 a,
Julia
Ertl
a,
Jürgen
Mut
a,
Juliane
Adelmann
a,
Thien Anh
Le
b,
Junwen
Shan
c,
Jörg
Teßmar
a,
Julia
Ertl
a,
Jürgen
Mut
a,
Juliane
Adelmann
a,
Thien Anh
Le
b,
Junwen
Shan
c,
Jörg
Teßmar
 c,
Andreas
Schlosser
d,
Bernd
Engels
b and
Jürgen
Seibel
c,
Andreas
Schlosser
d,
Bernd
Engels
b and
Jürgen
Seibel
 *a
*a
aInstitut für Organische Chemie, Universität Würzburg, Am Hubland, 97074 Würzburg, Germany. E-mail: seibel@chemie.uni-wuerzburg
bInstitut für Physikalische und Theoretische Chemie, Universität Würzburg, Emil-Fischer Strasse 42, 97074 Würzburg, Germany
cAbteilung für Funktionswerkstoffe der Medizin und der Zahnheilkunde, Universitätsklinikum Würzburg, Pleicherwall 2, D-97070 Würzburg, Germany
dRudolf-Virchow-Zentrum für Experimentelle Biomedizin, Universität Würzburg, Josef-Schneider Str. 2, Haus D15, 97080 Würzburg, Germany
First published on 22nd May 2018
Abstract
Carbohydrate processing enzymes are sophisticated tools of living systems that have evolved to execute specific reactions on sugars. Here we present for the first time the site-selective chemical modification of exposed tyrosine residues in SacB, a levansucrase from Bacillus megaterium (Bm-LS) for enzyme engineering purposes via an ene-type reaction. Bm-LS is unable to sustain the synthesis of high molecular weight (HMW) levan (a fructose polymer) due to protein–oligosaccharide dissociation events occurring at an early stage during polymer elongation. We switched the catalyst from levan-like oligosaccharide synthesis to the efficient production of a HMW fructan polymer through the covalent addition of a flexible chemical side-chain that fluctuates over the central binding cavity of the enzyme preventing premature oligosaccharide disengagement.
Introduction
Substantial efforts are devoted to customizing enzyme properties not only with the aim of optimizing a given reaction in a laboratory-scale but considering the needs of the industrial processes in which the biocatalysts may be employed.1 In this direction molecular engineering was successfully applied to a number of enzymes aiming for activity, specificity and stability improvements.2 Installation and functionalization of unnatural amino acids in proteins built on the pioneering work of Schultz3,4 result in a much wider range of functionalities outside the natural evolutionary boundaries of the 20 canonical amino acids.5–7Albeit providing multiple possibilities for fundamental research, incorporation of unnatural amino acids is not customary and requires specialized expression systems.8 In this regard chemical modification of native proteins remains an alternative to gain access to sufficient amounts of protein conjugates for large-scale applications. Unlike nucleophilic amino acids that are traditionally chosen for protein bioconjugation (i.e. cysteine, threonine, serine and lysine),9 aromatic residues tyrosine and tryptophan are suitable candidates for achieving selectivity, since they occur less frequently in proteins and are often entirely or partially buried when localized at protein surfaces.10–15
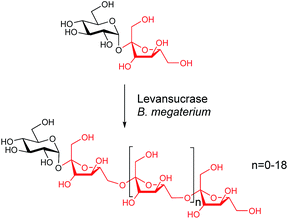
Here we explore the tyrosine-specific chemical engineering of a levansucrase from Bacillus megaterium (Bm-LS). Bm-LS (from glycoside hydrolase family GH68)16 synthesizes a wide distribution of fructooligosaccharides of various lengths using sucrose as a donor/acceptor of the fructosyl moiety.17 The fructose units are connected via β(2→6) linkages producing levan-type oligosaccharides (fructans). Levan is a functional polymer produced by a reduced number of plants and various microorganisms.18 Application of levan and levan-like oligosaccharides in the cosmetic, medical and food industries is contingent on their molecular weight.18–21 Contrary to most Gram-positive GH68 enzymes the signal peptide deleted, heterologously expressed levansucrase from Bacillus megaterium produces exclusively small oligosaccharides with a degree of polymerization (DP) of 2–20 fructose units (Scheme 1).22
While site-directed mutagenesis proved successful at narrowing the number and size of synthesized products,23 structural manipulation of oligosaccharide synthesizing GH68 enzymes to promote the synthesis of a HMW polymer is, to the best of our knowledge, unexplored.
Previous experiments involving GH68 enzymes payed particular attention to the residues involved in contacts with sucrose or with short polymerization starters (oligosaccharide chains of up to DP9), which grow in a clockwise motion.17,23 These residues are localized in the so-called first and second shells of the central pocket of GH68 enzymes (Fig. 1A), and mutations at these positions lead to changes in activity and/or oligosaccharide size.17,19,22,24 We hypothesized that altering residues of the third shell might have an impact on the “assembly line” of larger polysaccharide products. Amino acids from the second and third shells are the subject of this paper, with the focus on native tyrosines 196 and 247 (Fig. 1A).
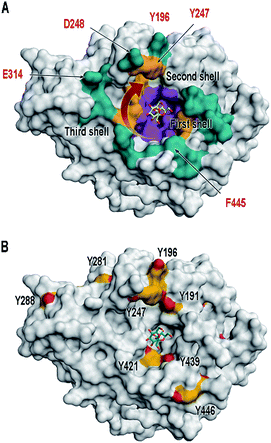 | ||
| Fig. 1 Surface illustration of Bm-LS (PDB 3om2). (A) The central pocket of Bm-LS coloured by substrate/product binding regions. Residues in contact with sucrose and short oligosaccharide products correspond to the first and second shells (purple and gold, respectively). Residues expected to bind larger oligosaccharides are depicted in cyan (third shell). A red arrow shows the direction of oligosaccharide elongation.17,23 Residues labelled in red were submitted to mutagenesis and were chemically modified in this paper. (B) Tyrosine residues in Bm-LS: Front view. Tyrosine residues are depicted in yellow and the substrate sucrose from PDB 1pt2 is shown in the active site. | ||
Results and discussion
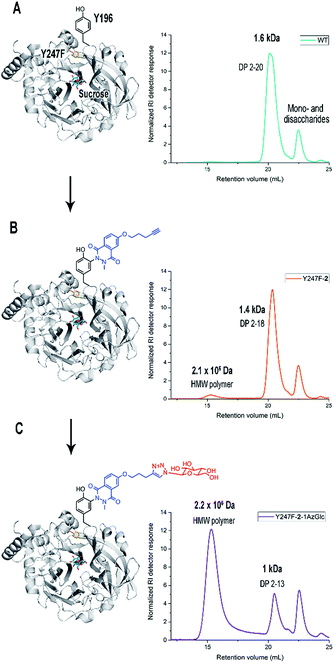
Bm-LS contains 25 tyrosine residues with different solvent accessibility (Fig. 1B and ESI, S1†). Residues 41, 196, 247, 281 and 446 are the most solvent exposed tyrosines, with Y247 taking part in oligosaccharide elongation.23 Accessibility, location and structural function make chemical modification of positions 196 and 247 appealing for studying and manipulating oligosaccharide/polymer elongation. Third shell solvent-exposed amino acids D248 and E314 as well as F445 were exchanged by tyrosine and the effect of their chemical modification was also studied in variants containing the double mutation Y196F/Y247F.Cyclic diazodicarboxamides such as 4-phenyl-3H-1,2,4-triazole-3,5(4H)-dione (PTAD) 1 and its derivatives are powerful electrophile modification reagents that can react in an ene-type reaction with tyrosine residues (Scheme 2A).12,25,26 The reactivity and stability of these compounds are strongly dependent on the R group and their oxidation immediately prior to use is needed.27 We followed the strategy based on tyrosine modification reported by Sato et al.,27 which relies on in situ oxidation of the luminol derivatives (diazodicarboxyamides) in the presence of hemin and hydrogen peroxide under ambient aqueous conditions.
