Open Access Article
Open Access ArticleMulticolor monitoring of cellular organelles by single wavelength excitation to visualize the mitophagy process†
Fang
Hu‡
 ac,
Xiaolei
Cai‡
a,
Purnima Naresh
Manghnani
a,
Kenry
ac,
Xiaolei
Cai‡
a,
Purnima Naresh
Manghnani
a,
Kenry
 a,
Wenbo
Wu
a,
Wenbo
Wu
 ad and
Bin
Liu
ad and
Bin
Liu
 *ab
*ab
aDepartment Chemical and Biomolecular Engineering, National University of Singapore, 4 Engineering Drive 4, Singapore 117585, Singapore. E-mail: cheliub@nus.edu.sg
bInstitute of Materials Research and Engineering, Agency for Science, Technology and Research (A*STAR), Singapore
cNanoscience and Nanotechnology Institute, National University of Singapore, 2 Engineering Drive 3, 117581, Singapore
dDepartment of Materials Science and Engineering, National University of Singapore, 7 Engineering Drive 1, 117574, Singapore
First published on 2nd February 2018
Abstract
Multiplexed cellular organelle imaging using single wavelength excitation is highly desirable for unravelling cellular functions but remains challenging. This requires the design of organelle specific fluorophores with distinct emission but similar absorption. Herein, we present two unique aggregation-induced emission (AIE) probes to track mitochondria and lysosomes simultaneously with emission colors that can be distinguished from that of the nucleus stain Hoechst 33342 upon single wavelength excitation. Compared to conventional organelle stains, the two AIE probes have larger Stokes shifts and higher photostability, which endow them with the capability to monitor bioprocesses, such as mitophagy with strong and sustained fluorescent signals. Moreover, both probes can also stain intracellular organelles in zebrafish larvae with good cell-penetrating capabilities, showing their great potential to monitor bioprocesses in vivo.
Introduction
Mitochondria are the powerhouses of cells, and they also play important roles in calcium signaling and cell apoptosis.1 However, damaged mitochondria tend to generate excessive reactive oxygen species, which is highly undesirable.2 In fact, the accumulation of dysfunctional mitochondria is believed to be associated with a range of human pathologies.3 Fortunately, the physiological system is able to maintain its health by regulating cellular metabolism through a specific mechanism known as mitophagy. In general, mitophagy serves as a key process to control mitochondrial quality as well as to facilitate autophagic processes to degrade damaged mitochondria.4 Specifically, during the mitophagy process, damaged mitochondria are driven into autophagosomes and are then delivered to lysosomes to be degraded and recycled.To investigate the mitophagy process, fluorescent probes that can stain mitochondria and lysosomes have been tested.5–7 Among all the fluorescent moieties, organic dyes are the most promising8,9 because of their non-invasiveness and small size, although they share the drawbacks of having small Stokes shifts, being easily photobleached10 and undergoing facile aggregation-caused quenching (ACQ).11 In addition, two wavelengths are often needed to excite both dyes to emit different colors. Therefore, the wavelength of the excitation laser has to be constantly switched, or two lasers with different excitation wavelengths have to be used simultaneously during mitophagy monitoring.
To minimize the complexity of fluorescence detection and simplify instrumentation requirements, probes that can be excited by a single-excitation wavelength to emit different colors are highly attractive.12 Specifically, two probes which are highly specific to mitochondria and lysosomes with different light emission upon single wavelength excitation would provide a convenient way of visualizing the mitophagy process. However, to date, dyes that can be excited with a single wavelength to generate different emission colors are quite limited. Although quantum dots (QDs) with broad excitation spectra13 and dual-luminophore-doped nanoparticles with fluorescence resonance energy transfer (FRET) characteristics14 are able to emit distinguishable fluorescence under single wavelength excitation, they are not suitable for organelle tracking or mitophagy monitoring. This is because it is highly complex to endow them with organelle-targeting abilities and they generally have low performance in organelle imaging.15
Recently, fluorogens with aggregation-induced emission (AIEgens) characteristics have been developed for numerous biological assays, including organelle imaging,16 specific protein/enzyme detection,17 bacteria identification,18 and many others.19,20 In contrast to traditional ACQ dyes, AIEgens show strong emission in an aggregated state. The twisted molecular structures of AIEgens endow them with short absorption wavelengths. Their emissions are highly tunable from the ultraviolet to near-infrared (NIR) regions,21 so that AIEgens generally show large Stokes shifts (>100 nm) with great potential to emit distinguishable colors upon single wavelength excitation. In addition, AIEgens can also be conveniently modified to exhibit organelle-targeting abilities.16 Therefore, AIE probes are highly promising for organelle tracking and mitophagy monitoring under single wavelength excitation.
