DOI:
10.1039/C8SC00098K
(Edge Article)
Chem. Sci., 2018,
9, 3563-3569
A pre-protective strategy for precise tumor targeting and efficient photodynamic therapy with a switchable DNA/upconversion nanocomposite†
Received
9th January 2018
, Accepted 28th February 2018
First published on 5th March 2018
Abstract
Tumor-specific targeting based on folic acid (FA) is one of the most common and significant approaches in cancer therapy. However, the expression of folate receptors (FRs) in normal tissues will lead to unexpected targeting and unsatisfactory therapeutic effect. To address this issue, we develop a pre-protective strategy for precise tumor targeting and efficient photodynamic therapy (PDT) using a switchable DNA/upconversion nanocomposite, which can be triggered in the acidic tumor microenvironment. The DNA/upconversion nanocomposite is composed of polyacrylic acid (PAA) coated upconversion nanoparticles (UCNPs), the surface of which is modified using FA and chlorin e6 (Ce6) functionalized DNA sequences with different lengths. Initially, FA on the shorter DNA was protected by a longer DNA to prevent the bonding to FRs on normal cells. Once reaching the acidic tumor microenvironment, C base-rich longer DNA forms a C-quadruplex, resulting in the exposure of the FA groups and the bonding of FA and FRs on cancer cell membranes to achieve precise targeting. Simultaneously, the photosensitizer chlorin e6 (Ce6) gets close to the surface of UCNPs, enabling the excitation of Ce6 to generate singlet oxygen (1O2) under near infrared light via Förster resonance energy transfer (FRET). In vivo experiments indicated that higher tumor targeting efficiency was achieved and the tumor growth was greatly inhibited through the pre-protective strategy.
Introduction
Cancer has become a major killer that threatens human life and health because of the rapid proliferation and metastasis of cancer cells.1,2 With the deeper understanding of cancer in the past decades, targeted cancer therapy has been one of the main choices in clinical trials.3–6 Based on the overexpression of the receptors on cancer cell membranes, employing corresponding ligands to improve the targeting efficiency was the most common method.7–9 By conjugating a series of targeting groups (e.g. FA, TAT peptide, c(RGDyC) peptide, and aptamers) on the nanoparticles, tumor-specific targeting can be realized.10–14 Among these approaches, the FA group has been widely used for tumor targeting due to its easy modification and good targeting efficiency.15–17 However, there is a key issue that has always been ignored, that is, folate receptors are not only overexpressed at high levels on the membrane of nonmalignant proliferative cells (e.g. bone marrow cells and follicle cells), but also expressed at low levels in most normal tissues, which tremendously reduces the targeting efficiency and therapeutic effect due to the large population of normal cells.18–20 Therefore, an effective strategy that can protect the targeting groups and decrease unexpected bonding is urgently needed.
As an important treatment for cancer, PDT possesses superior advantages over other therapeutic methods due to its minimal invasiveness, fast healing process and potential repeatability.21–27 A photosensitizer is indispensable for the application of PDT in cancer therapy, however, traditional photosensitizers lack active targeting ability. When the photosensitizer circulates in the body, the retention of agents in the tumor tissues is limited. In this case, the therapeutic effect of PDT without tumor targeting will inevitably be greatly reduced.28,29 Besides, the poor penetration of ultraviolet or visible excitation light in conventional PDT also severely limits its clinical application. Improving the penetration of excitation light is also a key factor for enhancing the treatment effects of PDT.30–34 Thus, precise tumor targeting accompanied by NIR excitation would be extremely favorable to improve the therapeutic effect of PDT.
Herein, we present a pre-protective strategy for precise tumor targeting and enhanced PDT by constructing a dynamic DNA/UCNP nanocomposite. Two kinds of DNA sequences with different lengths and modifications are anchored on the surface of polyacrylic acid (PAA) coated UCNPs. In normal tissues, FA groups on the shorter DNA sequences are first covered and protected by the longer DNA sequences to separate from FRs, which could avoid unexpected uptake. Once the nanocomposites reach the tumor region, C base-rich longer DNA would fold in the acidic tumor microenvironment and shorten.35,36 Thus, FA groups were exposed and further bond to FRs on the membrane of cancer cells, thereby achieving the precise targeting of nanocomposites. At the same time, the photosensitizer Ce6 on the longer DNA gets close to the surface of UCNPs, and can be excited to generate 1O2 under NIR via FRET due to the fluorescence spectra match,37–42 which can be employed to destroy tumor cells for cancer therapy. The construction of the nanocomposite (UCNPs@PAA–DNA) and details of precise tumor targeting and specific PDT are illustrated in Scheme 1.
