Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A halogen bond-mediated highly active artificial chloride channel with high anticancer activity†
Changliang
Ren
 a,
Xin
Ding
a,
Arundhati
Roy
a,
Xin
Ding
a,
Arundhati
Roy
 a,
Jie
Shen
a,
Shaoyuan
Zhou
b,
Feng
Chen
a,
Sam Fong
Yau Li
a,
Jie
Shen
a,
Shaoyuan
Zhou
b,
Feng
Chen
a,
Sam Fong
Yau Li
 c,
Haisheng
Ren
c,
Haisheng
Ren
 b,
Yi Yan
Yang
b,
Yi Yan
Yang
 a and
Huaqiang
Zeng
a and
Huaqiang
Zeng
 *a
*a
aInstitute of Bioengineering and Nanotechnology, 31 Biopolis Way, The Nanos, Singapore 138669. E-mail: hqzeng@ibn.a-star.edu.sg
bCollege of Chemical Engineering, Sichuan University, Chengdu, China 610065
cNUS Environmental Research Institute, Department of Chemistry, National University of Singapore, 3 Science Drive 3, Singapore 117543
First published on 15th March 2018
Abstract
Chloride-selective transmembrane carriers or channels might have possible uses in treating channelopathies or cancers. While chloride carriers have been extensively investigated, the corresponding chloride channels have remained limitedly studied. Moreover, all hitherto reported channel systems lack clearly definable and readily modifiable positions in their structures for the reliable construction and combinatorial optimization of their ion transport properties. As a result, the existing channels are limited by their large molecular weight, weak activity or low anion selectivity. In this report, we describe a readily accessible and robust monopeptide-based scaffold for the reliable construction of halogen bond-mediated artificial anion channels via directional assembly of electron-deficient iodine atoms, which create a transmembrane pathway for facilitating anion transport. The high intrinsic modularity of the backbone of the scaffold, which enables the rapid and combinatorial optimization of the transport activity and selectivity of channels, effectively delivers a highly active chloride channel A10. Such high activity in chloride transport subsequently leads to an excellent IC50 value of 20 μM toward inhibiting the growth of human breast cancer cells (BT-474), an anticancer activity that is even higher than that of the well-known anticancer agent cisplatin.
Introduction
Tightly regulated by varying families of ion channels, precise maintenance of ion concentration gradients across biological membranes is crucial for many biological and cellular processes. Chloride is the most abundant anion in the human body due to its important roles in controlling the membrane potential, cell volume, cellular pH balance, secretion of transepithelial fluid and electrolytes, etc.1 Misregulation of chloride ions could lead to a variety of life-shortening ‘channelopathies’ including cystic fibrosis, Bartter syndrome and Dent's disease. Accordingly, significant research efforts have been made recently to coherently develop artificial chloride carriers2 or channels3 as possible replacements for natural chloride channels for channelopathies.4 On the other hand, artificial anion transporters might also disrupt pH gradients4f,5a,b or ion homeostasis5c that may induce cell death, and therefore may have medical applications as anticancer agents.4f,5Recently, Matile et al. reported self-assembled chloride carriers2b,c or channels,3i elegantly exploiting halogen bonding interaction as the driving force for anion transport. Halogen bonds offer good bond strength and also good anion-induced directionality.6 Therefore, artificial anion transporters comprising simple structural motifs based on halogen–anion interactions may indeed offer a new horizon of prospects to comprehend complex chloride transport phenomena, often manifested in protein channels. Nevertheless, most documented artificial chloride channels purposefully harness various non-covalent interactions such as electrostatic, H-bonding, anion-π and halogen bonding forces. Channels that primarily rely on halogen bonds for facilitating anion transport still remain limitedly investigated with just one precedent.3i Such a rare occurrence presumably results from the lack of suitable molecular scaffolds for aligning electron-deficient iodine atoms into a well-defined one-dimensional (1D) array. Moreover, the majority of existing chloride channels3a–m suffer from relatively complex molecular structures and/or low selectivity in anion recognition, greatly restricting their application in drug discovery.