Herein, we report the synthesis and mitophagy tracking application of two AIE probes, AIE-Red and AIE-Green (Scheme 1). Upon excitation at 405 nm, AIE-Red can stain mitochondria with red signals and AIE-Green can stain lysosomes with green signals in HeLa cells. Together with the blue signals in the nucleus from Hoechst 33342, three different organelles are monitored simultaneously by 405 nm excitation in living HeLa cells. Owing to the mitochondrion specificity of AIE-Red and lysosome specificity of AIE-Green, the mitophagy process induced by rapamycin is conveniently investigated upon single laser excitation. In addition, AIE-Red and AIE-Green are also able to simultaneously target and stain mitochondria and lysosomes, respectively, inside the skin cells of zebrafish.
Results and discussion
Design, synthesis and photoproperties of AIE-Red and AIE-Green
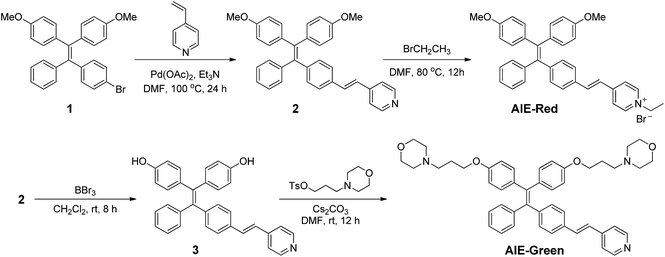
AIE-Red and AIE-Green were designed based on the following considerations. Tetraphenylethylene (TPE) is a typical AIE molecule, which has an excitation wavelength located in the ultraviolet region and blue emission under physiological conditions.22 Unfortunately, these optical characteristics are not suitable for bioimaging. To address this issue, we designed our AIE probes based on the following considerations. Firstly, upon the introduction of electron-donating alkoxyl groups and different electron-accepting moieties (e.g., vinyl pyridinium or vinyl pyridine) to TPE,23,24AIE-Red and AIE-Green are expected to have red and green emissions, respectively. Secondly, the excitation wavelengths of AIE-Red and AIE-Green are expected to red-shift to the visible light region with reasonable overlap to allow single wavelength excitation. Thirdly, the positively charged vinyl pyridinium is expected to endow AIE-Red with mitochondrion specificity,25 while the morpholine groups are anticipated to endow AIE-Green with lysosome specificity.26 The organelle specificity design is inspired by the special structures and properties of mitochondria and lysosomes. The membrane potential across outer and inner mitochondrial membranes is as high as −180 mV, which is higher by several orders of magnitude than that in other organelles and offers an opportunity to design positively charged dyes for mitochondria targeting. The positively charged triphenylphosphonium (TPP)27 and pyridinium moiety are commonly used to deliver probes to mitochondria. Normally, lysosomes have an acidic inner environment. Ligands such as N,N-dimethyl amino and morpholine are only partially protonated in neutral environments to permeate cell membranes. After cell penetration, they are found to interact with acid lysosomes in the cytoplasm.The synthetic routes to obtain AIE-Red and AIE-Green are shown in Scheme 2. Compound 2 was obtained by Heck reaction between 1 and 4-vinylpyridine. After that, AIE-Red was conveniently produced by heating a mixture of 2 and bromoethane in dimethyl formamide. Starting from compound 2, upon demethoxylation by boron tribromide, 3 was obtained in 99% yield. Morpholine groups were subsequently introduced through the reaction between 3-morpholinopropyl 4-methylbenzenesulfonate and compound 3 to yield AIE-Green. The experimental details are provided in the ESI.† The chemical structures of the intermediates and target compounds were characterized by NMR spectroscopy and mass spectrometry (Fig. S1–S8†) to reveal their correct structures with high purity.