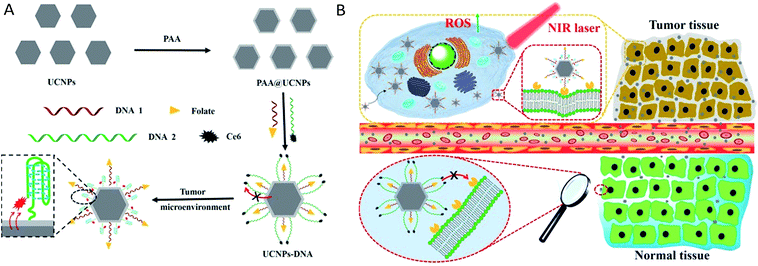 |
| | Scheme 1 (A) Synthesis process of UCNPs@PAA–DNA. (B) The details of precise tumor targeting and specific PDT for cancer. | |
Results and discussion
Synthesis and characterization of the nanocomposite
β-Phase UCNPs NaYF4 doped with 20% Yb3+ and 0.2% Er3+ (NaYF4:Yb3+,Er3+) were first prepared through an established solvothermal method according to a previous study with some modifications.43,44 These as-synthesized oleic acid (OA)-conjugated UCNPs were then coated with polyacrylic acid (PAA) to improve the water solubility and biocompatibility, and to facilitate subsequent DNA conjugation as well.45 As shown in the high resolution transmission electron microscopy (HRTEM) images in Fig. 1, OA–UCNPs exhibit a uniform hexagonal morphology with a diameter of about 25 nm, and disperse well in cyclohexane. After PAA coating, UCNPs@PAA possesses good monodispersity in water and the size of the nanoparticles increases to 35 nm. The zeta potentials change from −0.32 ± 0.04 mV to −14.7 ± 0.75 mV due to the introduced carboxyl groups of PAA on the surface of the nanoparticles (Fig. 2A). Finally, FA and Ce6 conjugated DNA sequences with different lengths (Table S1†) at a particular ratio were anchored on the nanoparticles via amido bonds, which further improves the biocompatibility after DNA decoration.46 From the absorption spectra in Fig. 2B, obvious absorption peaks of Ce6 at around 400 nm and 655 nm appear after DNA modifications, which provides further evidence for the successful fabrication of UCNPs@PAA–DNA.
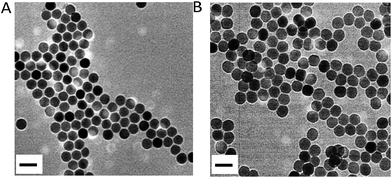 |
| | Fig. 1 HRTEM images of OA–UCNPs in cyclohexane (A) and UCNPs@PAA in the aqueous phase (B). Scale bars are 50 nm. | |
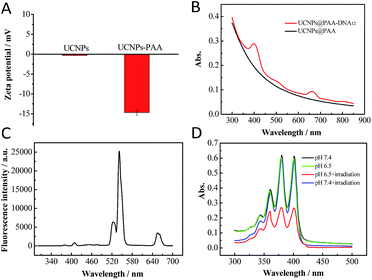 |
| | Fig. 2 (A) Zeta potential of UCNPs and UCNPs@PAA. (B) Absorption spectra of UCNPs@PAA and UCNPs@PAA–DNA1/2. (C) Fluorescence spectrum of UCNPs@PAA. The characteristic emission peaks appeared at 521 nm, 541 nm and 655 nm due to the Er3+ doping. (D) Absorption spectra of ABMD with UCNPs@PAA–DNA1/2 under different conditions: pH 7.4 without irradiation, pH 6.5 without irradiation, pH 7.4 with irradiation and pH 6.5 with irradiation. The laser was operated at a power of 1.5 W cm−2 for 5 min. | |
Acid triggered 1O2 generation
Acid environment triggered C-rich DNA folding was verified. A C-rich DNA sequence modified with Cy3 and BHQ1 was designed to demonstrate the C-quadruplex i-motif structure formation of C-rich DNA at pH 6.5 through fluorescence analysis (DNA4, Table S1†). As shown in Fig. S1 in the ESI,† the fluorescence intensity of Cy3 decreased obviously when DNA4 was at pH 6.5, which indicated that Cy3 was quenched by BHQ1 and further confirmed the DNA folding. Then we verified whether the nanocomposite could generate 1O2 in the mild acid environment under NIR irradiation. As shown in Fig. 2C, there are three characteristic emission peaks in the fluorescence spectrum of the UCNPs NaYF4:Yb3+,Er3+ at 521 nm, 541 nm and 655 nm, which corresponded to 2H11/2–4H15/2, 4S3/2–4S15/2, and 4F19/2–4I15/2 transformations of Er3+, respectively.47,48 Combined with the absorption spectrum of UCNPs@PAA–DNA1/2, we found that the absorption band of Ce6 around 655 nm overlapped well with the UCNP emission, which meets the needs of FRET. A special probe, 9,10-anthracenediyl-bis(methylene)dimalonic acid (ABMD), was employed for the detection of 1O2. As shown in Fig. 2D, the sharp decrease in the absorption of ABMD provided compelling evidence for the 1O2 generation when UCNPs@PAA–DNA2 was irradiated with 980 NIR light at pH 6.5, which was due to the acid environment triggering the C-quadruplex i-motif structure formation and FRET occurrence. The kinetic study showed that 1O2 was produced continuously with increasing irradiation time (Fig. S2, ESI†). As a control, when the pH was 7.4, there was no change in the absorption spectrum, which revealed no 1O2 generation and no side effects in the neutral normal tissues. This is because the C-rich longer DNA still maintains the original state under neutral conditions so that Ce6 molecules remain far away from the UCNP surface. These above results indicated that the acid environment could trigger the structure switch of the C base-rich DNA sequence and the nanocomposite could then generate 1O2 under NIR irradiation.
Optimization of the DNA ratio
To maximize the therapeutic effect, the ratio of shorter and longer DNAs per nanocomposite was optimized. Considering that more FA conjugated DNA1 would allow more cellular uptake and more Ce6 conjugated DNA2 would result in more 1O2 generation, the ratio of these two kinds of DNAs was of great importance. The total amount of DNAs on the nanocomposite surface was determined using fluorescence quantitative analysis. Different amounts of longer DNA2 first reacted with a certain amount of UCNPs@PAA for 12 h, and then the fluorescence of Ce6 in the supernatant was measured. As shown in Fig. S3 in the ESI,† when the amount of longer DNA2 reached 1.7 nmol, an obvious emission peak appeared in the fluorescence spectra, which indicated that the threshold amount of total DNA was 1.5 nmol. Subsequently, nanocomposites with various shorter and longer DNA ratios were prepared. The optimal ratio was based on the viability of cancer cells. We chose human breast cancer cells (MCF-7) as a cell model due to their high FR level (Fig. S4, ESI†). After incubation at pH 6.5 for 12 h, human breast cancer cells (MCF-7) were irradiated with 980 nm NIR for 5 min. Then we conducted MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assays to evaluate their ability in inducing cell apoptosis49,50 and further made the optimal choice on the DNA ratio. As shown in Fig. 3A, the nanocomposite with the DNA ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 treated MCF-7 cells exhibited the minimum value in viability, demonstrating that this DNA ratio was the best. Thus, the nanocomposite with the DNA ratio of 2
8 treated MCF-7 cells exhibited the minimum value in viability, demonstrating that this DNA ratio was the best. Thus, the nanocomposite with the DNA ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 was chosen for further use. What's more, to demonstrate the precise targeting and therapeutic ability of our designed nanocomposite, we constructed UCNPs@PAA–DNA1/3 as a negative control, in which C base-rich DNA2 was replaced by a scrambled sequence DNA3. As DNA3 could not switch its structure in an acid environment, FA was protected all the time and Ce6 remained far away from the UCNP surface, and both the targeting and therapeutic functions were restricted. After incubation with UCNPs@PAA–DNA1/3, most of the cells were still alive and displayed high viability up to about 90%. However, the viability of cancer cells incubated with UCNPs@PAA–DNA1/2 at pH 6.5 decreased markedly, suggesting excellent therapeutic effects (Fig. 3B).
8 was chosen for further use. What's more, to demonstrate the precise targeting and therapeutic ability of our designed nanocomposite, we constructed UCNPs@PAA–DNA1/3 as a negative control, in which C base-rich DNA2 was replaced by a scrambled sequence DNA3. As DNA3 could not switch its structure in an acid environment, FA was protected all the time and Ce6 remained far away from the UCNP surface, and both the targeting and therapeutic functions were restricted. After incubation with UCNPs@PAA–DNA1/3, most of the cells were still alive and displayed high viability up to about 90%. However, the viability of cancer cells incubated with UCNPs@PAA–DNA1/2 at pH 6.5 decreased markedly, suggesting excellent therapeutic effects (Fig. 3B).
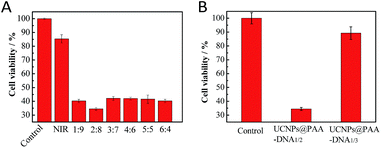 |
| | Fig. 3 MTT assays. The cell viability when incubated with UCNPs@PAA–DNA1/2 with different ratios of DNA1 and DNA2 (A), UCNPs@PAA–DNA1/2 and UCNPs@PAA–DNA1/3 (B) at pH 6.5 under 5 min irradiation. | |
FA mediated cellular uptake
FRs are generally overexpressed on the membrane of cancer cells and are expressed at low levels on normal cells. The presence of FA on the nanoparticles leads to the feature of specific recognition. The FR mediated uptake of the nanocomposite was investigated by employing a confocal laser scanning microscope (CLSM). As shown in Fig. 4, a bright red fluorescence signal of Ce6 was observed in the MCF-7 cells incubated with the UCNPs@PAA–DNA1/2. However, it was invisible when MCF-7 cells were pre-treated with FA before incubation with the nanocomposite, which was due to the competitive manner of free FA molecules.51 And a similar trend was observed in the fluorescence quantification analysis (Fig. S5, ESI†). Imaging flow cytometry (IFC) experiments were also carried out to confirm the FR mediated cell uptake. As shown by confocal images and statistical data, cells without pre-treated FA molecules exhibited stronger fluorescence intensity than FA treated cells (Fig. S6, Table S2, ESI†). The results verified that the nanocomposite is internalized via FRs. As reported in previous studies,52 FA conjugated nanoparticles would be finally located in lysosomes when they are internalized via FRs, which is beneficial for C-quadruplex i-motif structure formation and 1O2 generation.
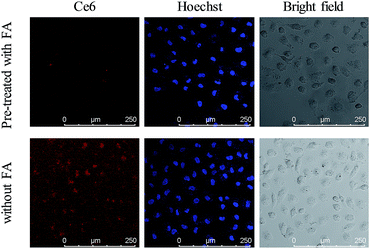 |
| | Fig. 4 Confocal images of MCF-7 cells incubated with UCNPs@PAA–DNA1/2 at pH 6.5 after cells were pre-treated with FA (top) or not (bottom). | |
In vitro targeting experiment
Acid microenvironment triggered specific targeting of cancer cells was then verified in MCF-7 cells using CLSM. As shown in the confocal images in Fig. 5, there was a bright red signal when MCF-7 cells were treated with the nanocomposite at pH 6.5, which resulted from the exposure of FA to the FRs on the cancer cell membrane. However, as neutral pH would not trigger the structural change of longer DNAs and the FA groups on the shorter DNAs were still protected, the signal was very weak when the incubation pH was 7.4. In addition, due to the low expression level of FRs on the membrane of normal human breast cells (MCF-10A), the cells exhibited a weak signal of Ce6 both at pH 6.5 and 7.4. Interestingly, the signal from MCF-10A cells at pH 6.5 was a little stronger than that at pH 7.4, indicating that the FA conjugated nanoparticles could target normal cells and may cause side effects if the FA remained exposed without protection. And the same results in fluorescence quantification further confirmed the ability of specific targeting of cancer cells and avoiding entrance into normal cells (Fig. S7, ESI†). IFC images and corresponding statistical data also evidenced the acid triggered specific targeting of cancer cells by the nanocomposite (Fig. S8, ESI†).
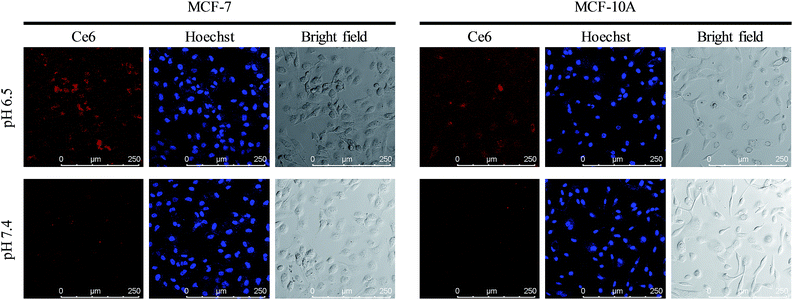 |
| | Fig. 5 Precise targeting experiments. Confocal images of MCF-7 cells and MCF-10A cells incubated with UCNPs@PAA–DNA1/2 at pH 6.5 and 7.4. | |
In vivo targeting experiment
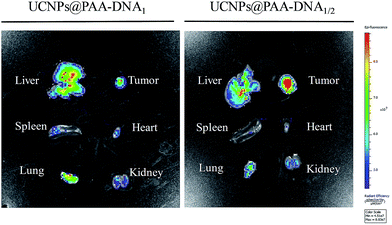
As the overexpression of FRs on the membrane of normal cells would sharply reduce the targeting efficiency and cause unexpected side effects, precise targeting of the tumor tissue by FA conjugated nanoparticles is of great significance for cancer therapy. To demonstrate the superior targeting ability of the nanocomposite, we conducted in vivo tumor targeting experiments in Balb/c mice with xenograft tumors. The mice were injected with the nanocomposite (4 mg kg−1) via intravenous administration. To highlight the superiorities of our designed nanocomposite as a precise targeting therapeutic agent, we chose UCNPs@PAA–DNA1(Ce6) as the control by modifying equal amount of Ce6 on the surface of UCNPs@PAA–DNA1, which represented the traditional FA conjugated nanoparticles. As shown in Fig. 6, besides the tumor region, UCNPs@PAA–DNA1(Ce6) are distributed in all the major organs (heart, liver, spleen, lungs, and kidneys), and only a small percentage of the injection dose reached the tumor tissue. However, as for UCNPs@PAA–DNA1/2, the fluorescence signal in tumor tissues was greatly enhanced, and the amount of nanocomposite in five major organs decreased sharply at the same time. The above results indicated that FRs on the membrane of normal cells would indeed interfere with the targeting effect of the traditional FA conjugated nanoparticles on the tumor tissue and our designed nanocomposite could achieve precise tumor targeting and reduced side effects on normal tissues. The targeting effect of UCNPs@PAA–DNA1/2 on tumors was quantified to be 12.1 ± 0.7% ID per g using inductively coupled plasma-atomic emission spectrometry (ICP-AES), which is higher than that of UCNPs@PAA–DNA1(Ce6) (4.1 ± 0.4% ID per g) (Fig. S9, ESI†). The metabolism kinetic study of UCNPs@PAA–DNA1/2 was also carried out by collecting the feces and urine. As shown in Fig. S10 in the ESI,† the retention content of nanoparticles in the body decreased to less than 5% at 72 h post injection. And 1O2 generation of UCNPs@PAA–DNA1/2 at pH 6.5 with human serum albumin (HSA, 10 mg mL−1) was detected by ABMD. As shown in Fig. S11 in the ESI,† the absorption of ABMD decreased obviously and was similar to that without HSA, which indicated that the protein has a negligible influence on our designed nanoparticles.
 |
| | Fig. 6
In vivo precise targeting. In vivo imaging of five major organs harvested from a mouse at 8 h post-injection with UCNPs@PAA–DNA1(Ce6) (left) or UCNPs@PAA–DNA1/2 (right). | |
Tumor growth inhibition in vivo
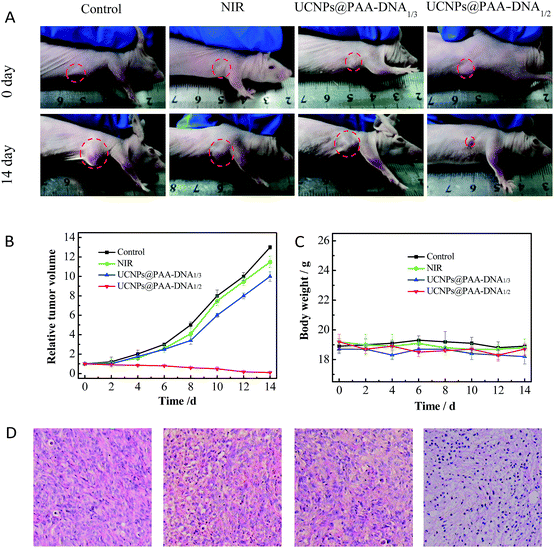
Encouraged by the outstanding therapeutic effect in vitro and good targeting ability in vivo, we then evaluated the efficacy in tumor growth inhibition. Nude mice were subcutaneously injected with MCF-7 cells to form a xenograft tumor. When the solid tumor reached about 150 mm3, the mice received different treatments: PBS, NIR irradiation only, UCNPs@PAA–DNA1/3 with NIR irradiation, and UCNPs@PAA–DNA1/2 with NIR irradiation. The nanoparticles were injected via intravenous administration only once and NIR irradiation was conducted for 5 min (1.5 W cm−2) at 8 h post injection. The tumor volume was monitored every other day in a period of 14 days without extra irradiation. As shown in Fig. 7, the volume of tumors in mice treated with PBS or NIR only increased greatly and was about 12 fold larger than its initial volume. As DNA3 could not change its structure in an acid environment, FA groups could be protected and Ce6 could not get close to the surface of UCNPs, resulting in low targeting efficiency and little 1O2 generation. Similarly, the tumor volume was found to increase nearly 10-fold over this period. Remarkably, the tumor was almost eliminated after being treated with our designed nanocomposite, due to the acid microenvironment triggered precise targeting, which demonstrated its excellent therapeutic effect (Fig. 7A and B). In addition, H&E staining on tumor sections provided further evidence for the treatment efficacy of inducing tumor cell apoptosis. As shown in H&E staining images, UCNPs@PAA–DNA1/2 treated tumors exhibited extensive apoptotic cells, which was reflected in a large amount of nuclear shrinkage and fragmentation (Fig. 7D). Meanwhile, no obvious body weight change was observed during the treatments, suggesting that the nanocomposite would not induce negative system toxicity (Fig. 7C). H&E staining on five major organs was also conducted at the end of the treatment period and no histopathological abnormalities appeared in all organs of mice with different treatments (Fig. S12, ESI†).
 |
| | Fig. 7
In vivo therapeutic effects. (A) Photographs of mice with different treatments on day 0 (top) and day 14 (bottom). The laser was operated at a power of 1.5 W cm−2 for 5 min. Tumor growth curves (B) and mice body weight curves (C) of different groups of tumor-bearing mice. (D) H&E staining of tumors in mice that received different treatments. | |
Conclusions
In summary, a pre-protective strategy was introduced for precise tumor targeting and efficient PDT with a switchable DNA/upconversion nanocomposite. The nanocomposite consists of PAA coated UCNPs that are conjugated with FA and Ce6 functionalized DNA sequences. In normal tissues, the FA group of the shorter DNA sequence was protected by the longer DNA sequence to prevent the conjugation to FRs. Once reaching the tumor region, C base-rich longer DNA would form C-quadruplex and FA groups will be exposed to bond FRs on the membrane of cancer cells, enabling precise tumor targeting. Simultaneously, the photosensitizer Ce6 on the longer DNA moved exactly close to the surface of UCNPs and could be activated to produce 1O2 under NIR via FRET. In vivo experiments demonstrated that nanocomposites accumulated in the tumor region and the tumor growth was severely inhibited, suggesting that the targeting efficiency and therapeutic effect of PDT were greatly improved. We anticipate that this pre-protective strategy could provide new insights for precise targeting and highly efficient cancer therapy.
Conflicts of interest
The authors declare no competing financial interest.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (21390411, 21535004, 91753111, and 21775094) and Natural Science Foundation of Shandong Province (JQ201503, ZR2015BQ003). All animal experiments were carried out according to the Principles of Laboratory Animal Care (People's Republic of China) and the Guidelines of the Animal Investigation Committee, Biology Institute of Shandong Academy of Science, China.
References
- D. Hanahan and R. A. Weinberg, Cell, 2011, 144, 646–674 CrossRef CAS PubMed.
-
B. Alberts, A. Johnson, J. Lewis, M. Raff, K. Roberts and P. Walter, Molecular biology of the cell, Garland Science, New York, 2008 Search PubMed.
- M. You, G. Zhu, T. Chen, M. J. Donovan and W. Tan, J. Am. Chem. Soc., 2015, 137, 667–674 CrossRef CAS PubMed.
- S. Maiti, N. Park, J. H. Han, H. M. Jeon, J. H. Lee, S. Bhuniya, C. Kang and J. S. Kim, J. Am. Chem. Soc., 2013, 135, 4567–4572 CrossRef CAS PubMed.
- K. Ulbrich, K. Holá, V. Šubr, A. Bakandritsos, J. Tuček and R. Zbořil, Chem. Rev., 2016, 116, 5338–5431 CrossRef CAS PubMed.
- J. Shi, Z. Xiao, N. Kamaly and O. C. Farokhzad, Chem. Res., 2011, 44, 1123–1134 CrossRef CAS PubMed.
- E.-K. Lim, T. Kim, S. Paik, S. Haam, Y.-M. Huh and K. Lee, Chem. Rev., 2015, 115, 327–394 CrossRef CAS PubMed.
- H. Koo, M. S. Huh, I.-C. Sun, S. H. Yuk, K. Choi, K. Kim and I. C. Kwon, Acc. Chem. Res., 2011, 44, 1018–1028 CrossRef CAS PubMed.
- P. S. Low, W. A. Henne and D. D. Doorneweerd, Acc. Chem. Res., 2008, 41, 120–129 CrossRef CAS PubMed.
- A. Oseledchyk, C. Andreou, M. A. Wall and M. F. Kircher, ACS Nano, 2017, 11, 1488–1497 CrossRef CAS PubMed.
- R.-H. Wang, J. Bai, J. Deng, C.-J. Fang and X. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 5828–5837 CAS.
- F. Danhier, A. L. Breton and V. Préat, Mol. Pharmaceutics, 2012, 9, 2961–2973 CrossRef CAS PubMed.
- Y.-H. Lao, K. K. L. Phua and K. W. Leong, ACS Nano, 2015, 9, 2235–2254 CrossRef CAS PubMed.
- H. Kobayashi, B. Turkbey, R. Watanabe and P. L. Choyke, Bioconjugate Chem., 2014, 25, 2093–2100 CrossRef CAS PubMed.
- W. Xia and P. S. Low, J. Med. Chem., 2010, 53, 6811–6824 CrossRef CAS PubMed.
- I. R. Vlahov and C. P. Leamon, Bioconjugate Chem., 2012, 23, 1357–1369 CrossRef CAS PubMed.
- Z. Farka, T. Juřík, D. Kovář, L. Trnková and P. Skládal, Chem. Rev., 2017, 117, 9973–10042 CrossRef CAS PubMed.
- C. Chen, J. Ke, X. E. Zhou, W. Yi, J. S. Brunzelle, J. Li, E.-L. Yong, H. E. Xu and K. Melcher, Nature, 2013, 500, 486–489 CrossRef CAS PubMed.
- C. P. Leamon and J. A. Reddy, Adv. Drug Delivery Rev., 2004, 56, 1127–1141 CrossRef CAS PubMed.
- L. E. Kelemen, Int. J. Cancer, 2006, 119, 243–250 CrossRef CAS PubMed.
- S. S. Lucky, K. C. Soo and Y. Zhang, Chem. Rev., 2015, 115, 1990–2042 CrossRef CAS PubMed.
- L. Cheng, C. Wang, L. Feng, K. Yang and Z. Liu, Chem. Rev., 2014, 114, 10869–10939 CrossRef CAS PubMed.
- Q. Yuan, Y. Wu, J. Wang, D. Lu, Z. Zhao, T. Liu, X. Zhang and W. Tan, Angew. Chem., Int. Ed., 2013, 52, 13965–13969 CrossRef CAS PubMed.
- Z. Yu, Q. Sun, W. Pan, N. Li and B. Tang, ACS Nano, 2015, 9, 11064–11074 CrossRef CAS PubMed.
- E. Ju, K. Dong, Z. Chen, Z. Liu, C. Liu, Y. Huang, Z. Wang, F. Pu, J. Ren and X. Qu, Angew. Chem., Int. Ed., 2016, 55, 11467–11471 CrossRef CAS PubMed.
- E. J. G. J. Dolmans, D. Fukumura and P. K. Jain, Nat. Rev. Cancer, 2003, 3, 380–387 CrossRef PubMed.
- Y. Ma, X. Li, A. Li, P. Yang, C. Zhang and B. Tang, Angew. Chem., Int. Ed., 2017, 56, 13752–13756 CrossRef CAS PubMed.
- J. Park, Q. Jiang, D. Feng, L. Mao and H.-C. Zhou, J. Am. Chem. Soc., 2016, 138, 3518–3525 CrossRef CAS PubMed.
- Z. Zhen, W. Tang, Y.-J. Chuang, T. Todd, W. Zhang, X. Lin, G. Niu, G. Liu, L. Wang, Z. Pan, X. Chen and J. Xie, ACS Nano, 2014, 8, 6004–6013 CrossRef CAS PubMed.
- S. Cui, D. Yin, Y. Chen, Y. Di, H. Chen, Y. Ma, S. Achilefu and Y. Gu, ACS Nano, 2013, 7, 676–688 CrossRef CAS PubMed.
- A. V. Kachynski, A. Pliss, A. N. Kuzmin, T. Y. Ohulchanskyy, A. Baev, J. Qu and P. N. Prasad, Nat. Photonics, 2014, 8, 455–461 CrossRef CAS.
- P. Vijayaraghavan, C. H. Liu, R. Vankayala, C. S. Chiang and K. C. Hwang, Adv. Mater., 2014, 26, 6689–6695 CrossRef CAS PubMed.
- M. Wang, Z. Chen, W. Zheng, H. Zhu, S. Lu, E. Ma, D. Tu, S. Zhou, M. Huang and X. Chen, Nanoscale, 2014, 6, 8274–8282 RSC.
- Y. Liu, Y. Liu, W. Bu, C. Cheng, C. Zuo, Q. Xiao, Y. Sun, D. Ni, C. Zhang, J. Liu and J. Shi, Angew. Chem., Int. Ed., 2015, 54, 8105–8109 CrossRef CAS PubMed.
- T. Tørring, R. Toftegaard, J. Arnbjerg, P. R. Ogilby and K. V. Gothelf, Angew. Chem., Int. Ed., 2010, 49, 7923–7925 CrossRef PubMed.
- J. Choi, S. Kim, T. Tachikawa, M. Fujitsuka and T. Majima, J. Am. Chem. Soc., 2011, 133, 16146–16153 CrossRef CAS PubMed.
- F. Wang, Y. Han, C. S. Lim, Y. Lu, J. Wang, J. Xu, H. Chen, C. Zhang, M. Hong and X. Liu, Nature, 2010, 463, 1061–1065 CrossRef CAS PubMed.
- F. Wang, R. Deng, J. Wang, Q. Wang, Y. Han, H. Zhu, X. Chen and X. Liu, Nat. Mater., 2011, 10, 968–973 CrossRef CAS PubMed.
- Z. Yu, W. Pan, N. Li and B. Tang, Chem. Sci., 2016, 7, 4237–4244 RSC.
- J. Xu, L. Xu, C. Wang, R. Yang, Q. Zhuang, X. Han, Z. Dong, W. Zhu, R. Peng and Z. Liu, ACS Nano, 2017, 11, 4463–4474 CrossRef CAS PubMed.
- X. Qin, X. Liu, W. Huang, M. Bettinelli and X. Liu, Chem. Rev., 2017, 117, 4488–4527 CrossRef CAS PubMed.
- X. Ai, C. J. H. Ho, J. Aw, A. B. E. Attia, J. Mu, Y. Wang, X. Wang, Y. Wang, X. Liu, H. Chen, M. Gao, X. Chen, E. K. L. Yeow, G. Liu, M. Olivo and B. Xing, Nat. Commun., 2016, 7, 1432–10440 Search PubMed.
- L. Zhao, J. Peng, Q. Huang, C. Li, M. Chen, Y. Sun, Q. Lin, L. Zhu and F. Li, Adv. Funct. Mater., 2014, 24, 363–371 CrossRef CAS.
- W. Li, J. Yang, Z. Wu, J. Wang, B. Li, S. Feng, Y. Deng, F. Zhang and D. Zhao, J. Am. Chem. Soc., 2012, 134, 11864–11867 CrossRef CAS PubMed.
- C. Wang, L. Cheng, X. Ma, X. Wang, Y. Liu, Z. Deng, Y. Li and Z. Liu, Adv. Funct. Mater., 2013, 23, 3077–3086 CrossRef CAS.
- L.-L. Li, P. Wu, K. Hwang and Y. Lu, J. Am. Chem. Soc., 2013, 135, 2411–2414 CrossRef CAS PubMed.
- R. Deng, J. Wang, R. Chen, W. Huang and X. Liu, J. Am. Chem. Soc., 2016, 138, 15972–15979 CrossRef CAS PubMed.
- J. Peng, A. Samanta, X. Zeng, S. Han, L. Wang, D. Su, D. T. B. Loong, N.-Y. Kang, S.-J. Park, A. H. All, W. Jiang, L. Yuan, X. Liu and Y.-T. Chang, Angew. Chem., Int. Ed., 2017, 56, 4165–4169 CrossRef CAS PubMed.
- N. Li, C. Chang, W. Pan and B. Tang, Angew. Chem., Int. Ed., 2012, 51, 7426–7430 CrossRef CAS PubMed.
- Y. Li, N. Li, W. Pan, Z. Yu, L. Yang and B. Tang, ACS Appl. Mater. Interfaces, 2017, 9, 2123–2129 CAS.
- S. K. Choi, T. Thomas, M.-H. Li, A. Kotlyar, A. Desai and J. R. Baker, Chem. Commun., 2010, 46, 2632–2634 RSC.
- J. Tian, L. Ding, H.-J. Xu, Z. Shen, H. Ju, L. Jia, L. Bao and J.-S. Yu, J. Am. Chem. Soc., 2013, 135, 18850–18858 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8sc00098k |
|
| This journal is © The Royal Society of Chemistry 2018 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article ,
Yegang
Ge
,
Yegang
Ge
 ,
Qiaoqiao
Sun
,
Qiaoqiao
Sun
 ,
Wei
Pan
,
Wei
Pan
 ,
Xiuyan
Wan
,
Xiuyan
Wan
 ,
Na
Li
,
Na
Li
 * and
Bo
Tang
* and
Bo
Tang
 *
*


![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 treated MCF-7 cells exhibited the minimum value in viability, demonstrating that this DNA ratio was the best. Thus, the nanocomposite with the DNA ratio of 2
8 treated MCF-7 cells exhibited the minimum value in viability, demonstrating that this DNA ratio was the best. Thus, the nanocomposite with the DNA ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 was chosen for further use. What's more, to demonstrate the precise targeting and therapeutic ability of our designed nanocomposite, we constructed UCNPs@PAA–DNA1/3 as a negative control, in which C base-rich DNA2 was replaced by a scrambled sequence DNA3. As DNA3 could not switch its structure in an acid environment, FA was protected all the time and Ce6 remained far away from the UCNP surface, and both the targeting and therapeutic functions were restricted. After incubation with UCNPs@PAA–DNA1/3, most of the cells were still alive and displayed high viability up to about 90%. However, the viability of cancer cells incubated with UCNPs@PAA–DNA1/2 at pH 6.5 decreased markedly, suggesting excellent therapeutic effects (Fig. 3B).
8 was chosen for further use. What's more, to demonstrate the precise targeting and therapeutic ability of our designed nanocomposite, we constructed UCNPs@PAA–DNA1/3 as a negative control, in which C base-rich DNA2 was replaced by a scrambled sequence DNA3. As DNA3 could not switch its structure in an acid environment, FA was protected all the time and Ce6 remained far away from the UCNP surface, and both the targeting and therapeutic functions were restricted. After incubation with UCNPs@PAA–DNA1/3, most of the cells were still alive and displayed high viability up to about 90%. However, the viability of cancer cells incubated with UCNPs@PAA–DNA1/2 at pH 6.5 decreased markedly, suggesting excellent therapeutic effects (Fig. 3B).