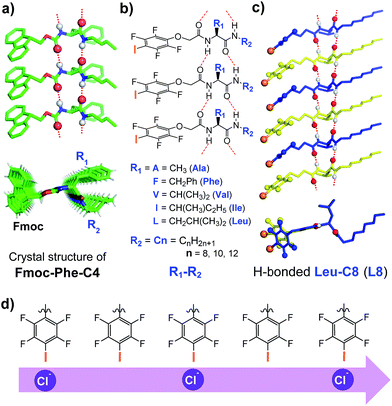
We recently described a novel class of readily accessible monopeptide-based scaffolds capable of self-assembling into 1D columnar structures through highly directional intermolecular H-bonding forces, subsequently enabling rapid room-temperature gelation of diverse types of crude oils (Fig. 1a).7a–c Revealed by the crystal structure of F-Phe-C4 (Fig. 1a), one unique feature of this monopeptide-based scaffold is that the same type of side chains such as Fmoc, R1 and R2 are always aligned to the same side as a result of the high directionality of H bonds (Fig. 1a).7a On this basis, we envisioned that replacing the Fmoc group with a chloride-binding unit such as the tetrafluoroiodobenzyl group may lead to an interesting class of artificial anion channels with anion transport mediated by halogen bonds (Fig. 1b–d).3i,7d A high intrinsic modularity involving R1 and R2 groups in the backbone should allow for rapid and combinatorial optimization of transport activity and selectivity of channels. In this report, we demonstrate that this indeed is the case, with the identified chloride channel exhibiting a high activity in chloride transport with respect to many other types of anions. We further demonstrate that human breast cancer cells (BT-474) are sensitive to a sodium chloride concentration gradient across the membrane, and this feature can be utilized by artificial chloride channels to inhibit cancer cell growth for their potential uses in cancer chemotherapy.4f,5
Results and discussion
Combinatorial identification of highly active anion-transporting artificial channels
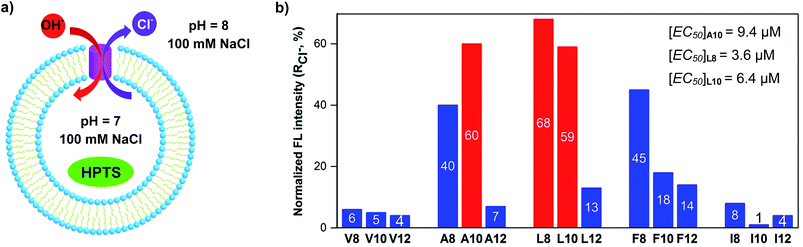
We synthesized a total of 15 possible channel molecules (five amino acids × three alkyl chains of different lengths, Fig. 1b) with the tetrafluoroiodobenzyl group as the chloride-binding and -transporting unit. Corroborated by the crystal structure of F-Phe-C4 (Fig. 1a) and the computationally optimized structure of L8 (Fig. 1c), the ability of these 15 molecules to pile up to form a H-bonded 1D structure can be convincingly demonstrated by the ability of A10, L8 and L10 to efficiently congeal in n-hexane via the formation of a 3D entangled fibrous network (ESI, Fig. S1†). The ion transport activities of the synthesized channels were then evaluated using the pH-sensitive HPTS (8-hydroxypyrene-1,3,6-trisulfonic acid) assay (Fig. 2 and S2†).8 In a typical experiment, large unilamellar vesicles (LUVs), encapsulating HPTS (100 μM) and NaCl (100 mM) at pH = 7.0, were diluted into the same buffer at pH 8.0 to generate a pH gradient across LUVs. After the addition of channel molecules, changes in the fluorescence intensity of HPTS were monitored over 5 min. Fig. 2b shows that, among all 15 channels tested at 10 μM, A10, L8 and L10 display the highest ion transport capacities with fractional activities (RCl−) of 60%, 68% and 59%, respectively. The corresponding EC50 values, which are the concentrations required to reach 50% transmembrane activity, were determined via Hill analyses to be 9.4, 3.6 and 6.4 μM, respectively (Fig. S3†).Cl−/OH− as the major transport species
Except for the presence of arrayed electron-deficient iodine atoms that can bind the Cl− anion,2b,c,3i all peptide molecules carry no other readily accessible functional groups for strongly binding either cationic or anionic species. Further, varying extravesicular metal chloride salts from LiCl to CsCl produces near-identical changes in fluorescence intensity (Fig. 3a and S4†), suggesting the inability of L8 or A10 to transport any of the five alkali metal ions. These combined structural features and experimental evidence led us to believe that, rather than the H+/M+ antiport mechanism, either the OH−/Cl− antiport (Fig. 2a) or H+/Cl− symport mechanism may largely account for the observed increases in the fluorescence intensity of HPTS.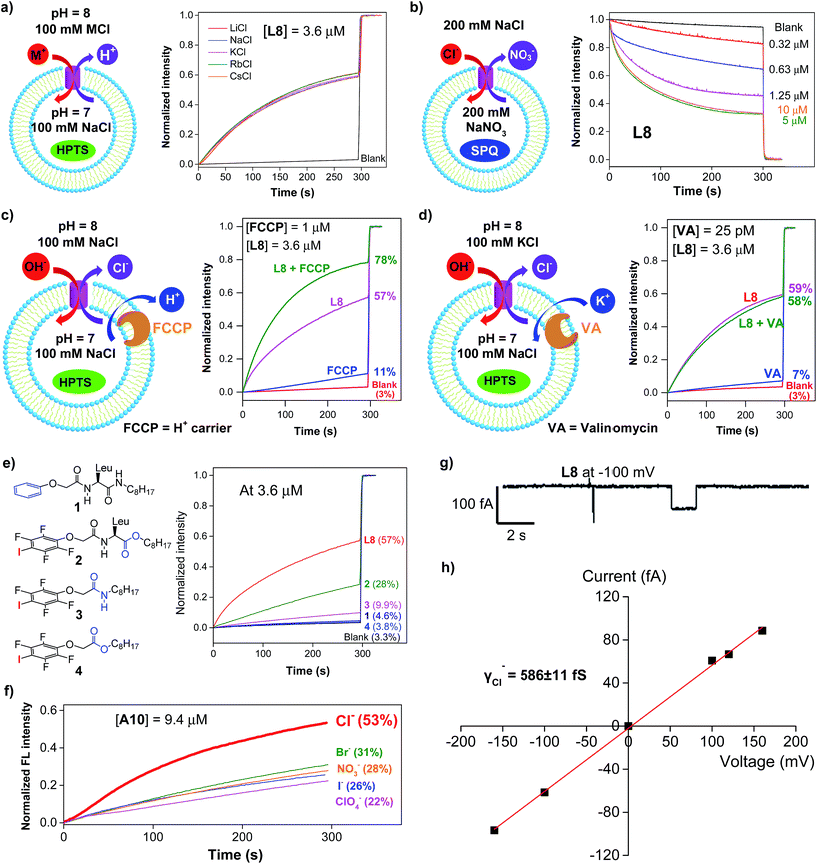 | ||