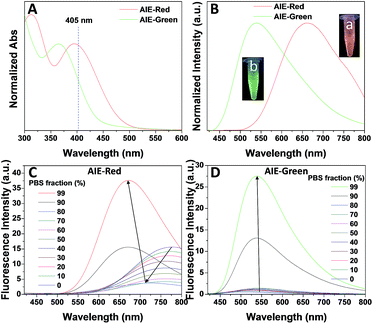
Fig. 1 shows the photoproperties of AIE-Red and AIE-Green. The absorption maxima of AIE-Red and AIE-Green are located at 367 nm and 394 nm, respectively. Both absorption spectra are red-shifted compared to that of unsubstituted TPE, and are partially located in the visible region (Fig. 1A). Importantly, the molar absorption coefficients of AIE-Red and AIE-Green in phosphate-buffered saline (PBS) at 405 nm are reasonably similar (εAIE-Red = 7.85 × 103 L mol−1 cm−1 and εAIE-Green = 5.35 × 103 L mol−1 cm−1), which provides the opportunity to excite the two probes using a 405 nm laser. The fluorescence spectra of the two AIE probes in PBS buffer are shown in Fig. 1B. The emission maxima of AIE-Red and AIE-Green are located at 665 nm and 538 nm, respectively, under the same 405 nm excitation, with red (inset (a) in Fig. 1B) and green (inset (b) in Fig. 1B) emissions, respectively. The large difference in the emission spectra but small difference in the absorption spectra between AIE-Red and AIE-Green come from the rational design of the donor–acceptor structure based on the TPE structure. It is reported that the introduction of different electron donating and accepting groups to the TPE structure can conveniently tune the emission color of AIE molecules because of the different degrees of intramolecular charge transfer (ICT).28 Molecules with stronger ICT generally show longer emission wavelengths. The electron-donating groups of AIE-Red and AIE-Green are methoxy and alkoxy, which have similar electron-donating abilities. The electron-accepting group of AIE-Red, vinyl pyridinium, is more electron-deficient than that of AIE-Green, vinyl pyridine, resulting in stronger ICT in AIE-Red than in AIE-Green. Therefore, AIE-Red has a much more red-shifted emission spectrum as compared to AIE-Green. However, the ICT effect has less impact on the ground state than the excited state due to the smaller dipole moment.29,30 Therefore, the introduction of different ICT does not lead to big differences in the absorption spectra of AIE-Red and AIE-Green. Upon incorporation of different electron-accepting groups to AIE-Red (vinyl pyridinium) and AIE-Green (vinyl pyridine), the difference of their emission maxima (127 nm) is much larger than that of their absorption maxima (27 nm). This enables the excitation of the two probes with a single wavelength for different emission colors. These unique photoproperties originate primarily from the large Stokes shifts of AIE-Red (271 nm) and AIE-Green (171 nm).
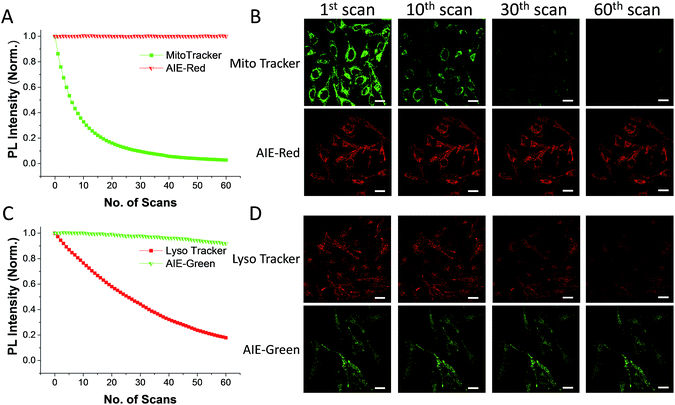
The AIE behaviors of AIE-Red and AIE-Green were investigated in a dimethyl sulfoxide (DMSO, good solvent) and PBS (poor solvent) mixture. As shown in Fig. 1C, AIE-Red exhibits red and NIR emission with a maximum at 775 nm in a dilute DMSO solution. The emission intensity decreased gradually with the increment of the PBS fraction from 0 to 60%, due to the twisted intramolecular charge transfer (TICT) effect. With higher PBS fractions, the emission is recovered with a much higher intensity and a blue-shift with a maximum at 665 nm. This fluorescence enhancement is attributed to the restriction of intramolecular motions, which is activated by the formation of nanoparticles of AIE-Red in aqueous media. This is different to the case of AIE-Green, which was almost non-emissive in the dilute DMSO solution (Fig. 1D). The emission intensity increases gradually as the PBS fraction increases from 0 to 80%. Then, a sharp intensity increase is noted as the PBS fraction goes up from 80% to 99%. As shown in Fig. S9,† the average hydrodynamic sizes of AIE-Red and AIE-Green in solutions of PBS/DMSO (99/1, v/v) are 160 and 400 nm, respectively, measured by dynamic light scattering (DLS). Owing to the AIE properties, both AIE-Red and AIE-Green can be used in high concentrations with reliable fluorescent signals. The fluorescence quantum yields (QYs) of AIE-Red and AIE-Green in PBS are 1.8% and 5.2%, respectively, which