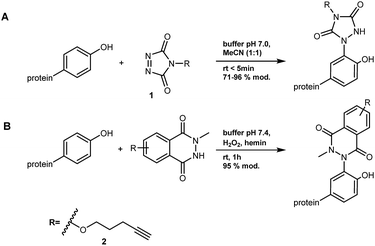 | ||
| Scheme 2 Bioconjugation of tyrosine with (A) PTAD 1 and (B) A luminol derivative 2.12,27 | ||
The alkyne 2 was synthesized and used in protein modification experiments. The alkyne group can be further extended via Cu(I)-catalyzed Huisgen-Meldal-Sharpless 1,3-dipolar cycloaddition (“click reaction”) for additional functionalization (Scheme 2B).28
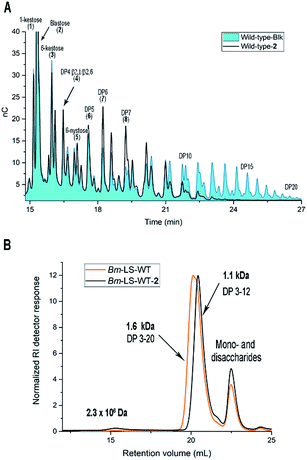
Time course samples of the tyrosine modification reaction containing wild-type Bm-LS (Bm-LS-WT), hemin, H2O2 and the alkyne 2 in phosphate buffer were analysed for enzymatic activity (ESI, Fig. S2A†). Time dependent chemical modification was assessed by LC-MS analysis of the full-length protein (ESI, Fig. S2B and S3†). A blank sample containing all the components except the luminol derivative 2 was analysed in parallel to study the mere influence of oxidative damage on Bm-LS activity and product specificity.
The tyrosine modification reaction proceeded almost to completion within 4 h with only approximately 5% of unmodified protein remaining after this time period (ESI, Fig. S2B†). After 24 h 61 and 39% of the enzyme was modified once or twice with compound 2, respectively. Extended reaction times increased tyrosine modification and reduced the enzymatic activity most likely due to methionine oxidation (ESI, Fig. S2A†).
NanoLC-MS/MS analysis of elastase digested Bm-LS-WT-2 indicates that modification mainly took place on Y196 and Y247 after 24 h reaction time. Other residues such as Y41, Y109, Y439 and Y446 were modified to a lesser extent and residues Y224, Y281 and Y288 were not significantly modified (ESI, Fig. S4†). Oxidation of methionine residues was also confirmed.
Tyrosine residues could be potentially modified in up to four modes since isomers of compound 2 can react with C3 or C5 of the tyrosine aromatic ring (Pose A and B, respectively. ESI, Scheme S1†).
Chemical modification with molecule 2 stimulates the synthesis of marginal amounts of HMW levan and restricts the size of oligosaccharides (Fig. 2). Bm-LS-WT-2 synthesizes levan of 2.3 × 106 Da and fructans with an average molecular weight of 1.1 kDa (DP 2–12), while the unmodified enzyme produces only oligosaccharides of DP 2–20. Transfer/hydrolysis partition remains unaffected in this process.
Because chemical modification preferentially happens at residues Y196 and Y247, the contribution of such modification to the product profile of Bm-LS-WT-2 was investigated. For this purpose, mutants Y196F, Y247F and Y196F/Y247F were constructed and the products of unmodified and modified variants were compared (ESI, Fig. S5†). This confirmed that changes in oligosaccharide/levan synthesis can indeed be attributable to the exclusive modification of both residues. Changes in the fructan profile of Bm-LS-WT-2 are the consequence of two different events. Modification of Y196 with alkyne 2 (variant Y247F-2) stimulates the synthesis of large polymer, which avoids the accumulation of products of DP 12–20 (Fig. 2 and ESI, S5B and S6†). On the contrary modification of Y247 (variant Y196F-2) hampers oligosaccharide growth, accumulating short oligosaccharides more efficiently than both its unmodified counterpart and variant Y247-2 (ESI, Fig. S5A†). The latter is the predominant behaviour observed for the modified wild-type enzyme. As expected the product profile of the double variant Y196F/Y247F-2 remains unchanged (ESI, Fig. S5C†). Because protein conjugation with molecule 2 has only a minor effect on the reaction products of Bm-LS-WT (Fig. 2 and ESI, S5†) we decided to investigate whether derivatization of the alkyne group of compound 2 would enhance the effect of the luminol derivative on the catalysis of Bm-LS.
To rule out the effect of chemical modification on residue Y247 experiments were carried out with single variant Y247F. In a two-step modification process this variant was modified with alkyne 2 at residue Y196 followed by a “click reaction” with 1-azido-1-deoxy-β-D-glucopyranoside (1AzGlc) (Fig. 3 and ESI, Fig. S7†). The glucopyranoside moiety was expected to interact with both the growing polymer and the protein, the latter due to the versatility of the plus subsites (beyond subsite +1) to accommodate glucose and fructose efficiently.
Y247F-2-1AzGlc showed in fact a dramatic shift from oligosaccharide formation to the production of a HMW polymer (2.3 × 106 Da) (Fig. 3), which was accompanied by a concomitant 1.5-fold increase in the transfer/hydrolysis partition compared to Y247F (ESI, Table S1†). The polymer was identified as levan by NMR analysis (ESI, Table S2 and Fig. S8–S11†). Gel Permeation Chromatography (GPC) profiles of products synthesized by Y247F-2-1AzGlc and B. subtilis levansucrase (B. subtilis-LS), an extensively characterized GH68 enzyme, are presented for comparison (ESI, Fig. S12†). Bioconjugation of variant Y247F (at position Y196) with alkyne 2 does not change the global KM for sucrose (Table 1) and changes in activity may obey the modification of other tyrosine residues or to methionine oxidation. The extension of compound 2 with 1AzGlc increased the KM of the variant by almost 2-fold and its kcat value was slightly improved (Table 1). Chemical modification did not affect the optimal pH of the enzymes (ESI, Fig. S13†).
| Enzyme | k cat [s−1] | K M [mM] | k cat/KM [mM s−1] |
|---|---|---|---|
| WT-control | 247.8 ± 6.9 | 21.2 ± 2.7 | 11.7 |
| WT-2 | 131.0 ± 5.0 | 19.2 ± 3.6 | 6.8 |
| Y247F-control | 254.2 ± 3.5 | 21.9 ± 1.3 | 11.6 |
| Y247F-2 | 153.6 ± 3.7 | 24.5 ± 2.6 | 6.26 |
| Y247F-2-1AzGlc | 172.1 ± 2.8 | 43.1 ± 2.8 | 3.9 |
| Y196F-control | 285.9 ± 2.8 | 29.8 ± 1.2 | 9.6 |
| Y196F-2 | 173.3 ± 3.4 | 48.1 ± 3.6 | 3.6 |
| Y247F/Y196F-control | 308.7 ± 6.7 | 16.7 ± 1.7 | 18.4 |
| Y247F/Y196F-2 | 204.9 ± 4.8 | 17.6 ± 1.9 | 11.6 |
Double variant Y196F/Y247F-2 was modified with 1AzGlc and its reaction products analysed by GPC. The product profile of this variant remained unaltered, confirming that modification of Y196 and not that of other tyrosine residues is responsible for HMW levan synthesis (ESI, Fig. S14†).
Binding of oligosaccharides to the protein surface is a pivotal factor in levan synthesis as demonstrated with wild-type levansucrases from Zymomonas mobilis and B. subtilis.29,30 The ability of soluble Z. mobilis levansucrase to synthesize oligosaccharides can be altered through the pH-driven formation of active protein microfibrils. The new scaffold creates additional anchorage sites for growing acceptors and enables the synthesis of large levan.30 Conversely the binding of growing oligosaccharides to the surface of other B. subtilis-LS molecules present in the reaction plays a detrimental role in polymer elongation. The inter-molecular competition of enzyme surfaces for non-productive oligosaccharide/levan binding results in mixtures of low molecular weight (LMW) and HMW levan. Accordingly only extremely low enzyme concentrations would decrease the occurrence of protein–protein encounters favouring the specific production of a large polymer.29
Although some protein aggregation occurs during the chemical modification process attributable to the interaction of Bm-LS with hemin, isolation and analysis of stable monomers allowed us to establish that neither protein aggregation nor protein concentration plays a role in the specificity of Y247, Y247F-2 and Y247F-2-1AzGlc for the synthesis of either oligosaccharides or levan (ESI, Fig. S15 and S16†).
To gain insights into the nature of the protein–ligand interactions of chemically modified proteins, the apparent dissociation constants (Kd) of variants Y247F-2 and Y247F-2-1AzGlc for LMW levan (average MW = 1.4 kDa, average DP = 9) were determined by Differential Scanning Fluorimetry (DSF).31 The low affinity character of the binding made attempts to measure interactions by isothermal titration calorimetry (ITC) unsuccessful. Binding constants obtained by DSF are based on changes in the apparent protein melting temperature, which is ligand concentration dependent (ESI, Fig. S17†).31
The Kd of both enzymes for LMW levan is around 8 mM, indicating a weaker binding although in the same order of magnitude as that of Bm-LS-WT (4.1 mM) and B. subtilis-LS (1.8 mM), the latter for levan with an average DP of 16.29 Although the binding between variant Y247F-2 and LMW levan was seemingly not modified after the “click” reaction, the possibility that little but significant changes in binding may have contributed to the formation of HMW levan by Y247F-2-1AzGlc cannot be dismissed.
It is worth mentioning that the kind of interactions resulting from incubating already formed levan with the levansucrase may not resemble the protein–product interactions taking place during in situ synthesis. Once oligosaccharides leave the binding pocket, their structure may undergo conformational changes that could prevent them from rebinding in a productive manner and thus grow to HMW levan.29
Relatively stable contacts between elongating oligosaccharides and the enzyme are essential at early stages of product extension if HMW levan is to be synthesized.29Bm-LS-WT and single variants modified only with compound 2 are unable to support the synthesis of a large polymer due to premature protein–oligosaccharide disengagement events. Such events should only occur due to insufficient binding of growing products within or near the catalytic pocket, since non-productive protein–product interactions do not appear to be responsible for the inability of these enzymes to produce HMW levan (ESI, Fig. S15†).
Installing a chemical side-chain in the variant Y247F-2-1AzGlc may generate intra polar contacts between 2-1AzGlc and the protein as observed by Molecular Dynamics (MD) simulations.
MD simulations of Y247F-2-1AzGlc (both isomers of alkyne 2 in pose A, ESI, Fig. S1†) were obtained with and without 6-kestose bound to the active site (ESI, Fig. S18†). Trajectories and interactions of -2-1AzGlc are similar regardless of the absence/presence of the ligand, indicating that chemical modification does not disturb the binding of short polymerization starters in the active site. Isomer 1 of -2-1AzGlc fluctuates back and forth over the binding pocket exhibiting short-lived interactions with the protein. On the contrary, the flexibility of isomer 2 is reduced due to the polar intermolecular interaction of -2-1AzGlc with the main and side-chains of I242, D243 and E244 (ESI, Fig. S18†). These results suggest that flexible -2-1AzGlc may act as a chemical lid introducing steric hindrance, which would prevent the size-increasing oligosaccharides from leaving the binding pocket too early. The synthesis of small amounts of short oligosaccharides may result from an incomplete “click” reaction or from side-chain -2-1AzGlc having more interaction with the protein, thus “opening” the lid and preventing steric interference.
1,2,3-Triazole ring systems are C–H hydrogen bond donors with peptidomimetic characteristics, with a similar size, planarity and dipolar character to an amide linkage.32 To gain further knowledge on the contribution of the 1,2,3-triazole ring and other azides to polymer production sodium azide, 2-azido-1-(3-hydroxyphenyl)ethan-1-one (AN28) and 5-FAM-azide were also tested. HMW levan production is observed notwithstanding the nature of the modifying azide, however the presence of triazole alone is not enough to completely shift the reaction from LMW to HMW polymer (ESI, Fig. S19†) revealing the need for larger azides to hinder oligosaccharide–enzyme dissociation.
The modification of triple variants Y196F/Y247F/D248Y, Y196F/Y247F/E314Y and Y196F/Y247F/F445Y seems to support the previous assumption. D248, E314 and F445 are located at the periphery (third shell) of the central pocket (Fig. 1A). While the GPC profiles of Y247F-2-1AzGlc and Y196F/Y247F/E314Y-2-1AzGlc are similar, variants Y196F/Y247F/D248Y-2-1AzGlc and Y196F/Y247F/F445Y-2-1AzGlc show significant differences (ESI, Fig. S20†). Of all variants, tyrosine introduced at position 248 would be the most exposed. Variant Y196F/Y247F/D248Y-2-1AzGlc synthesizes almost entirely HMW levan producing only traces of LMW oligosaccharides. Variant Y196F/Y247F/F445Y-2-1AzGlc, however, presents a lower HMW/LMW levan ratio compared to other “clicked” variants which is in agreement with the position of residue 445. Depending on the orientation of the chemical side-chain in variant Y196F/Y247F/F445Y-2-1AzGlc, it may keep oligosaccharide–protein interactions or hinder elongation of nascent oligosaccharides if oriented towards the active site.
Our results suggest that the change in the product specificity of chemically modified levansucrases may obey the restriction of oligosaccharide mobility triggered by the presence of the chemical side-chain -2-azide. This may impede early protein–oligosaccharide dissociation events. Interactions among enzymes, oligosaccharides and chemical tags, however, merit a thorough investigation.
Conclusions
Site directed mutagenesis has been used to modify the products of a number of GH68 enzymes.17,19,22–24 Such manipulations are often focused on reducing the number of synthesized oligosaccharides; consequently, information regarding how oligosaccharide elongation and polymer formation progress is scarce.29,30 Herein we demonstrate in an unprecedented manner that the approach of modifying strategically situated tyrosine residues can be applied to manipulate the product specificity of Bm-LS to create novel polymer products while increasing the transfer/hydrolysis partition. Chemically and genetically modified Bm-LS variants switched from the production of a wide oligosaccharide distribution to the synthesis of a HMW polymer maintaining both protein integrity and functionality. Chemical tyrosine modification via the Ene-reaction seems to be an attractive strategy especially for tailoring the scaffold of carbohydrate processing enzymes, which deal with polymers as substrates or products. The fact that a correctly folded enzyme produced under standard culture procedures can be post-translationally modified with luminol derivatives carrying any chemical residue of interest adds to the appeal of this technology.Experimental
Calculation of the accessible surface areas (ASA)
The ASA of tyrosine residues was calculated using the crystallographic structure of the levansucrase PDB 3om2 with Proteins, Interfaces, Structures and Assemblies (PISA) server (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html).33Gene constructs and mutagenesis
The gene of the levansucrase from B. megaterium (GenBank: ADF38395.1) was cloned into the vector pETM11 between the restriction sites NcoI and XhoI. Mutagenesis reactions were performed by using the QuikChange Lightning Site-Directed Mutagenesis and QuikChange Lightning Multi Site-Directed Mutagenesis Kits according to the provider's instructions or by whole plasmid amplification using phosphorylated primers and introducing the mutated codon only in the forward primer. A vector containing the wild-type B. megaterium levansucrase was used as a template for generating variants Y196F, Y247F and the double variant Y196F/Y247F. The latter was used as a template to create triple variants Y196F/Y247F/D248Y and Y196F/Y247F/D248Y. The resulting sequences were confirmed by DNA sequencing.Primers employed in PCR are as indicated in Table 2.
| Variant | Primer sequence (5′ to 3′ direction) |
|---|---|
| a PCR performed with the QuikChange Lightning Site-Directed Mutagenesis Kit using forward and reverse primers. b PCR performed with the QuikChange Lightning Multi Site-Directed Mutagenesis Kit using only forward primers. c Variant Y196F/Y247F was used as a template. d Only the forward primer contains the new mutation and whole plasmid amplification was performed with phosphorylated primers. Mutations are underlined. | |
| Y196Fa |
|
|
|
| Y247Fa |
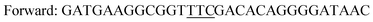
|
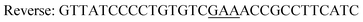
|
|
| Y196F/Y247Fb | Forward primers Y196F and Y247F |
| Y196F/Y247F/D248Yc,d |
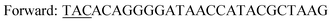
|
| Reverse: GAAACCGCCTTCATCAATAAACTGC | |
| Y196F/Y247F/D248Ya,c |
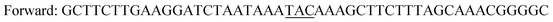
|
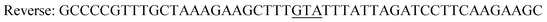
|
|
| Y196F/Y247F/F445Yb |
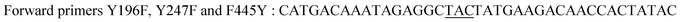
|
Protein expression and purification
One single colony of freshly transformed E. coli bearing selected plasmids was used to inoculate 5 mL LB-medium containing 50 μg mL−1 kanamycin. Pre-cultures were incubated overnight at 37 °C and used to inoculate 250 mL LB-medium with an appropriate antibiotic. Expression of levansucrase was induced at an OD600 of around 0.6 by adding IPTG at a final concentration of 0.5 mM. Cultures were incubated overnight at 20 °C. Cells were harvested by centrifugation and resuspended in 7 mL of lysis buffer (20 mM NaHPO4, 20 mM Na2HPO4, 20 mM imidazole, 0.5 M NaCl, pH 8.0). After sonication, extracts were cleared by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g. For Immobilized Metal Affinity Chromatography (IMAC), cleared lysates were loaded onto a 1 mL HisTrap FF column (GE healthcare Life Sciences) incorporated onto an Äkta prime plus purification system and levansucrase eluted with a linear gradient of elution buffer (20 mM NaHPO4, 20 mM Na2HPO4, 0.25 M imidazole, 0.5 M NaCl, pH 8.0). Purification fractions containing pure protein were pooled and concentrated.
000 × g. For Immobilized Metal Affinity Chromatography (IMAC), cleared lysates were loaded onto a 1 mL HisTrap FF column (GE healthcare Life Sciences) incorporated onto an Äkta prime plus purification system and levansucrase eluted with a linear gradient of elution buffer (20 mM NaHPO4, 20 mM Na2HPO4, 0.25 M imidazole, 0.5 M NaCl, pH 8.0). Purification fractions containing pure protein were pooled and concentrated.
The enzyme contains a TEV-protease cleavable N-terminal hexahistidine-tag. Digestion with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 TEV/levansucrase ratio in 20 mM Tris–HCl buffer pH 7.5 and 0.1 mM EDTA was allowed to proceed over 48 h. Levansucrase was separated from the cleaved peptide and the protease via IMAC and size exclusion chromatography over a HiLoad 16/600 Superdex 200 pg column (GE healthcare Life Sciences) in 50 mM NaHPO4, 20 mM Na2HPO4, 20 mM imidazole, 0.5 M NaCl, pH 8.0.
100 TEV/levansucrase ratio in 20 mM Tris–HCl buffer pH 7.5 and 0.1 mM EDTA was allowed to proceed over 48 h. Levansucrase was separated from the cleaved peptide and the protease via IMAC and size exclusion chromatography over a HiLoad 16/600 Superdex 200 pg column (GE healthcare Life Sciences) in 50 mM NaHPO4, 20 mM Na2HPO4, 20 mM imidazole, 0.5 M NaCl, pH 8.0.
An extinction coefficient of 51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 014 M−1 cm−1 and a path length of 1 cm were used to calculate the concentration of purified enzymes at 280 nm.
014 M−1 cm−1 and a path length of 1 cm were used to calculate the concentration of purified enzymes at 280 nm.
Tyrosine specific modification reactions
10 μM purified protein was incubated in 0.1 M phosphate buffer, pH 7.5 containing 1 mM H2O2, 10 μM hemin and 1 mM luminol derivative 2.27 Hemin and 2 were prepared in 15 and 100% DMSO (w/v), respectively. H2O2 was diluted immediately before use.Blank reactions were prepared under similar reaction conditions excluding the luminol derivatives. Modification reactions were allowed to proceed over 72 h and time course samples of enzymes were assayed for activity on sucrose.
Enzymes used for oligosaccharide/levan synthesis and kinetic determinations were modified over 22 h. Subsequent to tyrosine modification with compound 2 (alkyne modifier) the enzyme was separated from H2O2, hemin and the excess of alkyne 2 by buffer exchange against 50 mM Sorensen's buffer, pH 6.9, using a 5 mL HiTrap Desalting column (GE healthcare Life Sciences).
Copper-catalysed azide–alkyne cycloaddition (CuAAC) reaction (click reaction)
The enzyme modified with 2 was concentrated after buffer exchange and incubated at a final concentration of 10 μM in 0.1 M phosphate buffer containing 100 μM CuSO4, 0.5 mM THPTA and 50 μM of the azide (1-azido-1-deoxy-β-D-glucopyranoside, sodium azide, 2-azido-1-(3-hydroxyphenyl)ethan-1-one (AN28) or 5-FAM-azide). The click reaction was started by addition of 5 mM sodium ascorbate. Blank reactions were prepared excluding the azide.The click reaction was stopped after 3 h by addition of 1 mM EDTA prior to buffer exchange against 50 mM Sorensen's buffer, pH 6.9.
Determination of optimal pH
Reactions for determining pH-dependent activity profiles contained 1 U mL−1 enzyme and 0.5 mM sucrose in 50 mM acetate or Sorensen's buffer ranging from pH 5–8, with an increase of 0.5 pH units. Acetate buffer was employed in reactions performed at pH 5.0 and 5.5. For the remaining pH values Sorensen's buffer was used. The global activities were calculated from the release of reducing sugars by using the DNS (3,5-dinitro-salicylic acid method)19,34 method.Activity assay and determination of kinetic parameters
Initial rates were determined from the release of reducing sugars (DNS)19,34 from 0.5 mM sucrose in 50 mM Sorensen's buffer pH 6.9 containing the unmodified or tyrosine-modified enzyme over the time course.Catalytic parameters for the global reaction (hydrolysis and transfer) were obtained from reactions performed in 50 mM Sorensen's buffer pH 6.9 containing purified enzyme and varying concentrations of sucrose. KM and kcat values were calculated by fitting the data to the Michaelis–Menten equation using a non-linear curve in OriginPro (OriginLab).
HPAEC-PAD analysis of products from sucrose
Reactions containing 0.5 M sucrose and an enzymatic activity of 3 U mL−1 in 50 mM Sorensen's buffer pH 6.9 were employed to determine the transfer/hydrolysis partition and product profile of modified and unmodified levansucrases (wild-type and variants). HPAEC-PAD analysis of oligosaccharides was performed with a Dionex ICS-5000+ SP system utilizing a Carbopac PA10 column with pulsed amperometric detection. Eluents were 100 mM NaOH (A), 100 mM NaOH, and 1 M NaOAc (B). For quantification of glucose and fructose a multistep gradient was programmed as follows: 0–5 min 100% A, 5–30 min 0–50% B, and 30–45 min 100% A. The release of glucose reflects the global activity (hydrolysis and transfructosylation), while fructose only results from hydrolysis. The difference between glucose and fructose concentrations corresponds to transfructosylation.Gel permeation chromatography (GPC)
Products from modified and unmodified enzymes were obtained from reactions performed in 50 mM Sorensen's buffer pH 6.9 with 0.5 M sucrose and 3 U mL−1 enzymatic activity unless stated otherwise. Reactions were stopped with 50 mM NaOH after 48 h.Reactions with 0.1, 1–3 and 10 U mL−1 enzymatic activity were stopped after 120, 48 and 5 h, respectively.
Products of the wild-type levansucrase and its variants were purified by ethanol precipitation (5 volumes of cold ethanol by 1 volume of the reaction medium after approximately 90% sucrose conversion). Levan and/or oligosaccharides were recovered by centrifugation and freeze-dried.
Gel permeation chromatography of the polysaccharides was performed with an aqueous GPC system from Malvern (Herrenberg, Germany). The system includes a Viscotek GPCmax (in-line degasser, 2-piston-pump and autosampler), a column oven (35 °C), a refractive index (RI) detector (Viscotek VE3580) and Viscotek A-columns (Guard column and two A6000M analytical columns, length = 50 mm + resp. 300 mm, width = 8 mm, porous poly(methyl methacrylate), and particle size 13 μm). An aqueous solution of Millipore water with 8.5 g L−1 NaNO3 and 0.2 g L−1 NaN3 was used as a solvent and eluent. The samples were dissolved overnight and filtered with a 0.45 μm membrane. A volume of 100 μL was injected into the GPC system and eluted with an elution rate of 0.7 mL min−1. The molecular weights of the polysaccharides were calculated using a conventional calibration obtained from dextran standards (SIGMA, Germany).
Size exclusion chromatography (SEC)
Modified and unmodified levansucrases were subjected to size exclusion chromatography to analyse their monomeric/oligomeric status.0.5 mL of protein with a concentration of 4 mg mL−1 was applied to a HiLoad 16/600 Superdex 200 column (GE Healthcare, Germany) coupled to an Äkta purification system (GE Healthcare, Germany) and eluted with 50 mM Sorensen's buffer pH 6.9. The molecular weight of proteins was determined by comparing their elution volume with that of known SEC protein standards (SIGMA, Steinheim, Germany).
Differential scanning fluorimetry (DSF)
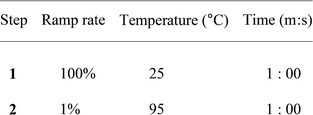
150 μL samples containing a final concentration of 2 μM protein (monomers of Y247F-2 or Y247F-2-1AzGlc), 50 mM Sorensen's buffer pH 6.9, 5× Sypro Orange (Thermo Fisher, Germany) and 0, 0.5, 1.0, 2.0, 4.0, 8.0, 15.0, 22.0, 35.0 or 50.0 mM levan with an average DP of 9 were prepared in 1.5 mL Eppendorf tubes. Control experiments without protein were also included. Aliquots of 25 μL of each sample were applied 4 times on a 96 well plate that was later sealed with a transparent adhesive film and centrifuged at 500 × g for 1 minute.The well plate was placed in a HT/7900 fast real time PCR system (Applied Biosystems, Germany) and changes in fluorescence over a temperature range were recorded using the ROX filter and the following program.
Fluorescence data were processed in OriginPro to obtain the melting temperature at each levan concentration by fitting the data to the Boltzmann function.
Melting temperature values of two independent measurements with four replicates each were used to calculate the apparent dissociation constant Kd using the single site ligand binding equation with the GraphPad Prism software.31
Molecular dynamic simulations
The trajectories and interactions of compounds 2 and 2-1AzGlc were investigated by MD simulations of the covalent complexes in which the compounds were attached to Y196 of Bm-LS variant Y247F. Simulations were performed using the AMBER14 program package.35 To achieve the wild-type of the protein we used the D257A mutant (pdb code 3OM2) and replaced Ala257 by Asp257. The covalent attachment of 2 on Y196 was performed with the Avogadro program package (http://avogadro.cc/).36 considering the enzyme surrounding (5 Å radius of the corresponding tyrosine residue). The positioning of sucrose was achieved by aligning the X-ray structure of Bm-LS complexed with sucrose (pdb code 1PT2) with wild-type modification (of 3OM2). 6-Kestose was inserted by aligning the fructose unit of sucrose and 6-kestose. The protein was described by the Amber14 force field ffl4SB.37 For 2, 2-1AzGlc, sucrose and 6-kestose the generalized AMBER force field (GAFF) was applied using Antechamber.38,39 To include the environment in an appropriate manner an octahedral solvation shell with periodic boundaries was used. A minimization containing 1000 steps was carried out with restraints on the protein–ligand complex followed by 2500 steps without restraints allowing minimization of the entire system. Gradual heating from 0 to 300 K was performed with a duration of 100 ps including restraints on the protein–inhibitor complex. Afterwards a MD simulation of 10 ns without restraints was carried out.Analysis of full length unmodified and modified proteins by LC-MS
The time course samples of the tyrosine modification reaction were analysed by LC-MS on a micrOTOF-Q III (Bruker Daltonics) equipped with an ESI Ion Source (Apollo II) and coupled to an Agilent 1100 HPLC System. The Q-TOF MS instrument has a resolution of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and a mass accuracy of 5 ppm with external calibration. Full length proteins were loaded on a reversed phase column (YMS-Pack C4, 15 cm × 4.6 mm ID, 5 μM, YMC) and separated with a flow of 0.5 mL min−1 over 30 minutes using a step gradient from 25% to 90% acetonitrile. The MS spectra were recorded continuously in the range m/z 200–3000 with the following settings: capillary voltage −4.5 kV, end plate voltage – 4 kV, nitrogen nebulizer pressure 3 bar, dry gas flow 12 L min−1, dry temperature 180 °C, funnel 1 and 2 RF 400 Vpp, Hexapole RF 200 Vpp, Quadrupole ion energy 5 eV, Quadrupole low mass 200 m/z, collision energy 7 eV, collision RF 1000 Vpp, transfer time 100 μs, and pre pulse storage 10 μs. The instrument was calibrated with the MMI-L Low Concentration Tuning Mix (Agilent) prior to measurement of the samples. The accuracy of the calibration masses was re-checked by measuring the Tuning Mix after the sample measurements. The spectra were recorded with the Compass Software (Bruker Daltonics) containing the OTOF Control 3.4 and Hystar 3.2 software for controlling the MS instrument and the HPLC. For spectra deconvolution and data evaluation the Data Analysis software DA 4.2 (Bruker Daltonics) was used.
000 and a mass accuracy of 5 ppm with external calibration. Full length proteins were loaded on a reversed phase column (YMS-Pack C4, 15 cm × 4.6 mm ID, 5 μM, YMC) and separated with a flow of 0.5 mL min−1 over 30 minutes using a step gradient from 25% to 90% acetonitrile. The MS spectra were recorded continuously in the range m/z 200–3000 with the following settings: capillary voltage −4.5 kV, end plate voltage – 4 kV, nitrogen nebulizer pressure 3 bar, dry gas flow 12 L min−1, dry temperature 180 °C, funnel 1 and 2 RF 400 Vpp, Hexapole RF 200 Vpp, Quadrupole ion energy 5 eV, Quadrupole low mass 200 m/z, collision energy 7 eV, collision RF 1000 Vpp, transfer time 100 μs, and pre pulse storage 10 μs. The instrument was calibrated with the MMI-L Low Concentration Tuning Mix (Agilent) prior to measurement of the samples. The accuracy of the calibration masses was re-checked by measuring the Tuning Mix after the sample measurements. The spectra were recorded with the Compass Software (Bruker Daltonics) containing the OTOF Control 3.4 and Hystar 3.2 software for controlling the MS instrument and the HPLC. For spectra deconvolution and data evaluation the Data Analysis software DA 4.2 (Bruker Daltonics) was used.
Protein digestion and analysis by mass spectrometry
Tyrosine-modified and unmodified enzymes were analysed by SDS-PAGE applying 5 μg of reduced and alkylated proteins onto 12% acrylamide gels.Excised gel bands were destained with 30% acetonitrile in 0.1 M NH4HCO3 (pH 8), shrunk with 100% acetonitrile, and dried in a vacuum concentrator (Concentrator 5301, Eppendorf, Germany). Digests were performed with 0.1 μg elastase per gel band overnight at 37 °C in 0.1 M NH4HCO3 (pH 8). After removing the supernatant, peptides were extracted from the gel slices with 5% formic acid, and extracted peptides were pooled with the supernatant.
NanoLC-MS/MS analyses were performed on an Orbitrap Fusion (Thermo Scientific) equipped with a PicoView Ion Source (New Objective) and coupled to an EASY-nLC 1000 (Thermo Scientific). Peptides were loaded on capillary columns (PicoFrit, 30 cm × 150 μm ID, New Objective) self-packed with ReproSil-Pur 120 C18-AQ, 1.9 μm (Dr Maisch) and separated with a 30 minute linear gradient from 3% to 40% acetonitrile and 0.1% formic acid and a flow rate of 500 nL min−1.
Both MS and MS/MS scans were acquired in the Orbitrap analyzer with a resolution of 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 for MS scans and 15
000 for MS scans and 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 for MS/MS scans. A mixed ETD/HCD method was used. HCD fragmentation was applied with 35% normalized collision energy. For ETD calibrated charge-dependent ETD parameters were applied. A top speed data-dependent MS/MS method with a fixed cycle time of 3 seconds was used. Dynamic exclusion was applied with a repeat count of 1 and an exclusion duration of 10 seconds; singly charged precursors were excluded from selection. The minimum signal threshold for precursor selection was set to 50
000 for MS/MS scans. A mixed ETD/HCD method was used. HCD fragmentation was applied with 35% normalized collision energy. For ETD calibrated charge-dependent ETD parameters were applied. A top speed data-dependent MS/MS method with a fixed cycle time of 3 seconds was used. Dynamic exclusion was applied with a repeat count of 1 and an exclusion duration of 10 seconds; singly charged precursors were excluded from selection. The minimum signal threshold for precursor selection was set to 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. Predictive AGC was used with an AGC target value of 2 × 105 for MS scans and 5 × 104 for MS/MS scans. EASY-IC was used for internal calibration.
000. Predictive AGC was used with an AGC target value of 2 × 105 for MS scans and 5 × 104 for MS/MS scans. EASY-IC was used for internal calibration.
PEAKS Studio 8.0 was used for raw data processing and database searching with the following parameters: parent mass error tolerance: 7.0 ppm, fragment mass error tolerance: 0.015 Da, enzyme: none, variable modifications: carbamidomethyl (C) (57.02 Da), oxidation (M) (15.99 Da), acetylation (protein N-terminal) (42.01), pyro-Glu (N-terminal Q) (−17.03 Da), 2 (labelled as J2) (Y) (256.08 Da), and 2-1AzGlc (labelled as J2-azido-glucopyranoside) (Y) (461.15 Da). A small custom-made database (about 200 proteins) containing sequences of wild-type Bm-LS and its variants was applied. The results were filtered with 1% FDR at the PSM level.
Synthesis of 4-hydroxyphthalic acid (4)
In a 1 L-three-necked piston with inside a thermometer and KPG-stirrer a solution of 50% aqueous 4-sulphophthalic acid (591 g, 1.20 mol) was stirred at room temperature and NaOH (577 g, 14.4 mol) was added in small portions. The reaction mixture was stirred and heated for 1 h at 180 °C. After cooling the reaction to room temperature, 1 L water and 1 L conc. HCl were added to precipitate the solid product. The filtrate was extracted with AcOEt by using a perforator, the combined organic layer was dried over Na2SO4 and the solvent was removed under reduced pressure. Product 4 (50.7 g, 23%) was obtained as a white solid.1H-NMR (400 MHz, DMSO-d6): δ = 12.81 (s, 2H, COOH), 10.39 (s, 1H, OH), 7.64 (d, 1H, 3J = 8.52 Hz, H-1), 6.87 (dd, 1H, 3J = 8.42 Hz, 4J = 2.50 Hz, H-2), 6.84 (d, 1H, 4J = 2.31 Hz, H-3) ppm.
13C-NMR (100 MHz, DMSO-d6): δ = 169.6 (C-8), 167.4 (C-7), 160.2 (C-3), 137.5 (C-5), 131.6 (C-1), 120.8 (C-6), 116.3 (C-2), 114.3 (C-4) ppm (ESI, Fig. S21†).
HRMS (ESI pos., m/z): calculated C8H6O5 [M + Na]+ 205.01129, measured 205.01074, Δppm: 1.46.
Synthesis of dimethyl-4-hydroxyphthalate (5)
4-Hydroxyphthalic acid (15.4 g, 84.6 mmol) was dissolved under a protective gas atmosphere in 160 mL dry methanol. The solution was cooled to 0 °C and 18 mL thionyl chloride (248 mmol) was added dropwise. The solvent was removed under reduced pressure and the residue was redissolved in 50 mL AcOEt. The organic layer was washed with 30 mL water, 30 mL brine and dried over MgSO4. Product 5 (16.2 g, 91%) was obtained as a white solid by evaporating the solvent under reduced pressure.1H-NMR (400 MHz, CDCl3): δ = 7.76 (d, 1H, 3J = 8.53 Hz, H-1), 7.02 (d, 1H, 4J = 2.56 Hz, H-2), 6.93 (dd, 1H, 3J = 8.54 Hz, 4J = 2.60 Hz, H-4), 3.91–3.87 (s, 6H, H-9/10) ppm.
13C-NMR (100 MHz, CDCl3): δ = 169.5 (C-8), 167.2 (C-7), 159.2 (C-3), 135.8 (C-5), 132.1 (C-1), 121.9 (C-6), 117.4 (C-2), 115.4 (C-4), 53.1 (C-10), 52.7 (C-9) ppm (ESI, Fig. S22†).
HRMS (ESI pos., m/z): calculated C10H10O5 [M + Na]+ 233.04204, measured 233.04222, Δppm: 0.74.
Synthesis of 5-bromo-1-pentin (7)
Pent-4-yn-1-ol (1.27 g, 15.0 mmol) and tetrabromide (7.73 g, 23.3 mmol) were dissolved in 20 mL DCM and cooled to 0 °C. After adding triphenylphosphine (6.31 g, 24.1 mmol) in small portions the reactions mixture was stirred for 1 h at room temperature. Complete precipitation of the product was obtained by adding 10 mL cyclohexane and storing the suspension at 4 °C overnight. The product was filtered and washed with cyclohexane. The solvent was removed under reduced pressure and the product was purified by silica gel column (n-pentane) providing 7 (1.10 g, 50%) as a colourless oil.1H-NMR (400 MHz, CDCl3): δ = 3.54 (t, 2H, 3J = 6.4 Hz, H-1); 2.40 (dt, 2H, 3J = 8 Hz, 4J = 2.7 Hz, H-3); 2.06 (quin, 2H, 3J = 6.6 Hz, H-2); 1.99 (t, 1H, 4J = 2.7 Hz, H-5) ppm.
13C-NMR (100 MHz, CDCl3): δ = 82.5 (C-4), 69.5 (C-5), 32.3 (C-1), 31.3 (C-2), 17.2 (C-3) ppm (ESI, Fig. S23†).
Synthesis of dimethyl-4-(pent-4-yn-1-yloxy)phthalate (8)
To a solution of compound 5 (1.05 g, 5.0 mmol) and K2CO3 (1.05 g, 7.7 mmol) in DMF (10 mL) compound 7 (1.59 g, 7.72 mmol) was added to deliver compound 8. After stirring the mixture for 16 h at 60 °C the reaction was quenched with AcOEt (40 mL) and H2O (40 mL). The product was extracted with AcOEt and the combined organic layer was washed with brine and dried over MgSO4. The solvent was evaporated under reduced pressure and the product was purified using a silica gel column (Cy![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EE = 5
EE = 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) providing 8 (0.67 g, 51%) as a colourless oil.
1) providing 8 (0.67 g, 51%) as a colourless oil.
1H-NMR (400 MHz, CDCl3): δ = 7.80 (d, 1H, 3J = 8.6 Hz, H-1); 7.07 (d, 1H, 4J = 2.6 Hz, H-4); 6.99 (dd, 1H, 3J = 8.7 Hz, 4J = 2.6 Hz, H-2); 4.13 (t, 2H, 3J = 6.1 Hz, H-1′); 3.91 (s, 3H, H-9/10); 3.87 (s, 3H, H-9/10); 2.41 (dt, 2H, 3J = 6.9 Hz, 3J = 2.6 Hz, H-3′); 2.04–1.99 (m, 2H, H-2′); 1.98 (t, 1H, 4J = 2.6 Hz, H-5′) ppm.
13C-NMR (100 MHz, CDCl3): δ = 169.0 (C-8), 166.9 (C-7), 161.5 (C-3), 135.8 (C-5), 131.7 (C-1), 122.2 (C-6), 116.2 (C-2), 114.1 (C-4), 83.1 (C-4′), 69.3 (C-5′), 66.7 (C-1′), 52.9 (C-10), 52.5 (C-9), 27.9 (C-2′), 15.1 (C-3′) ppm (ESI, Fig. S24†).
HRMS (ESI pos., m/z): calculated C15H16O5 [M + Na]+ 299.08899, measured 299.08968, Δppm: 2.28 ppm.
Synthesis of (4-(pent-4-yn-1-yloxy)phthalic acid) (9)
To a solution of compound 8 (0.67 g, 2.43 mmol) in 10 mL THF and MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) a solution of 5 mL NaOH (5 m) was added and stirred for 16 h at room temperature. The reaction mixture was then acidified by adding 12 mL 1 M HCl (pH = 1) and the product was extracted with AcOEt. The combined organic layer was washed with brine, dried over MgSO4 and the solvent was removed under reduced pressure to obtain 9 (0.57 g, 2.30 mmol, 95%) as a white solid.
1) a solution of 5 mL NaOH (5 m) was added and stirred for 16 h at room temperature. The reaction mixture was then acidified by adding 12 mL 1 M HCl (pH = 1) and the product was extracted with AcOEt. The combined organic layer was washed with brine, dried over MgSO4 and the solvent was removed under reduced pressure to obtain 9 (0.57 g, 2.30 mmol, 95%) as a white solid.
1H-NMR (400 MHz, DMSO-d6): δ = 13.0 (s, 2H, COOH); 7.72 (d, 1H, 3J = 8.6 Hz, H-1); 7.08 (dd, 1H, 3J = 8.5 Hz, 3J = 2.6 Hz, H-2); 7.05 (d, 1H, 4J = 2.5 Hz, H-4); 4.12 (t, 2H, 3J = 6.16 Hz, H-1′); 2.83 (t, 1H, 4J = 2.6 Hz, 5′); 2.32 (dt, 2H, 3J = 7.1 Hz, 4J = 2.6 Hz, H-3′); 1.90 (quin, 2H, 3J = 6.7 Hz, H-2′) ppm.
13C-NMR (100 MHz, DMSO-d6): δ = 169.2 (C-8), 167.3 (C-7), 160.6 (C-3), 137.1 (C-5), 131.3 (C-1), 122.4 (C-6), 115.5 (C-2), 113.5 (C-4), 83.6 (C-4′), 71.8 (C-5′), 66.6 (C-1′), 27.5 (C-2′), 14.4 (C-3′) ppm (ESI, Fig. S25†).
HRMS (ESI pos., m/z): calculated C13H12O5 [M + Na]+ 271.05824, measured 271.05754.
Synthesis of 2-methyl-6-(pent-4-yn-1-yloxy)-2,3-dihydrophthalazine-1,4-dione (2) (isomer mixture)
To a solution of compound 9 (0.52 g, 2.11 mmol) in 20 mL dry THF 2.2 mL (23.3 mmol) Ac2O was added and the reaction mixture was stirred for 3 h under reflux conditions. After removing the solvent under reduced pressure, the residue was dissolved in 20 mL ethanol and 0.7 mL methylhydrazine (13.4 mmol) was added. The mixture was stirred again for 3 h under reflux conditions. The solution was cooled to room temperature and quenched with 50 mL sat. NH4Cl and 50 mL AcOEt. The product was extracted with AcOEt and the combined organic layer was washed with brine, dried over MgSO4 and the solvent was evaporated under reduced pressure. Detailed synthesis of 2 is described in ESI Scheme S2.† The purification was carried out using a silica gel column (Cy![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EE = 1
EE = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) providing 2 (0.53 g, 96%) as a white solid.
1) providing 2 (0.53 g, 96%) as a white solid.
1H-NMR (400 MHz, DMSO-d6): δ = 11.6 (s, 1H + 1.15H, NH); 8.12 (d, 1H, 3J = 8.8 Hz, H-5*); 7.79 (d, 1.15H, 3J = 8.8 Hz, H-8); 7.59 (d, 1.15H, 4J = 2.6 Hz, H-5); 7.45 (dd, 1.15H, 3J = 8.8 Hz, 4J = 2.7 Hz, H-7); 7.42 (dd, 1H, 3J = 8.8 Hz, 4J = 2.6 Hz, H-6*); 7.29 (d, 1H, 4J = 2.4 Hz, H-8*); 4.20 (m, 2H + 2 × 1.15H, H-1′); 3.55 (s, 3 × 1.15H, H-2*); 3.53 (s, 3H, H-2); 2.84 (m, 1H + 1.15H, H-5′); 2.39–2.34 (m, 2H + 2 × 1.15H, H-3′); 1.98–1.91 (m, 2H + 2 × 1.15H, H-2′) ppm (ESI, Fig. S26†).
NMR spectra of levan
NMR spectra (1H-NMR, 13C-NMR, DEPT135, COSY, HMBC, and HSQC) were measured on a BRUKER AVANCE 400 FT-NMR or a BRUKER AVANCE DMX 600 FT-NMR spectrometer at 25 °C. Proton chemical shifts (δ scale) are expressed in parts per million (ppm) and were determined relative to a residual protic solvent as an internal reference (CDCl3: δ = 7.26 ppm, MeOD: δ = 3.31 ppm). Data for 1H-NMR spectra are listed as follows: chemical shift (δ ppm) (multiplicity, integration, coupling constants (Hz), and assigned protons). Couplings are indicated as: s = singlet, d = doublet, t = triplet, and m = multiplet. Carbon nuclear magnetic resonance (13C-NMR) spectra were recorded with the same BRUKER spectrometers at 100.9 and 150.9 MHz, respectively. Carbon chemical shifts (δ scale) are indicated in ppm and calibrated to the carbon resonance of the respective solvent (CDCl3: δ = 77.16 ppm, MeOD: δ = 49.00 ppm, DMSO-d6: δ = 2.50 ppm) (ESI, Fig. S8–S11 and Table S2†).Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support by the TRR SFB 225 (397988206) Biofabrikation and the Bundesministerium für Bildung und Forschung (01DN16034) for the project “Synthesis of tailor-made modular hybrid functional oligosaccharides (Hy-OS)” is gratefully acknowledged. We would like to thank Anne Neumann for technical support.References
- S. G. Burton, D. A. Cowan and J. M. Woodley, Nat. Biotechnol., 2002, 20, 37–45 CrossRef PubMed
.
- M. T. Reetz, J. Am. Chem. Soc., 2013, 135, 12480–12496 CrossRef PubMed
.
- C. J. Noren, S. J. Anthonycahill, M. C. Griffith and P. G. Schultz, Science, 1989, 244, 182–188 Search PubMed
.
- V. W. Cornish, K. M. Hahn and P. G. Schultz, J. Am. Chem. Soc., 1996, 118, 8150–8151 CrossRef
.
- I. Kwon and B. Yang, Ind. Eng. Chem. Res., 2017, 56, 6535–6547 CrossRef
.
- T. H. Wright, B. J. Bower, J. M. Chalker, G. J. L. Bernardes, R. Wiewiora, W. L. Ng, R. Raj, S. Faulkner, M. R. J. Vallee, A. Phanumartwiwath, O. D. Coleman, M. L. Thezenas, M. Khan, S. R. G. Galan, L. Lercher, M. W. Schombs, S. Gerstberger, M. E. Palm-Espling, A. J. Baldwin, B. M. Kessler, T. D. W. Claridge, S. Mohammed and B. G. Davis, Science, 2016, 354, aag1465 CrossRef PubMed
.
- A. Yang, S. Ha, J. Ahn, R. Kim, S. Kim, Y. Lee, J. Kim, D. Soll, H. Y. Lee and H. S. Park, Science, 2016, 354, 623–626 CrossRef PubMed
.
- K. Wals and H. Ovaa, Front. Chem., 2014, 2 DOI:10.3389/fchem.2014.00015
.
- C. D. Spicer and B. G. Davis, Nat. Commun., 2014, 5 DOI:10.1038/ncomms5740
.
- J. M. Antos, J. M. McFarland, A. T. Iavarone and M. B. Francis, J. Am. Chem. Soc., 2009, 131, 6301–6308 CrossRef PubMed
.
- K. L. Seim, A. C. Obermeyer and M. B. Francis, J. Am. Chem. Soc., 2011, 133, 16970–16976 CrossRef PubMed
.
- H. Ban, J. Gavrilyuk and C. F. Barbas, J. Am. Chem. Soc., 2010, 132, 1523–1525 CrossRef PubMed
.
- S. Meunier, E. Strable and M. G. Finn, Chem. Biol., 2004, 11, 319–326 CrossRef PubMed
.
- S. D. Tilley and M. B. Francis, J. Am. Chem. Soc., 2006, 128, 1080–1081 CrossRef PubMed
.
- N. S. Joshi, L. R. Whitaker and M. B. Francis, J. Am. Chem. Soc., 2004, 126, 15942–15943 CrossRef PubMed
.
- V. Lombard, H. G. Ramulu, E. Drula, P. M. Coutinho and B. Henrissat, Nucleic Acids Res., 2014, 42, D490–D495 CrossRef PubMed
.
- A. Homann, R. Biedendieck, S. Gotze, D. Jahn and J. Seibel, Biochem. J., 2007, 407, 189–198 CrossRef PubMed
.
- E. T. Öner, L. Hernandez and J. Combie, Biotechnol. Adv., 2016, 34, 827–844 CrossRef PubMed
.
- M. E. Ortiz-Soto, M. Rivera, E. Rudino-Pinera, C. Olvera and A. Lopez-Munguia, Protein Eng., Des. Sel., 2008, 21, 589–595 CrossRef PubMed
.
- G. M. T. Calazans, R. C. Lima, F. P. de Franca and C. E. Lopes, Int. J. Biol. Macromol., 2000, 27, 245–247 CrossRef PubMed
.
- N. Elvassore, A. Bertucco and P. Caliceti, J. Pharm. Sci., 2001, 90, 1628–1636 CrossRef PubMed
.
- M. E. Ortiz-Soto, C. Possiel, J. Gorl, A. Vogel, R. Schmiedel and J. Seibel, Glycobiology, 2017, 27, 755–765 CrossRef PubMed
.
- C. P. Strube, A. Homann, M. Gamer, D. Jahn, J. Seibel and D. W. Heinz, J. Biol. Chem., 2011, 286, 17593–17600 CrossRef PubMed
.
- L. K. Ozimek, S. Kralj, T. Kaper, M. J. E. C. van der Maarel and L. Dijkhuizen, FEBS J., 2006, 273, 4104–4113 CrossRef PubMed
.
- H. Ban, M. Nagano, J. Gavrilyuk, W. Hakamata, T. Inokuma and C. F. Barbas, Bioconjugate Chem., 2013, 24, 520–532 CrossRef PubMed
.
- R. Dahl, K. K. Baldridge and N. S. Finney, Synthesis, 2010, 13, 2292–2296 Search PubMed
.
- S. Sato, K. Nakamura and H. Nakamura, ACS Chem. Biol., 2015, 10, 2633–2640 CrossRef PubMed
.
- H. C. Kolb, M. G. Finn and K. B. Sharpless, Angew. Chem., Int. Ed., 2001, 40, 2004–2021 CrossRef PubMed
.
- E. Raga-Carbajal, E. Carrillo-Nava, M. Costas, J. Porras-Dominguez, A. Lopez-Munguia and C. Olvera, Glycobiology, 2016, 26, 377–385 CrossRef PubMed
.
- D. Goldman, N. Lavid, A. Schwartz, G. Shoham, D. Danino and Y. Shoham, J. Biol. Chem., 2008, 283, 32209–32217 CrossRef PubMed
.
- M. Vivoli, H. R. Novak, J. A. Littlechild and N. J. Harmer, J. Visualized Exp., 2014 DOI:10.3791/51809
.
- Y. R. Hua and A. H. Flood, Chem. Soc. Rev., 2010, 39, 1262–1271 RSC
.
- E. Krissinel and K. Henrick, J. Mol. Biol., 2007, 372, 774–797 CrossRef PubMed
.
- G. L. Miller, Anal. Chem., 1959, 31, 426–428 CrossRef
.
-
D. A. Case, V. Babin, J. T. Berryman, R. M. Betz, Q. Cai, D. S. Cerruti, T. E. Cheatham, T. A. Darden, R. E. Duke, H. Gohlke, A. W. Goetz, S. Gusarov, N. Homeyer, P. Janowski, J. Kaus, I. Kolossváry, A. Kovalenko, T. S. Lee, S. LeGrand, T. Luchko, R. Luo, B. Madej, K. M. Merz, F. Paesani, D. R. Roe, A. Roitberg, C. Sagui, R. Salomon-Ferrer, G. Seabra, C. L. Simmerling, W. Smith, J. Swails, R. C. Walker, J. Wang, R. M. Wolf, X. Wu and P. A. Kollman, AMBER 14, University of California, San Francisco, 2014 Search PubMed
.
- M. D. Hanwell, D. E. Curtis, D. C. Lonie, T. Vandermeersch, E. Zurek and G. R. Hutchison, J. Cheminf., 2012, 4, 17 Search PubMed
.
- J. A. Maier, C. Martinez, K. Kasavajhala, L. Wickstrom, K. E. Hauser and C. Simmerling, J. Chem. Theory Comput., 2015, 11, 3696–3713 CrossRef PubMed
.
- J. Wang, W. Wang, P. A. Kollman and D. A. Case, J. Mol. Graphics Modell., 2006, 25, 247–260 CrossRef PubMed
.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8sc01244j |
| This journal is © The Royal Society of Chemistry 2018 |