| Fig. 3 (a) Transport selectivity of alkali metal ions by L8 (3.6 μM) obtained from the HPTS assay by varying the extravesicular MCl (M+ = Li+, Na+, K+, Rb+ and Cs+). (b) Concentration-dependent changes in the fluorescence intensity of the chloride-sensitive SPQ dye (λex = 360 nm, λem = 430 nm) after the addition of L8 at different concentrations. Inside LUV: 200 mM NaNO3 and 0.5 mM SPQ. Outside LUV: 200 mM NaCl. (c) Ion transport activities of L8 (3.6 μM) determined in the absence or presence of proton transporter FCCP (1 μM), indicating a preferential transport of Cl− over H+. (d) Ion transport activities of L8 (3.6 μM) determined in the absence or presence of a potassium carrier, valinomycin (VA, 25 pM), indicating a preferential transport of Cl− over OH−. (e) Ion transport activities of four control compounds 1–4 at 3.6 μM, suggesting a critical role of halogen bonds in mediating the transport of Cl− and OH−. (f) Anion selectivity assayed for A10 (For L8 and L10, see Fig. S10†). (g) Single current trace of L8 recorded at −100 mv with symmetric baths (cis chamber = trans chamber = 1 M KCl). (h) I–V curve for obtaining the Cl− conductance (γCl−) of L8. All the data were averaged over two measurements. | ||
To deduce the most likely transport species, we first carried out the SPQ assay9 using a chloride-sensitive SPQ dye (6-methoxy-N-(3-sulfopropyl)quinolinium). As illustrated in Fig. 3b and S5,† the addition of L8 or A10 results in a rapid quenching of SPQ fluorescence in a concentration-dependent manner. These results suggest that the influx of Cl− increases with increasing channel concentrations from 0.32 to 5 μM for L8 and from 2.5 to 20 μM for A10, thereby establishing the Cl− anion as one of the molecular species transported by both L8 and A10.
Next, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP, a well-known proton carrier) was employed (Fig. 3c). Compared to the transport efficiencies of 11% for FCCP alone and of 57% for L8 alone, an increased transport efficiency of 13% (e.g., 78–57% – (11–3%)) for L8 in the presence of FCCP indicates a cooperative action between L8 and FCCP. In the case of A10, an even larger enhancement of 25% was observed (Fig. S6†). These results suggest that the transport rate of Cl− is faster than that of H+.
The HPTS assay was then performed in the presence of valinomycin (VA, a K+-selective carrier, Fig. 3d) to compare the transport rate between OH− and Cl−. For this assay carried out under iso-osmolar K+ (extravesicular) vs. Na+ (intravesicular) and pH gradient, the VA-mediated influx of K+ ions will induce an anion channel-mediated influx of either OH− or Cl− in order to maintain overall charge equality. If the influx of OH− is faster than that of Cl−, an increase in fluorescence intensity is anticipated. Experimentally, valinomycin at 25 pM produces a small increment of fluorescence intensity (7%) compared to the blank (3%). In a similar way, the ion transport activities of L8 (3.6 μM) in the presence and absence of valinomycin were found to be near-identical (58% vs. 59%), which is indicative of a preferential transport of Cl− over OH− (e.g., Cl− > OH−). A similar result was also seen for A10 (Fig. S7†). Consistent with transport activities presented in Fig. 3a, these data also suggest that the H+/M+ antiport mechanism is unlikely for L8-mediated enhancement in fluorescence intensity.
A mere increase of 1.3% (4.6–3.3%, Fig. 3e) in fluorescence intensity at 3.6 μM for molecule 1, carrying a simple benzene group, strongly suggests a lack of readily accessible functional groups in the H-bonded peptidic backbone for interacting with either anions or protons, and thus a minor role played by the backbone in mediating chloride transport. Instead, it is halogen bonds formed between anions (e.g., Cl− or OH−) and electron-deficient iodine atoms that cause efficient exchanges between Cl− and OH− anions across LUVs as observed for L8 (57%), pointing to a high unlikelihood of having H+/Cl− as the transport species. The herein assumed formation of halogen bonds I⋯Cl− and I⋯OH− was recently observed in their corresponding crystal structures2c and can be further supported by 19F NMR titration experiments involving titrating 0–20 equiv. of tetrabutylammonium chloride into a D2O-saturated CDCl3 solution containing L8 at 1 mM (Fig. S8†). Relative to the internal standard (1,4-difluorobenzene, −120.50 ppm), increasing additions of up to 20 equiv. of TBACl led to increasing upfield shifts of up to 0.61 ppm in the chemical shift of 19F of L8, a fact that is consistent with earlier observations2b and indicates binding between the chloride anion and the acidic iodine atom.
Additional comparisons among L8 and 2–4 convincingly demonstrate the importance of (1) the co-existence of two amide bonds that likely allow for a tighter and more directional stacking of channel molecules (L8vs.2), (2) the side chain of leucine for more efficient stacking (2vs.3) and (3) the amide bond in forming a H-bonded structure for ordered spanning of the hydrophobic membrane region (3vs.4). These summative findings are also in accordance with the molecular dynamics (MD) simulation results (Fig. 4).
Anion selectivity in chloride transport
With high activity exhibited by these chloride transporters, our subsequent investigations focused on evaluating and comparing anion selectivity for the most efficient anion channels formed by A10, L8 and L10. For this purpose, both intra- and extravesicular anions were kept the same (100 mM NaX, X− = Cl−, Br−, I−, NO3− and ClO4−) with a proton gradient of pH 7 (inside) to pH 8 (outside). Data compiled in Fig. 3f, S9 and S10† show that A10 with an EC50 value of 9.4 μM exhibits good anion selectivity in chloride transport, and channels L8 and L10 likely selectively transport non-chloride anions (Fig. S10†).Chloride transport through a channel mechanism
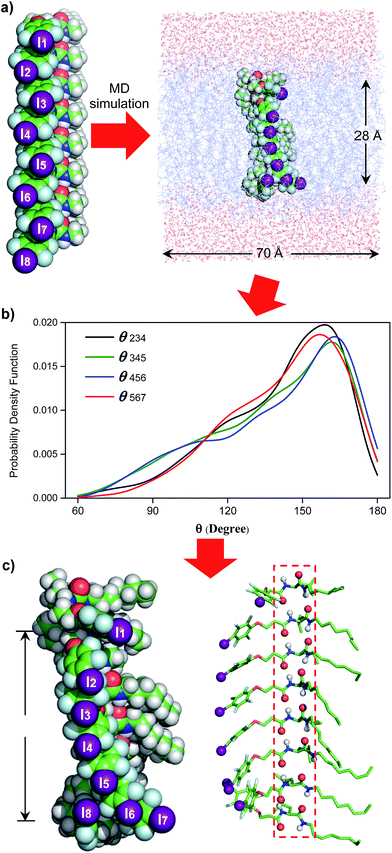
Single channel current traces for chloride transport, recorded in a planar lipid bilayer at various voltages including −100 mv in symmetric baths (cis chamber = trans chamber = 1 M KCl, Fig. 3g and S11†), unambiguously confirm that L8-mediated chloride transport occurs via a channel, rather than a carrier mechanism. On the basis of the fitted linear current–voltage (I–V) plot (Fig. 3h and S10†), the Cl− conductance (γCl−) of L8 was found to be 586 ± 11 fS.To shed some light on the possible structural features of one-dimensionally aligned channel molecules in the lipid membrane, MD simulation using the CHARMM program,10a–e PME method10f and SHAKE algorithm10g was performed on L8. In particular, the H-bonded 1D structure, which consists of eight molecules of L8 (528 atoms), was first computationally optimized using the COMPASS force field10h and then was embedded in a bilayer of 128 POPC molecules (17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 152 atoms) solvated on two sides by 2 × 2397 water molecules (Fig. 4a). This leads to a simulation system of 32
152 atoms) solvated on two sides by 2 × 2397 water molecules (Fig. 4a). This leads to a simulation system of 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 062 atoms with a dimension of 70 Å (w) × 70 Å (w) × 74 Å (h). After equilibration steps, the production run was carried out for 30 ns. The last 20 ns trajectories with 1000 structural snapshots were used for analysing both the hydrophobic thickness of the POPC membrane and the distribution probability of angle θ (e.g., the angle formed by the three immediately adjacent iodine atoms such as iodine atoms 2–4, Fig. 4a and b) using a probability density function.10i
062 atoms with a dimension of 70 Å (w) × 70 Å (w) × 74 Å (h). After equilibration steps, the production run was carried out for 30 ns. The last 20 ns trajectories with 1000 structural snapshots were used for analysing both the hydrophobic thickness of the POPC membrane and the distribution probability of angle θ (e.g., the angle formed by the three immediately adjacent iodine atoms such as iodine atoms 2–4, Fig. 4a and b) using a probability density function.10i
To estimate the hydrophobic membrane thickness, the Z-coordinates along the membrane normal of all 254 ester O-atoms from 64 POPC molecules located either at the top or on the bottom layers were averaged. The separation distance between the two averaged Z-coordinates of O-atoms from the top and bottom layers was calculated with deduction of a van der Waals diameter of 3.1 Å for the O-atom. The same calculation was performed for all 1000 structural snapshots to derive the probability distribution of the hydrophobic membrane thickness (Fig. S12†). From these computations, 633 structures have a hydrophobic thickness of 27 to 29 Å, and the average thickness over 1000 structures is 28.1 Å, a value that could be corroborated by the experimentally and computationally determined values of 27.1 and 27.8 Å, respectively, for the POPC membrane.11
Embedding eight molecules of L8 in the lipid membrane shows that the first and eighth molecules are not aligned well with the central six molecules (Fig. 4a and c). The most highly populated angles for θ234, θ345, θ456 and θ567 are 159°, 162°, 163° and 157°, respectively, with a representative structure, closely capturing these angles, shown in Fig. 4c. In this highly populated structure, the largest intermolecular separation distance along the Z axis among the eight iodine atoms occurs between the 1st and 7th iodine atoms (25 Å). Consistent with the intermolecular separation of 5.0 Å in Fmoc-Phe-C4 in the solid state (Fig. 1a)7a and of 4.9 Å in the computationally optimized H-bonded L8 (Fig. 1c), this separation distance of 25 Å suggests that six or seven molecules might be sufficient to span the hydrophobic core distance of 28.1 Å.
Through these MD simulations, the most important point to note is that, regardless of angle θ of varying magnitudes, all eight molecules of L8 remain H-bonded to each other in all 1000 structures surveyed (Fig. 4c). This demonstrates the reliability of intermolecular H-bonds in linking six or seven anion-binding molecules together to form a self-assembled 1D pathway, which spans the hydrophobic membrane region to facilitate highly efficient anion transport across the membrane.
High activity in chloride transport
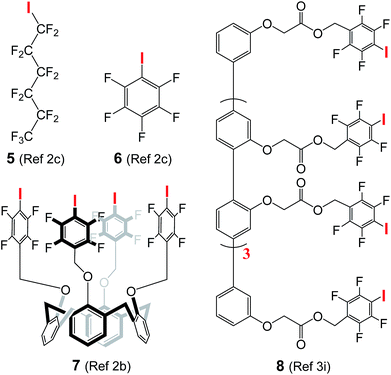
The use of halogen bonds to mediate anion transport was first explored by Matile,2b,c,3i with compounds 5 (EC50 = 3.1 μM)2c and 8 (EC50 = 0.88 μM in terms of chloride binding units)3i as the most active carrier and channel molecules among their own categories, respectively. These EC50 values, including those of 6 (260 μM) and 7 (68 μM) presented in Table 1, were determined using cholesterol-free LUVs with a total lipid concentration of 31.3 μM, while our assay conditions contain lipid and cholesterol at concentrations of 73.9 and 36.9 μM, respectively. For a fair comparison, we have therefore re-determined the EC50 values of A10, L8 and L10 using Matile's conditions (100 mM NaCl, 10 mM HEPES, pH gradient from 7 to 8, and the total lipid concentration without cholesterol = 31.3 μM).2b,c,3i| A10 | L8 | L10 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|
| a Determined using Matile's conditions (100 mM NaCl, 10 mM HEPES, pH gradient from 7 to 8, and the total lipid concentration without cholesterol = 31.25 μM; see ref. 2b, c and 3i). b EC50 value from ref. 2c is 3.1 μM with a Hill coefficient of 3.3. c From ref. 2c. d From ref. 2b. e From ref. 3i with the value normalized by the authors on the basis of eight halogen-binding units contained in channel 8. f Hill coefficient. | |||||||
| EC50 (μM) | 2.37 | 0.39 | 0.93 | 3.6 | 260 | 68 | 0.88 |
| n valuef | 3.5 | 3.6 | 3.1 | 3.3 | 3.6 | 1.9 | 0.8 |
To confirm that we are able to produce LUVs with properties similar to those prepared by Matile and his co-workers under the same assay conditions, we have performed Hill analysis on commercially available compound 5, and obtained an EC50 value of 3.6 μM with a Hill coefficient of 3.3 (Fig. S13†). These values are similar or identical to the reported values of EC50 (3.1 μM) and the Hill coefficient (3.3).2c Under these assay conditions, the EC50 values of A10, L8 and L10 were
In addition, chloride carrier 5, when evaluated under our assay conditions containing 33 mol% cholesterol that makes membrane less fluid,12 shows a moderate transport activity of 38% at 40 μM, and such moderate activity remains unchanged even when the concentration increases to 100 μM (Fig. S14†). These values suggest the ion transport activity of 5 to be at least 10 times lower than that of L8 having an EC50 value of 3.6 μM in the cholesterol-containing environment.
High anticancer activity
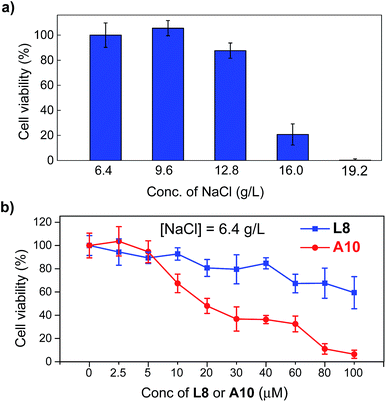
The above verified highly efficient chloride transport through a channel mechanism displayed by these artificial anion channels prompted us to examine the possibility of their uses in cancer chemotherapy.4f,5 Human breast cancer cells (BT-474, obtained from the American Type Culture Collection, USA) were cultured in Dulbecco's Modified Eagle Medium in the presence of up to 19.2 g L−1 NaCl (6.4 g L−1 is the typical concentration for cell growth) with concentrations of L8 or A10 varying from 0 to 100 μM (Fig. 5 and S15†). The viabilities of cells, determined after culturing the cells for 1 day at 37 °C with 5% CO2 in the absence of channel molecules, show that BT-474 cells are sensitive to the sodium chloride concentration gradient (Fig. 5a). That is, compared to cell viability at [NaCl] = 6.4 g L−1, 12, 79 and 99% fewer cells survive at [NaCl] of 12.8, 16.0 and 19.2 g L−1, respectively. Continued testing shows that A10 is considerably more potent than L8 in inhibiting cell growth across all three different concentrations of NaCl (e.g., 6.4, 9.6 and 12.8 g L−1) with an IC50 value of 20 μM for A10 at [NaCl] = 6.4 g L−1 (>100 μM for L8, Fig. 5b and S15†). For comparison, highly effective anticancer agent cisplatin has an IC50 value of 37 μM against the same BT-474 cells.13 Assuming all A10 molecules associate to form channels with each channel comprising six or more such molecules, the IC50 value in terms of effective channel concentration is lower than 3.3 μM.Conclusions
In conclusion, we have developed a class of artificial chloride channels with high activity and good selectivity via directional self-assembly of polarized electron-deficient iodine atoms. While these linearly arrayed acidic iodine atoms, which form a series of chloride-binding and -transporting sites, are responsible for facilitating chloride transport likely via a multi-ion jumping mode, the high modularity of the monopeptide backbone enables rapid optimization of the transport activity and selectivity of channels in a combinatorial format. The identified channels A10, L8 and L10 all turn out to be very active with the best EC50 values for chloride transport reaching 0.39 μM (1.2 mol% relative to lipid) and 3.6 μM (3.2 mol% relative to lipid/cholesterol) in cholesterol-free and -containing LUVs. In particular, the highly active A10 exhibits not only a fractional transport activity for chloride much better than other monovalent anions including bromide, but also an excellent inhibitory activity toward human breast cancer cells with an IC50 value of 20 μM. Further refinement to enhance the anion selectivity and cytotoxicity of channels toward cancer cells is possible and expected.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Institute of Bioengineering and Nanotechnology (Biomedical Research Council, Agency for Science, Technology and Research, Singapore) and the Singapore National Research Foundation under its Environment and Water Research Programme administered by PUB.Notes and references
- (a) R. Benz and R. E. W. Hancock, J. Gen. Physiol., 1987, 89, 275 CrossRef CAS PubMed; (b) C. Duran, C. H. Thompson, Q. Xiao and C. Hartzell, Annu. Rev. Physiol., 2010, 72, 95 CrossRef CAS PubMed; (c) N. Busschaert, C. Caltagirone, W. Van Rossom and P. A. Gale, Chem. Rev., 2015, 115, 8038 CrossRef CAS PubMed.
- (a) P. V. Santacroce, J. T. Davis, M. E. Light, P. A. Gale, J. C. Iglesias-Sánchez, P. Prados and R. Quesada, J. Am. Chem. Soc., 2007, 129, 1886 CrossRef CAS PubMed; (b) A. Vargas Jentzsch, D. Emery, J. Mareda, P. Metrangolo, G. Resnati and S. Matile, Angew. Chem., Int. Ed., 2011, 50, 11675 CrossRef CAS PubMed; (c) A. V. Jentzsch, D. Emery, J. Mareda, S. K. Nayak, P. Metrangolo, G. Resnati, N. Sakai and S. Matile, Nat. Commun., 2012, 3, 905 CrossRef PubMed; (d) H. Valkenier, L. W. Judd, H. Li, S. Hussain, D. N. Sheppard and A. P. Davis, J. Am. Chem. Soc., 2014, 136, 12507 CrossRef CAS PubMed; (e) A. Roy, D. Saha, A. Mukherjee and P. Talukdar, Org. Lett., 2016, 18, 5864 CrossRef CAS PubMed; (f) J. Gravel, J. Kempf and A. Schmitzer, Chem.–Eur. J., 2015, 21, 18642 CrossRef CAS PubMed; (g) E. B. Park and K.-S. Jeong, Chem. Commun., 2015, 51, 9197 RSC; (h) M. Lisbjerg, H. Valkenier, B. M. Jessen, H. Al-Kerdi, A. P. Davis and M. Pittelkow, J. Am. Chem. Soc., 2015, 137, 4948 CrossRef CAS PubMed.
- (a) V. Sidorov, F. W. Kotch, G. Abdrakhmanova, R. Mizani, J. C. Fettinger and J. T. Davis, J. Am. Chem. Soc., 2002, 124, 2267 CrossRef CAS PubMed; (b) P. H. Schlesinger, R. Ferdani, J. Liu, J. Pajewska, R. Pajewski, M. Saito, H. Shabany and G. W. Gokel, J. Am. Chem. Soc., 2002, 124, 1848 CrossRef CAS PubMed; (c) N. Madhavan, E. C. Robert and M. S. Gin, Angew. Chem., Int. Ed., 2005, 44, 7584 CrossRef CAS PubMed; (d) V. Gorteau, G. Bollot, J. Mareda, A. Perez-Velasco and S. Matile, J. Am. Chem. Soc., 2006, 128, 14788 CrossRef CAS PubMed; (e) J. D. Sadowsky, W. D. Fairlie, E. B. Hadley, H. S. Lee, N. Umezawa, Z. Nikolovska-Coleska, S. M. Wang, D. C. S. Huang, Y. Tomita and S. H. Gellman, J. Am. Chem. Soc., 2007, 129, 139 CrossRef CAS PubMed; (f) J. Mareda and S. Matile, Chem.–Eur. J., 2009, 15, 28 CrossRef CAS PubMed; (g) X. Li, B. Shen, X.-Q. Yao and D. Yang, J. Am. Chem. Soc., 2009, 131, 13676 CrossRef CAS PubMed; (h) C. R. Yamnitz, S. Negin, I. A. Carasel, R. K. Winter and G. W. Gokel, Chem. Commun., 2010, 46, 2838 RSC; (i) A. Vargas Jentzsch and S. Matile, J. Am. Chem. Soc., 2013, 135, 5302 CrossRef CAS PubMed; (j) T. Saha, S. Dasari, D. Tewari, A. Prathap, K. M. Sureshan, A. K. Bera, A. Mukherjee and P. Talukdar, J. Am. Chem. Soc., 2014, 136, 14128 CrossRef CAS PubMed; (k) T. Saha, A. Gautam, A. Mukherjee, M. Lahiri and P. Talukdar, J. Am. Chem. Soc., 2016, 138, 16443 CrossRef CAS PubMed; (l) X. Wei, G. Zhang, Y. Shen, Y. Zhong, R. Liu, N. Yang, F. Y. Al-mkhaizim, M. A. Kline, L. He, M. Li, Z.-L. Lu, Z. Shao and B. Gong, J. Am. Chem. Soc., 2016, 138, 2749 CrossRef CAS PubMed; (m) H. Behera and N. Madhavan, J. Am. Chem. Soc., 2017, 139, 12919 CrossRef CAS PubMed; (n) C. J. E. Haynes, J. Zhu, C. Chimerel, S. Hernández-Ainsa, I. A. Riddell, T. K. Ronson, U. F. Keyser and J. R. Nitschke, Angew. Chem., Int. Ed., 2017, 56, 15388 CrossRef CAS PubMed.
- (a) G. W. Gokel and N. Barkey, New J. Chem., 2009, 33, 947 RSC; (b) J. T. Davis, O. Okunola and R. Quesada, Chem. Soc. Rev., 2010, 39, 3843 RSC; (c) P. R. Brotherhood and A. P. Davis, Chem. Soc. Rev., 2010, 39, 3633 RSC; (d) S. Matile, A. Vargas Jentzsch, J. Montenegro and A. Fin, Chem. Soc. Rev., 2011, 40, 2453 RSC; (e) D. S. Kim and J. L. Sessler, Chem. Soc. Rev., 2015, 44, 532 RSC; (f) X. Wu, L. W. Judd, E. N. W. Howe, A. M. Withecombe, V. Soto-Cerrato, H. Li, N. Busschaert, H. Valkenier, R. Pérez-Tomás, D. N. Sheppard, Y.-B. Jiang, A. P. Davis and P. A. Gale, Chem, 2016, 1, 127 CrossRef CAS.
- (a) N. Busschaert, M. Wenzel, M. E. Light, P. Iglesias-Hernández, R. Pérez-Tomás and P. A. Gale, J. Am. Chem. Soc., 2011, 133, 14136 CrossRef CAS PubMed; (b) V. Soto-Cerrato, P. Manuel-Manresa, E. Hernando, S. Calabuig-Fariñas, A. Martínez-Romero, V. Fernández-Dueñas, K. Sahlholm, T. Knöpfel, M. García-Valverde, A. M. Rodilla, E. Jantus-Lewintre, R. Farràs, F. Ciruela, R. Pérez-Tomás and R. Quesada, J. Am. Chem. Soc., 2015, 137, 15892 CrossRef CAS PubMed; (c) S.-K. Ko, S. K. Kim, A. Share, V. M. Lynch, J. Park, W. Namkung, W. Van Rossom, N. Busschaert, P. A. Gale, J. L. Sessler and I. Shin, Nat. Chem., 2014, 6, 885 CrossRef CAS PubMed; (d) J. L. Sessler, L. R. Eller, W.-S. Cho, S. Nicolaou, A. Aguilar, J. T. Lee, V. M. Lynch and D. J. Magda, Angew. Chem., Int. Ed., 2005, 44, 5989 CrossRef CAS PubMed; (e) B. A. Smith, M. M. Daschbach, S. T. Gammon, S. Xiao, S. E. Chapman, C. Hudson, M. Suckow, D. Piwnica-Worms, G. W. Gokel and W. M. Leevy, Chem. Commun., 2011, 47, 7977 RSC; (f) S. N. Berry, V. Soto-Cerrato, E. N. W. Howe, H. J. Clarke, I. Mistry, A. Tavassoli, Y.-T. Chang, R. Perez-Tomas and P. A. Gale, Chem. Sci., 2016, 7, 5069 RSC; (g) T. Saha, M. S. Hossain, D. Saha, M. Lahiri and P. Talukdar, J. Am. Chem. Soc., 2016, 138, 7558 CrossRef CAS PubMed.
- L. C. Gilday, S. W. Robinson, T. A. Barendt, M. J. Langton, B. R. Mullaney and P. D. Beer, Chem. Rev., 2015, 115, 7118 CrossRef CAS PubMed.
- (a) C. Ren, G. H. B. Ng, H. Wu, K.-H. Chan, J. Shen, C. Teh, J. Y. Ying and H. Zeng, Chem. Mater., 2016, 28, 4001 CrossRef CAS; (b) C. Ren, F. Chen, F. Zhou, J. Shen, H. Su and H. Zeng, Langmuir, 2016, 32, 13510 CrossRef CAS PubMed; (c) C. L. Ren, J. Shen, F. Chen and H. Q. Zeng, Angew. Chem., Int. Ed., 2017, 56, 3847 CrossRef CAS PubMed; (d) C. L. Ren, J. Shen and H. Q. Zeng, J. Am. Chem. Soc., 2017, 139, 12338 CrossRef CAS PubMed.
- H. Zhao, S. Sheng, Y. Hong and H. Zeng, J. Am. Chem. Soc., 2014, 136, 14270 CrossRef CAS PubMed.
- A. S. Verkman, R. Takla, B. Sefton, C. Basbaum and J. H. Widdicombe, Biochemistry, 1989, 28, 4240 CrossRef CAS PubMed.
- (a) S. Jo, T. Kim, V. G. Iyer and W. Im, J. Comput. Chem., 2008, 29, 1859 CrossRef CAS PubMed; (b) E. L. Wu, X. Cheng, S. Jo, H. Rui, K. C. Song, E. M. Davila-Contreras, Y. Qi, J. Lee, V. Monje-Galvan, R. M. Venable, J. B. Klauda and W. Im, J. Comput. Chem., 2014, 35, 1997 CrossRef CAS PubMed; (c) S. Jo, J. B. Lim, J. B. Klauda and W. Im, Biophys. J., 2009, 97, 50 CrossRef CAS PubMed; (d) K. Vanommeslaeghe, E. Hatcher, C. Acharya, S. Kundu, S. Zhong, J. Shim, E. Darian, O. Guvench, P. Lopes, I. Vorobyov and A. D. MacKerell, J. Comput. Chem., 2010, 31, 671 CAS; (e) W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926 CrossRef CAS; (f) U. Essmann, L. Perera, M. L. Berkowitz, T. Darden, H. Lee and L. G. Pedersen, J. Chem. Phys., 1995, 103, 8577 CrossRef CAS; (g) G. J. Martyna, D. J. Tobias and M. L. Klein, J. Phys. Chem., 1994, 101, 4177 CrossRef CAS; (h) H. Sun, J. Phys. Chem. B, 1998, 102, 7338–7364 CrossRef CAS; (i) D. N. Bopege, M. Petrowsky, A. M. Fleshman, R. Frech and M. B. Johnson, J. Phys. Chem. B, 2012, 116, 71 CrossRef CAS PubMed.
- Y. Guo, S. Pogodin and V. A. Baulin, J. Chem. Phys., 2014, 140, 174903 CrossRef PubMed.
- (a) G. M. Cooper, The Cell: A Molecular Approach, Sinauer Associates, Sunderland, MA, 2nd edn, 2000 Search PubMed; (b) D. Marsh, Biochim. Biophys. Acta, 2009, 1788, 2114 CrossRef CAS PubMed.
- M. Buccioni, D. Dal Ben, C. Lambertucci, F. Maggi, F. Papa, A. Thomas, C. Santinelli and G. Marucci, Sci. World J., 2014, 2014, 264829 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8sc00602d |
| This journal is © The Royal Society of Chemistry 2018 |