are measured with reference to 4-(dicyanomethylene)-2-methyl-6-(p-dimethylaminostyryl)-4H-pyran (DCM). After addition of 10% fetal bovine serum (FBS) to mimic the living organism system, their QYs increased to 8.1% and 7.2%, respectively. The increased QYs in FBS solution are beneficial for cellular system tracking. The pyridine moiety in AIE-Green could be protonated in acidic lysosomes, which may affect the emission of AIE-Green. Therefore, the pH-dependent fluorescence spectra of AIE-Red and AIE-Green were measured (Fig. S10†). AIE-Red exhibits constant fluorescence, while AIE-Green exhibits gradually decreased fluorescence along with pH reduction. The fluorescence decrease of AIE-Green is ascribed to the partial protonation of the pyridine moiety. As the emission wavelength remains unchanged, it will not affect the multicolor tracking by single wavelength excitation. Additionally, the high concentration of these AIE probes leads to their enhanced photostability. As can be seen in Fig. 2, AIE-Red and AIE-Green are far more resistant to photobleaching as compared to the commercial mitochondria and lysosome trackers. Signals from MitoTracker® Green FM (Mito Tracker) disappeared almost completely at the 60th scan, while signals from AIE-Red remained stable (Fig. 2A and B). Similarly, the signal loss of LysoTracker® Red DND-99 (Lyso Tracker) was 82% after 60 scanning cycles, while the signal loss of AIE-Green was only 8% (Fig. 2C and D). Both AIE-Red and AIE-Green also have good biocompatibility even at high concentrations (Fig. S11†). Since AIE-Red is reported as a photosensitizer, which can produce toxic reactive oxygen species (ROS) upon white light irradiation, the cytotoxicity of AIE-Red under confocal laser irradiation is also studied. As shown in Fig. S12,† upon continuous irradiation of AIE-Red incubated HeLa cells by a confocal laser for 10 min, the cell morphology hardly changed, indicating negligible cytotoxicity. Its biocompatibility was further confirmed using a live/dead assay after continuous confocal laser irradiation for 10 min. In these images, the green signals from fluorescein diacetate (FDA) indicate living cells, and the lack of red signals from propidium iodide (PI) indicates that there are no dead cells. The HeLa cell viability remained above 80% after treatment with AIE-Red or AIE-Green at 15 μM for 24 h. The high photostability and excellent biocompatibility of AIE-Red or AIE-Green are highly attractive for monitoring cellular bioprocesses. As commercial organelle targeting dyes are commonly used at a very low concentration, we also compared the fluorescence intensities of AIE-Red and AIE-Green to the commercial Mito Tracker and Lyso Tracker at different concentrations (Fig. S13†). Mito Tracker (150 nM) has a sharp emission at around 525 nm in PBS, while AIE-Red (150 nM) is almost non-emissive. The negligible emission of AIE-Red at 150 nM is ascribed to its good water solubility and lack of AIE activation. After addition of 10% FBS to mimic the living organism system, the emission of AIE-Red (150 nM) is dramatically enhanced to the same level as that of Mito Tracker (150 nM) due to AIE activation by interaction with proteins. When the concentration of AIE-Red is increased to 5 μM, the emissions in PBS and FBS-containing PBS are both stronger than those of Mito Tracker (150 nM). AIE-Green (150 nm) has a much stronger emission at 525 nm than Lyso Tracker (150 nM), which is quite weak with a maximum at 590 nm in both PBS and FBS-containing PBS. The emissions of AIE-Green at 5 μM are even stronger. These results indicate that AIE-Red has almost the same fluorescence intensity as Mito Tracker at 150 nM in the FBS-mimicked living organism system, and AIE-Green has a much stronger fluorescence intensity than Lyso Tracker at 150 nM. However, AIE-Red and AIE-Green at 5 μM still have very good biocompatibility. The usage of AIE-Red and AIE-Green at high concentration is aimed at high photostability for continuous scanning.
Multicolor imaging of organelles and mitophagy monitoring
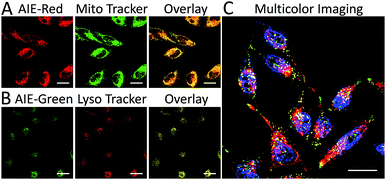
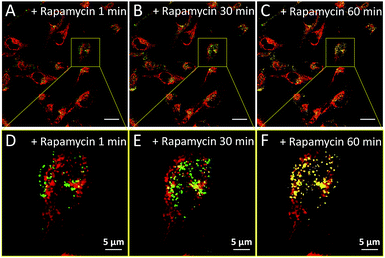
We subsequently studied the mitochondrion specificity of AIE-Red and lysosome specificity of AIE-Green in HeLa cells by the colocalization of both probes with commercial organelle dyes. As shown in Fig. 3A, AIE-Red and Mito Tracker were simultaneously incubated with HeLa cells for 15 min at 37 °C. The red emission signals from AIE-Red overlapped with the green signals from Mito Tracker very well, with a Pearson correlation coefficient of 0.94, indicating the outstanding targeting ability of AIE-Red towards the mitochondria of living HeLa cells. Similarly, the lysosome specificity of AIE-Green was well confirmed by its colocalization with Lyso Tracker, with a Pearson correlation coefficient of 0.95 (Fig. 3B). AIE-Red and AIE-Green also exhibit organelle targeting abilities toward other cell lines such as MCF-7 and MDA-MB-231 cells. As shown in Fig. S14,† the colocalizations with commercial organelle targeting dyes are good in MCF-7 and MDA-MB-231 cells. The Pearson correlation coefficients of AIE-Red with Mito Tracker are 0.73 and 0.92 for MCF-7 and MDA-MB-231 cells, respectively. The Pearson correlation coefficients of AIE-Green with Lyso Tracker are 0.81 and 0.82 for the MCF-7 and MDA-MB-231 cells, respectively. We then investigated the distinguishable emission colors of both probes based on the 405 nm excitation wavelength inside the HeLa cells. As shown in Fig. 3C, the organelle images with highly distinguishable emission colors were obtained inside the HeLa cells after simultaneous incubation with AIE-Red, AIE-Green and Hoechst 33342. The networks of mitochondria were clearly shown by the red signals while the morphology and distribution of lysosomes were portrayed well through green dots. These two distinguishable emission colors under the same visual field of the living cells afford the monitoring of mitophagy using only a single excitation laser. The blue nucleus signals originate from the commercial Hoechst 33342, which can also be excited by the 405 nm laser.With the advantages of distinguishable emission colors upon single wavelength excitation and high specificity to mitochondria and lysosomes, AIE-Red and AIE-Green were further utilized to monitor the mitophagy process in real time. In these experiments, rapamycin, which can promote the mitophagy process, was incubated with HeLa cells pre-treated with AIE-Red and AIE-Green. Subsequently, the confocal images of rapamycin-incubated HeLa cells were continuously collected under 405 nm excitation. As shown in Fig. 4A, after rapamycin treatment for 1 min, the red signals from the mitochondria hardly overlapped with the green signals from the lysosomes, with a Pearson correlation coefficient of 0.23. This indicates that the occurrence of the mitophagy process was not notable. As time went on, the Pearson correlation coefficient increased gradually, specifically, to 0.36 at 30 min and 0.46 at 60 min, indicating that the occurrence of the mitophagy process became more pronounced. Meanwhile, the evident increase in the amount and intensity of the yellow signals with a prolonged rapamycin treatment, as noted in the enlarged images, provide visual evidence of the mitophagy process (Fig. 4D–F). During these experiments, all fluorescent signals were collected upon excitation with a 405 nm laser, which is more convenient as compared to conventional methods.
Organelle imaging in zebrafish
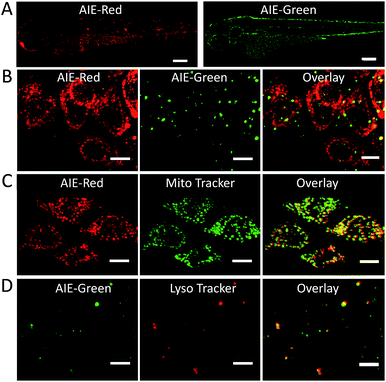
Since the zebrafish is a transparent and versatile in vivo model, multicolor imaging of different cellular organelles in zebrafish larvae was also explored. After soaking the larvae in the AIE-Red and AIE-Green solutions consecutively for 30 min, bright red and green signals were both observed on the epithelial cells under 405 nm excitation. This can be ascribed to their good cell-penetrating capability (Fig. 5A). Co-staining with Mito Tracker or Lyso Tracker in zebrafish larvae further confirmed the high specificity of AIE-Red and AIE-Green towards the zebrafish cellular mitochondria (Fig. 5C) and lysosomes (Fig. 5D), respectively. Fig. 5B shows the enlarged images of the skin cells of zebrafish larvae, in which the red mitochondria and green lysosomes can be easily distinguished in the same cells. Therefore, AIE-Red and AIE-Green are also promising for monitoring organelle-related bioprocesses in zebrafish systems.Conclusions
In summary, we have developed two AIE probes, AIE-Red and AIE-Green, which showed similar absorption maxima but completely different emission colors, highly beneficial for multicolor imaging through single wavelength excitation. The positively charged vinyl pyridinium in AIE-Red and morpholine groups in AIE-Green endow these AIE probes with high specificity towards mitochondria and lysosomes, respectively. Taking advantage of the organelle-targeting abilities, the mitophagy process, which is directly related to mitochondria and lysosomes, can be conveniently monitored in a real-time setting using AIE-Red and AIE-Green, together with the nucleus stain Hoechst 33342. Furthermore, AIE-Red and AIE-Green are also able to stain organelles in zebrafish larvae for single wavelength excitation-based multicolor imaging. In this regard, this work has demonstrated the development of a highly convenient and robust platform to monitor multicomponent-related bioprocesses with only one excitation laser.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Singapore National Research Foundation (R279-000-444-281 and R279-000-483-281), the National University of Singapore (R279-000-482-133), and the Institute of Materials Research and Engineering of Singapore (IMRE/13-8P1104).Notes and references
- C. V. Logan, G. Szabadkai, J. A. Sharpe, D. A. Parry, S. Torelli, A.-M. Childs, M. Kriek, R. Phadke, C. A. Johnson and N. Y. Roberts, Nat. Genet., 2014, 46, 188–193 CrossRef CAS PubMed.
- R. Scherz-Shouval and Z. Elazar, Trends Cell Biol., 2007, 17, 422–427 CrossRef CAS PubMed.
- S. Raha and B. H. Robinson, Trends Biochem. Sci., 2000, 25, 502–508 CrossRef CAS PubMed.
- R. J. Youle and D. P. Narendra, Nat. Rev. Mol. Cell Biol., 2011, 12, 9–14 CrossRef CAS PubMed.
- N. J. Dolman, K. M. Chambers, B. Mandavilli, R. H. Batchelor and M. S. Janes, Autophagy, 2013, 9, 1653–1662 CrossRef CAS PubMed.
- N. Sun, J. Yun, J. Liu, D. Malide, C. Liu, I. I. Rovira, K. M. Holmström, M. M. Fergusson, Y. H. Yoo and C. A. Combs, Mol. Cell, 2015, 60, 685–696 CrossRef CAS PubMed.
- Y. Liu, J. Zhou, L. Wang, X. Hu, X. Liu, M. Liu, Z. Cao, D. Shangguan and W. Tan, J. Am. Chem. Soc., 2016, 138, 12368–12374 CrossRef CAS PubMed.
- G. Despras, A. I. Zamaleeva, L. Dardevet, C. Tisseyre, J. G. Magalhaes, C. Garner, M. De Waard, S. Amigorena, A. Feltz, J. M. Mallet and M. Collot, Chem. Sci., 2015, 6, 5928–5937 RSC.
- Z. W. Xue, H. Zhao, J. Liu, J. H. Han and S. F. Han, Chem. Sci., 2017, 8, 1915–1921 RSC.
- D. Avnir, D. Levy and R. Reisfeld, J. Phys. Chem., 1984, 88, 5956–5959 CrossRef CAS.
- X. Ma, R. Sun, J. Cheng, J. Liu, F. Gou, H. Xiang and X. Zhou, J. Chem. Educ., 2015, 93, 345–350 CrossRef.
- A. K. Tong, Z. Li, G. S. Jones, J. J. Russo and J. Ju, Nat. Biotechnol., 2001, 19, 756–759 CrossRef CAS PubMed.
- X. Wu, H. Liu, J. Liu, K. N. Haley, J. A. Treadway, J. P. Larson, N. Ge, F. Peale and M. P. Bruchez, Nat. Biotechnol., 2003, 21, 41–46 CrossRef CAS PubMed.
- R. Hu, X. Zhang, Z. Zhao, G. Zhu, T. Chen, T. Fu and W. Tan, Angew. Chem., Int. Ed., 2014, 53, 5821–5826 CrossRef CAS PubMed.
- K. D. Wegner and N. Hildebrandt, Chem. Soc. Rev., 2015, 44, 4792–4834 RSC.
- F. Hu and B. Liu, Org. Biomol. Chem., 2016, 14, 9931–9944 CAS.
- G. Zhang, F. Hu and D. Zhang, Langmuir, 2014, 31, 4593–4604 CrossRef PubMed.
- W. Chen, Q. Li, W. Zheng, F. Hu, G. Zhang, Z. Wang, D. Zhang and X. Jiang, Angew. Chem., Int. Ed., 2014, 53, 13734–13739 CrossRef CAS PubMed.
- J. Mei, N. L. Leung, R. T. Kwok, J. W. Lam and B. Z. Tang, Chem. Rev., 2015, 115, 11718–11940 CrossRef CAS PubMed.
- Y. Yuan and B. Liu, Chem. Sci., 2017, 8, 2537–2546 RSC.
- G. Qian, B. Dai, M. Luo, D. Yu, J. Zhan, Z. Zhang, D. Ma and Z. Y. Wang, Chem. Mater., 2008, 20, 6208–6216 CrossRef CAS.
- Y. Hong, J. W. Lam and B. Z. Tang, Chem. Soc. Rev., 2011, 40, 5361–5388 RSC.
- P. Data, P. Pander, M. Okazaki, Y. Takeda, S. Minakata and A. P. Monkman, Angew. Chem., Int. Ed., 2016, 55, 5739–5744 CrossRef CAS PubMed.
- F. Hu, Y. Yuan, D. Mao, W. Wu and B. Liu, Biomaterials, 2017, 144, 53–59 CrossRef CAS PubMed.
- T. Ryan, Y. Chris, W. Jacky and B. Tang, Chem. Commun., 2015, 51, 9022–9025 RSC.
- M. Gao, Q. Hu, G. Feng, B. Z. Tang and B. Liu, J. Mater. Chem. B, 2014, 2, 3438–3442 RSC.
- M. Ross, G. Kelso, F. Blaikie, A. James, H. Cocheme, A. Filipovska, T. Da Ros, T. Hurd, R. Smith and M. Murphy, Biochemistry, 2005, 70, 222–230 CAS.
- X. Gu, J. Yao, G. Zhang, C. Zhang, Y. Yan, Y. Zhao and D. Zhang, Chem.–Asian J., 2013, 8, 2362–2369 CrossRef CAS PubMed.
- H. Zhu, M. Li, J. Hu, X. Wang, J. Jie, Q. Guo, C. Chen and A. Xia, Sci. Rep., 2016, 6, 24313 CrossRef CAS PubMed.
- W. Huang, Z. Xie, Y. Deng and Y. He, Sens. Actuators, B, 2018, 254, 1057–1060 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7sc04585a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |