Open Access Article
Open Access ArticleIsolation and characterization of a novel bacterium Pseudomonas aeruginosa for biofertilizer production from kitchen waste oil
Ying Li ,
Ting Cui,
Yaxin Wang and
Xizhen Ge*
,
Ting Cui,
Yaxin Wang and
Xizhen Ge*
Beijing Key Laboratory of Biomass Waste Resource Utilization, Biochemical Engineering College, Beijing Union University, Beijing 100023, China. E-mail: 20057003@buu.edu.cn
First published on 17th December 2018
Abstract
Kitchen waste oil is composed of long chain triglycerides (LCTs) that has high energy density. However, it is hard to be degraded by microbes, thereby leading to increasing levels of environmental pollution due to landfill disposition. In this study, we isolated and characterized a novel bacterium Pseudomonas aeruginosa PA-3 that could convert kitchen waste oil into biofertilizer. PA-3 could survive on trilaurin or kitchen waste oil as the sole carbon source, and 10 g L−1 trilaurin or kitchen waste oil was completely consumed within 7 days. Interestingly, the degradation products of kitchen waste oil can be used as biofertilizer in promoting cabbage growth. The plant height, leaf area and stem diameter of cabbage plants were all increased with the addition of kitchen waste oil cultivation products into the soil. Kitchen waste oil degradation products were analyzed by gas chromatography mass spectrometry (GC-MS), and short chain alcohols or fatty acids were observed to be the main products. To unravel the mechanism underlying the accelerated cabbage growth, bacterial diversity of the soil was investigated after using this biofertilizer. Results showed that agricultural probiotics accumulated with the addition of kitchen waste oil cultivation products. Finally, the whole genome of PA-3 was sequenced and analyzed, which showed the existence of a complete β-oxidation pathway in the genome of PA-3. To our knowledge, this is the first study on kitchen waste oil degradation and re-utilization by bacteria, which provides a new method for waste source re-utilization.
1. Introduction
Waste oil from restaurants is composed of long chain triglycerides (LCTs) that comes from vegetable oil or animal fats.1 The emission of LCTs causes environmental pollution because LCTs are difficult to be degraded by most of the environmental microorganisms except for a few bacteria and fungi that contain β-oxidation pathway.2,3 To avoid damage to human health and the environment, resource utilization was carried out to convert kitchen waste oil into animal feed or biodiesel.4,5 However, the prepared animal feed was of poor quality due to the harmful oxidation compounds formed during cooking. Meanwhile, biodiesel production from waste oil was impractical due to the high cost and secondary pollution.6 Therefore, a new method for the sustainable re-use of kitchen waste oil is still required.Biofertilizer is an ideal alternative for chemical fertilizer due to the efficiency in promoting plant growth and the safety to the environment.7,8 A biofertilizer is mainly composed of probiotic microorganisms and the fermentation products from low-cost substrates.9 Microbes such as Bacillus, Streptomyces, and Glomus were successfully applied as probiotics in promoting plant growth.10–12 The fermentation products of the microorganisms are directly dried and powdered into fertilizers without separation of the microbes, as the metabolites such as amino acids and vitamins are also beneficial to plants.13 Hence, the cost for biofertilizer production is mostly determined by the carbon source consumed in the fermentation process. The commonly used low-cost substrates are starch, rice bran, soy-bean cake and other castoffs of upstream production lines.14,15 However, the substrate cost is still an obstacle in biofertilizer production due to the poor energy density and the low conversion ratio of the substances. In contrast, waste cooking oil that contains high energy density is a feasible carbon source for fermentation.16
Triglyceride degradation microorganisms mostly consist of a few bacteria and fungi. The degradation of triglyceride is divided into two parts. First, triglyceride is hydrolyzed into fatty acid and glycerol by lipase secreted by microbes.17 Subsequently the glycerol and fatty acid are permitted into the cell and then they take part in the oxidation pathway for complete degradation.18 For LCTs, 6–8 cycles of oxidation are needed until they are completely oxidized into acetyl-CoA. Nevertheless, there is less research on metabolic pathways for bacterial triglyceride degradation. The advantage in the application of bacteria over fungi for the degradation of LCTs is the fast cell growth for biomass accumulation.19 Therefore, isolating a LCTs degradation bacterium is imperative for kitchen waste oil disposition or even production of valuable chemicals.
In this study, we aim to isolate a bacterium that is able to consume long chain triglyceride (LCT) and kitchen waste oil efficiently. In addition, the fermentation products are proposed to be a new biofertilizer for cabbage growth promotion. Furthermore, the degradation rate and degradation products were analyzed. In parallel, soil bacterial diversity was determined to reveal the plant growth-promoting mechanisms of the biofertilizer. Finally, genome sequencing was carried out to deduce the degradation pathway of kitchen waste oil.
2. Experimental
2.1 Isolation and identification of LCT-degrading strain
The soil samples that had been treated with LCT for 5 weeks were collected from the experimental field of Beijing Union University. To isolate LCT-degrading bacteria, continuous culture was carried out. The screening medium contained the following (g L−1): K2HPO4 0.05, KH2PO4 0.015, CaCO3 0.01, LCT 0.1. The LCT-treated soil sample (0.01 g L−1) was added at the first time of cultivation. A 250 mL shake flask was used and 100 mL medium was added to it. The shake flask was incubated at 37 °C and agitated at 150 rpm. After incubation for 3 days, 1 mL aliquot from each shake flask was transferred into a new flask containing the same medium, and it was again incubated at 37 °C with agitation. This process was repeated 5 times before the final culture was diluted onto a LCT solid medium. The LCT solid medium was the same as the screening medium except that 15 g L−1 agar powder was added. All plates were incubated at 37 °C for bacterial isolation. After continuous isolation for more than 5 generations, the pure bacterial cultures that could grow with LCT as the sole carbon source were selected, and then the strain was transferred onto Luria–Bertani (LB) solid medium (NaCl, 1%; peptone, 1%; yeast extract, 0.5%, agar powder, 1.5%) for preservation. Subsequently, 16S rDNA sequencing was conducted with two universal primers 27F and 1492R. DNA sequencing was accomplished in Beijing Biomed Company, and the phylogenetic tree was constructed by MEGA system.2.2 Kitchen waste oil identification and LCT degradation
Kitchen waste used in this study was obtained from the canteen of Beijing Union University. Each 200 g kitchen waste was diluted with 100 mL petroleum ether for kitchen waste oil extraction. After incubation and agitation for 3 min, the upper layer was transferred into a rotary evaporator to remove petroleum ether at 50 °C. In total, 1 kg of kitchen waste was used for extraction, and the extracted kitchen waste oil was preserved at 2 °C. The content of waste oil was analyzed by GC-MS after methylation. For the methylation reaction, 0.01 g kitchen waste oil was dissolved in 1 mL petroleum ether with 20 μL saturated KOH–methanol. After rotation, the supernatant was analyzed by GC or GC-MS. Lauric acid triglycerin ester (Trilaurin, CAS: 538-24-9) was supplied as LCT in degradation experiments.To study the LCT degradation ability of the isolated bacteria, the strain was firstly cultured in liquid LB medium for enrichment. After the OD600 value of the medium reached 1.0, 2 mL of the medium was taken out for biomass extraction by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. After washing 2 times with sterile ddH2O, the cell was added into a 250 mL shake flask containing LCT-degrading medium, and the initial LCT concentration was set at 10 g L−1. The flask was placed at 37 °C and shaken at 200 rpm. Samples were taken every 24 h to determine the LCT concentration. To determine the insoluble LCT in the medium, 20 mL petroleum ether was added into the flask to obtain the upper liquid layer. The mixture was then evaporated and the mass of LCT was determined. For soluble LCT concentration assay, 0.5 mL of the lower layer in the cultivation broth was taken out and mixed with 0.5 mL petroleum ether. After oscillation, the upper layer of the mixture was obtained for methylation followed by GC or GC-MS analysis. The soluble kitchen waste oil in the initial medium was set as 100%, and the relative content was defined by the peak area of methyl-octadecanoate alignments.
000 rpm for 10 min. After washing 2 times with sterile ddH2O, the cell was added into a 250 mL shake flask containing LCT-degrading medium, and the initial LCT concentration was set at 10 g L−1. The flask was placed at 37 °C and shaken at 200 rpm. Samples were taken every 24 h to determine the LCT concentration. To determine the insoluble LCT in the medium, 20 mL petroleum ether was added into the flask to obtain the upper liquid layer. The mixture was then evaporated and the mass of LCT was determined. For soluble LCT concentration assay, 0.5 mL of the lower layer in the cultivation broth was taken out and mixed with 0.5 mL petroleum ether. After oscillation, the upper layer of the mixture was obtained for methylation followed by GC or GC-MS analysis. The soluble kitchen waste oil in the initial medium was set as 100%, and the relative content was defined by the peak area of methyl-octadecanoate alignments.
2.3 Cabbage growth determination
To determine the plant growth-promoting effect of the biofertilizer, cabbage seeds were planted separately in nutrient soil and vermiculite. The two media were firstly sterilized at 115 °C for 20 min, then they were put into φ6 cm cultivation cups and the thickness of the medium was set at 8 cm. Five cabbage seeds were planted into each cup at 1 cm depth after they were soaked in ddH2O for 6 h. Then the cups were placed in an illumination incubator at 25 °C and 1000 lx light intensity with the humidity set at 40%. After cultivation for three days, the cups that showed similar height and area of the new buds were used for the following experiments. LCT degradation products on the 4th day were applied for drip irrigating the new buds, and the same amount of sterilized ddH2O was applied for the control group. Every 10 mL of the medium or water was used in one cup at three days of cabbage seed cultivation. From the 3rd day till the end, 3 mL of water was added into the medium every day to maintain the humidity of the soil. The height and stem diameter were measured using a vernier caliper, and the leaf area was automatically calculated in a TLC scanner by external standard method. Cabbage seeds, nutrient soil and vermiculite were purchased from LvBa-Agritech Company, Shandong.2.4 Bacterial diversity analysis
The effect of LCT degradation products on soil bacterial diversity was studied on the 12th day after planting. The determination of bacterial diversity was carried out according to the ref. 20. Briefly, 0.5 g soil sample at 2 cm depth was collected and diluted in ddH2O followed by genome extraction using a soil genome extraction kit from Beijing Biomed Company. Subsequently, 16S rDNAs of all the bacteria were amplified and gel recycled. About 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 PCR products of each sample were sequenced independently and the bacteria were classified at different levels. The heat-map image representing the bacterial diversity was plotted in Origin 8.0. DNA high-throughput sequencing and data analysis were accomplished at the Shanghai Hanyu Biotech Center.
000 PCR products of each sample were sequenced independently and the bacteria were classified at different levels. The heat-map image representing the bacterial diversity was plotted in Origin 8.0. DNA high-throughput sequencing and data analysis were accomplished at the Shanghai Hanyu Biotech Center.
2.5 Analytical methods
Fatty acid methyl ester concentration was measured in a PUXI-G5 GC system equipped with a 30 m capillary FFAP column and a FID detector.21 The mobile phase was N2 and the speed was 30 mL min−1. The column temperature program was set as follows: 60 °C for 3 min; 5 °C min−1 to 200 °C; 200 °C for 15 min. The injection volume was set at 1 μL for all the samples. For GC-MS analysis, the degradation compounds were extracted by petroleum ether before injection into an Agilent 5977B system, and the scanned range was set from m/z 0 to 1000.22 The agents used in this study were products of Adamas, Shanghai. Whole bacterial genome sequencing (WGS) was conducted in Yuanquanyike Tech. Beijing.23 The proteins were annotated in UniProtKB and then the pathways were classified by the KEGG pathway and GO analysis. Genes belonging to the fatty acid degradation pathway (ko00071) were selected.3. Results
3.1 Isolation and characterization of LCT degradation bacteria
A bacterial single colony that could grow with LCT as the sole carbon source was obtained after continuous purification. The 16S rDNA sequencing result suggested that the strain belonged to the Pseudomonas genus and it was 97% similar to Pseudomonas alcaligenes NEB 585. The strain was named as PA-3. The phylogenetic tree was constructed based on the 16S rDNA sequence, and it is displayed in Fig. 1A. It can be inferred that PA-3 showed a distant genetic relationship to other Pseudomonas, meaning that PA-3 was an unexploited strain for LCT degradation. At the same time, the contents of kitchen waste oil isolated from kitchen waste were analysed, and the results are shown in Table 1. There were 6 kinds of fatty acids in kitchen waste oil in total, and the most dominant contents were 16C and 18C saturated or unsaturated linear chain fatty acids. The content of unsaturated fatty acids occupied 49.95% of the total peak area.| RT (min) | Name | Formula | CAS | Relative area (%) |
|---|---|---|---|---|
| 17.5 | Methyl tetradecanoate | C15H30O2 | 124-10-7 | 0.80 |
| 21.8 | Methyl hexadec-11-enoate | C17H32O2 | 822-05-9 | 0.61 |
| 22.2 | Hexadecanoic acid, methyl ester | C17H34O2 | 112-39-0 | 28.53 |
| 24.8 | 9,12-Octadecadienoic acid methyl ester | C19H34O2 | 112-63-0 | 11.30 |
| 25.0 | 9-Octadecenoic acid methyl ester | C19H36O2 | 112-62-9 | 38.04 |
| 25.4 | Methyl stearate | C19H38O2 | 112-61-8 | 20.72 |
Subsequently, LCT and kitchen waste oil degradation were carried out in a shake flask, and the results are displayed in Fig. 2. It can be inferred that PA-3 was able to utilize LCT or kitchen waste oil as its sole carbon source. LCT was rapidly used by PA-3 and 10 g L−1 of the insoluble substrate was consumed within 5 days (Fig. 2A and B). Similarly, kitchen waste oil was also consumed by PA-3 after one day of adaption, and then rapidly utilized from the 2nd day to the 5th day (Fig. 2C and D). The whole 10 g L−1 kitchen waste oil was completely used up on the 6th day. The average consumption ratio of LCT and kitchen waste oil was 2 g L−1 d−1 and 1.67 g L−1 d−1, respectively, showing great potential in LCT degradation. Next, the degradation of LCT and kitchen waste oil by PA-3 with different initial concentrations of substrates were conducted, and the results are displayed in Fig. 2D and F. It can be inferred that 20 g L−1 LCT and kitchen waste oil was assimilated completely within 7 days. However, 1.22 g L−1 LCT and 0.49 g L−1 kitchen waste oil remained when 30 g L−1 of substrates were initially provided. For higher concentrations of substrates, 14.46 g L−1 LCT and 11.99 g L−1 kitchen waste oil were unused in the medium, which meant that the maximum degradation amount of both LCT and kitchen waste oil by PA-3 was nearly 28 g L−1 within 7 days of cultivation.
3.2 Cabbage growth promotion
The study of the effect of LCT degradation products with PA-3 on cabbage growth was conducted in nutrient soil and vermiculite, which represent richly and poorly fertilized soil, respectively. The results are displayed in Fig. 3. It can be inferred that the addition of the fermentation medium significantly accelerated cabbage growth under different circumstances. In vermiculite (Fig. 3A and B), the height of the cabbages with addition of LCT degradation products reached 7.83 cm in 12 days, while that of the control group was 5.17 cm. The leaf areas at 12th day and 15th day were 1.08 cm2 and 1.35 cm2, respectively, which were significantly increased compared to the control (0.56 cm2 and 0.71 cm2). The results of the nutrient soil group (Fig. 3C and D) were similar to that of vermiculite. Plant height and leaf area at 12 days were increased by 41.0% and 76.2%, respectively, compared to those of the control. However, when the cultivation broth was centrifugally separated into extracted PA-3 and extracted medium, no significant differences were found in plant growth compared to that of the control group. The addition of extracted medium slightly promoted plant height and leaf area, while extracted PA-3 displayed little effect on plant growth. Fig. 3E shows that the addition of LCT degradation products increased the stem diameter of cabbage in vermiculite from 0.069 cm to 0.106 cm, while for the nutrient soil group, the stem diameters of the experimental group and the control group were similar. The addition of LCT degradation products facilitated the significant increase of leaf area in nutrient soil, while plant height and stem diameter were more evidently promoted in vermiculite. Based on the above data, LCT degradation products by PA-3 functioned as biofertilizer that efficiently accelerated cabbage growth.3.3 Metabolite analysis
To reveal the LCT degradation metabolites, compounds in the cultivation broth were analyzed by GC-MS after pre-treatment, and the contents of the mixture are displayed in Table 2. It can be inferred by relative peak area determination that the most abundant component was butyl acetate. The other components that occupied nearly 13% of each peak area were identified as ethyl 2-hydroxyisobutyrate, diethylene glycol and triethylene glycol. Moreover, 1,2-ethanediol occupied 8.66% of the whole peak area. The remaining components were all defined as low contents that occupied less than 1% of the whole peak area. It can be concluded that LCT degradation products by PA-3 were mostly classified as short chain esters, alcohols and fatty acids. PA-3 successfully catalyzed the degradation of LCT into compounds with low-molecular weight that were rarely reported for biosynthesis, providing a new way for the biological production of these high-value platform compounds.24–26| RT (min) | Name | Formula | CAS | Relative area (%) |
|---|---|---|---|---|
| 3.77 | Butyl acetate | C6H12O2 | 123-86-4 | 35.19 |
| 5.50 | 1-Butanol | C4H10O | 71-36-3 | 0.34 |
| 7.13 | 2-Methoxy-1-propanol | C4H10O2 | 1589-47-5 | 0.54 |
| 7.44 | Ethyl 2-hydroxyisobutyrate | C6H12O3 | 80-55-7 | 13.79 |
| 16.06 | 1,2-Propanediol | C3H8O2 | 4254-15-3 | 0.61 |
| 16.92 | 1,2-Ethanediol | C2H6O2 | 107-21-1 | 8.66 |
| 17.03 | Butanoic acid | C4H8O2 | 107-92-6 | 0.17 |
| 22.46 | 2-(2-Hydroxypropoxy)-1-propanol | C6H14O3 | 106-62-7 | 0.69 |
| 24.37 | Diethylene glycol | C4H10O3 | 111-46-6 | 13.93 |
| 32.08 | Glycerol | C3H8O3 | 56-81-5 | 0.42 |
| 32.30 | Triethylene glycol | C6H14O4 | 112-27-6 | 12.82 |
Interestingly, theoretical 6C to 10C LCT degradation products were not detected during the whole fermentation process. As the degradation products of LCT, fatty acids were the indispensable intermediates. However, except for the low content of butanoic acid, no fatty acids or fatty acid CoA were detected with or without derivation of the products in the fermentation medium. Since LCT degradation in PA-3 was probably mediated by a similar β-oxidation cycle, this result suggested that 10C and shorter fatty acids were efficiently dissimilated in the cell cytoplasm of PA-3.
3.4 Soil bacterial diversity
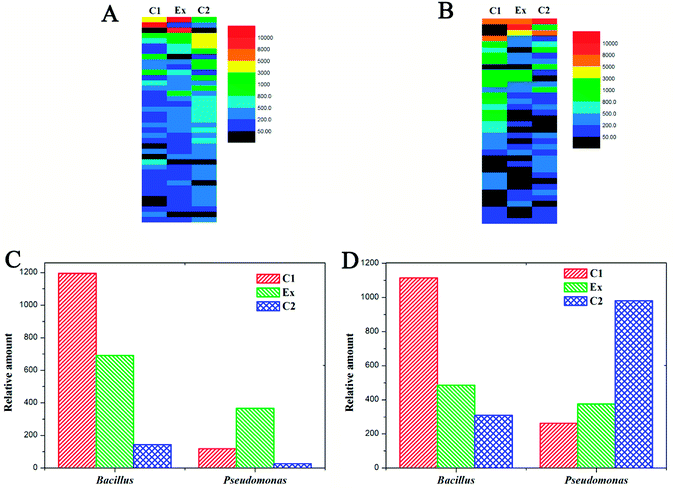
To discover the mechanism of LCT degradation products on promoting plant growth, the changes in soil bacterial diversity were researched, and the results are displayed in Fig. 4 and 5 at different levels. It can be seen that the soil bacterial diversity in different experimental groups (Fig. 4 (Ex)) were significantly changed compared to those of C1 (no planting) and C2 (planting without LCT degradation products added) groups. In nutrient soil (Fig. 4A), the most distinct differences were located in row 1, row 3 and row 16, representing Burkholderia, Providencia and Nocardioides genus, respectively. The contents of Burkholderia and Providencia were increased to a great extent, while the amount of Nocardioides was much less than in the control. There were no significant changes in the other bacterial genera in the experimental group. In parallel, the addition of LCT degradation products in vermiculite also led to the changes in soil bacterial diversity (Fig. 4B). The distinct increases were in row 2, row 10 and row 11, representing Providencia, Paenibacillus and Acinetobacter genus, respectively. Meanwhile, the significant decreases were observed in row 6, row 9 and row 13, representing Comamonas, Pedobacter and Pseudomonas genus, respectively. The amounts of Bacillus genus and Pseudomonas genus are displayed in Fig. 4C and D. It can be seen that the addition of LCT degradation products led to a 3-fold increase in the amount of Bacillus genus, which is an agricultural probiotic. However, the amount of Pseudomonas genus showed no significant increase even though it was previously added. In vermiculite, the amount of Pseudomonas was even lower compared to that of the control.At the phylum level (Fig. 5), the changes were also apparent with the addition of LCT degradation products. In nutrient soil, the amount of Proteobacteria increased from 62.82% to 83.74%, occupying the most abundant phylum of soil bacteria. In contrast, the amounts of bacteria in the other phyla were all decreased. In vermiculite, Proteobacteria were also the dominant content, and 88.69% of the soil bacteria belonged to this phylum. Moreover, the content of Firmicutes that includes Bacillus genus also increased from 3.1% to 8.31%, and this phenomenon was different from that in nutrient soil.
3.5 Whole genome sequencing
The whole genome of PA-3 was sequenced and the whole genome data was submitted to NCBI (Accession: CP033084.1). After whole genome alignment, the strain PA-3 was classified into Pseudomonas aeruginosa, which is different from 16S rDNA alignment. The genome size of PA-3 was 6.33 Mbp and the GC content of the genome was 66.51%. The genes belonging to fatty acid degradation (ko00071) were collected and are displayed in Fig. 6. It can be inferred that there exists a complete β-oxidation pathway in PA-3. However, the copies of the different genes were distinct. The first step of fatty acid degradation was catalyzed by Acyl-CoA synthetase (EC: 6.2.1.3), and 4 copies were detected in different areas of the genome. The acyl-CoA synthetase functioned in the oxidation of fatty acid into fatty acid CoA, and then evolved in no other reactions in the following steps. After the formation of hexadecanoyl-CoA, it was subsequently oxidized by acyl-CoA dehydrogenase (EC: 1.3.8.-), and various coding genes were found in this classification. However, the amount of genes evolved in the following step of fatty acid degradation was much less than those evolved in the others. Only 4 copies of Enoyl-CoA hydratase (EC: 4.2.1.17) coding gene were located in the genome. In comparison, the copies of genes belonging to the final 2 steps (CoA-dehydrogenase and CoA-acyltransferase) were much more than that in the other steps. Thus it can be deduced from the gene copies that the speed limiting step in PA-3 was probably concentrated in the hydration reaction catalyzed by Enoyl-CoA hydratase. | ||
| Fig. 6 Metabolic pathway in PA-3 for LCT degradation. The enzymes marked in the β-oxidation pathway are found in PA-3 through GO and KEGG pathway analysis. | ||
4. Discussion
Triglyceride, especially LCT, is difficult to be completely used by microorganisms. Previous reports on LCT degradation were concentrated on fungi, and the degradation was mediated by the β-oxidation pathway, which has been rarely discovered in bacteria.27,28 However, the five key enzymes including CoA-ligase, CoA-oxidase, CoA-hydratase, CoA-dehydrogenase and CoA-thiolase in the β-oxidation pathway for triglyceride degradation were all found in some bacteria such as E. coli and Klebsiella pneumoniae. These genes participated in the lipid biosynthesis process used in cell wall and cell membrane. Among the key enzymes, CoA-hydratase (EC: 1.3.3.6) is the most uncommon one in bacteria. Additionally, bacteria belonging to Pseudomonas were usually isolated for the high enzyme activity of lipase, so the Pseudomonas was the most probable LCT degradation bacteria.29–31 PA-3 was able to consume nearly 28 g L−1 LCT within 7 days, indicating that all the key enzymes in triglyceride esterification and β-oxidation participated in LCT degradation. Compared to LCT degradation in fungi, the degradation by PA-3 was faster for cell biomass accumulation. Thus, PA-3 showed great potential in biological treatment of wastes with high LCTs content.Biofertilizers mainly consist of plant growth-promoting microorganisms accompanied with organic compounds that facilitate microbial growth.32 Nowadays various bacteria and fungi are reported to be used as biofertilizers such as Bacillus, Streptomyces and Azotobacter. Compared to the other rhizobacteria that function as biofertilizers, PA-3 accompanied with LCT degradation products significantly promoted plant growth within the primary 2 weeks after planting,33 showing faster effect than other biofertilizers. LCT degradation products were harvested after 4 days of cultivation of 10 g L−1 LCT, and nearly 1 g L−1 LCT was left in the medium. Thus, the remaining LCT in the soil might function as the subsequent carbon source of PA-3. However, purified PA-3 failed to facilitate cabbage growth compared to the other plant growth-promoting Pseudomonas,34,35 meaning that LCT degradation products by PA-3 possessed distinct mechanisms compared to the previous reports.
Plant growth-promoting bacteria play a crucial role in providing supplements for plant roots. For example, biofixation of N2 by Azotobacter and phosphorus dissolution by phosphate dissolving bacteria provide essential elements for plant growth.36 Thus, the application of the purified functional bacteria can facilitate plant growth by increasing bacterial diversity in soil, which is different from the application of LCT degradation products. Besides, fermentation products of bacteria were also used in promoting plant growth by reducing plant diseases.37,38 However, the addition of both extracted LCT degradation products and the extracted PA-3 had limited effect on cabbage height and leaf area. Thus, the explanation for the plant growth promotion of LCT degradation products was mainly due to the changes in soil bacterial diversity with the joint effects of PA-3 and its metabolites. In nutrient soil, the natural preservation of the soil for 2 weeks led to the increased amount of Firmicutes phylum to a dominant level. However, with the planting of cabbage, the content of Firmicutes decreased largely from 59.82% to 3.45% (Fig. 5A and C), and that of Proteobacteria increased from 32.94% to 62.82% (Fig. 5D and F). Beneficially, the addition of LCT degradation products led to the further decrease in Firmicutes and increase in Proteobacteria (Fig. 5B and E). The increase of Proteobacteria was mainly concentrated in the enrichment of Burkholderia and Providencia genera, and both of the two genera were reported as plant growth-promoting bacteria for agricultural applications.39,40 The decrease was concentrated in Bacteroidetes at the phylum level because the amount of Sphingobacteriaceae family was 6 times lower compared to that of the control. The bacteria belonging to the Sphingobacteriaceae family such as Mucilaginibacter were reported as parasitic bacteria in disease-causing nematodes.41 Thus, the changes of bacteria in nutrient soil promoted soil probiotic concentrations, and harmful bacteria were inhibited.
Compared to the nutrient soil, vermiculite contained no useful compounds for plants and the bacterial diversity change was more convincing. After planting cabbage seeds for 3 days, PA-3 and degradation products were introduced. However, the amount of Pseudomonas was even lower than that of the control. This result indicated that the growth-promoting effect was not attained by the sole addition of PA-3, which was in accordance with the control experiment where extracted PA-3 was used (Fig. 3). At the genus level, the amount of Providencia was also increased, which was identical to that in nutrient soil. Moreover, the other two agricultural probiotics Paenibacillus and Acinetobacter also increased. At phylum level, the amounts of Proteobacteria and Firmicutes increased. The increased amount of Proteobacteria was also caused by the sharp change in Providencia genus, and the increased Firmicutes amount was led by the joint increase in Paenibacillus and Alicyclobacillus genera, which are also agricultural probiotics.42,43 In conclusion, the addition of LCT degradation products resulted in the changes in soil bacterial diversity, and several agricultural probiotics were enriched in the soil, which further facilitated cabbage growth.
In bacteria, genes that evolved in fatty acid degradation usually existed in the genome used for lipids biosynthesis and degradation because lipids are the essential components of cell wall and cell membrane. For instance, genes belonging to the ko00071 pathway were found in E. coli K12, but this strain is unable to survive on LCT as carbon source. In fact, it is difficult for bacteria or even fungi to degrade LCT or kitchen waste oil due to the overlong carbon chain in LCTs. The enzymes used for LCTs degradation function consistently from long chain fatty acids to short chain fatty acids until the LCTs were completely consumed. Thus, the specificity of the enzyme probably restricts the utilization of LCT and its metabolites. However, PA-3 contains all the 5 essential genes of the β-oxidation pathway, and PA-3 efficiently degrades nearly 28 g L−1 LCT or kitchen waste oil within 7 days, meaning that these essential genes efficiently function in various saturated and unsaturated LCTs degradation. LCT is a high density carbon source, and final LCTs degradation product was acetyl-CoA, which participated in the TCA cycle in bacterial cytoplasm. Nevertheless, the valuable compounds (butanediol, and 1,2-propanediol) detected in the fermentation broth of PA-3 with LCT as substrate was too low for industrial biosynthesis. At the same time, kitchen waste oil was mostly catalyzed into acetyl-CoA, which was oxidized into CO2. The further genetic manipulations on PA-3, which draw metabolic flux to valuable chemicals will lead to a more efficient consumption of LCT and kitchen waste oil.
5. Conclusions
In this study, we isolated and characterized a novel LCT degradation bacteria P. aeruginosa PA-3. PA-3 was able to use LCT or kitchen waste oil as its sole carbon source, and 10 g L−1 LCT was completely degraded into short chain valuable compounds within 5 days. Moreover, kitchen waste oil degradation products acted as biofertilizer in promoting cabbage growth. Soil bacterial diversity analysis indicated that LCT degradation products led to the increase in the amount of agricultural probiotics. The whole genome sequencing of PA-3 revealed existence of essential genes evolved in the β-oxidation pathway in the genome of PA-3.Conflicts of interest
There are no conflicts to declare.Abbreviations
| LCT | Long chain triglyceride |
| LCTs | Long chain triglycerides |
| GC-MS | Gas chromatography mass spectrometry |
| WGS | Whole bacterial genome sequencing |
| LB | Luria–Bertani |
| Trilaurin | Lauric acid triglycerin ester |
| PA-3 | Isolated Pseudomonas aeruginosa |
Acknowledgements
This study was financially supported by Joint Funding of Beijing Municipal Natural Science Foundation–Beijing Municipal Education Commission, National Key R&D Program of China [2017YFD0201105] and Premium Funding Project for Academic Human Resources Development in Beijing Union University [BPHR2017DZ07].Notes and references
- M. J. Ramos, C. M. Fernández, A. Casas, L. Rodríguez and A. Pérez, Bioresour. Technol., 2009, 100, 261–268 CrossRef CAS PubMed.
- A. B. Chhetri, K. C. Watts and M. R. Islam, Energies, 2008, 1, 3–18 CrossRef CAS.
- S. Harayama, Y. Kasai and A. Hara, Curr. Opin. Biotechnol., 2004, 15, 205–214 CrossRef CAS PubMed.
- M. G. Kulkarni and A. K. Dalai, Ind. Eng. Chem. Res., 2006, 45, 2901–2913 CrossRef CAS.
- M. Lapuerta, J. M. Herreros, L. L. Lyons, R. García-Contreras and Y. Briceño, Fuel, 2008, 87, 3161–3169 CrossRef CAS.
- A. Galadima, O. Muraza, J. Bundschuh, T. Yusaf, J. P. Maity, E. Nelson, R. Mamat and T. M. I. Mahlia, Energy, 2014, 78, 72–83 CrossRef CAS.
- B. Lugtenberg and F. Kamilova, Annu. Rev. Microbiol., 2009, 2009, 541–556 CrossRef PubMed.
- M. E. Leggett and S. C. Gleddie, Adv. Plant Pathol., 1995, 11, 59–74 Search PubMed.
- J. D. Flores-Félix, M. Marcos-García, L. R. Silva, E. Menéndez, E. Martínez-Molina, P. F. Mateos, E. Velázquez, P. García-Fraile, P. Andrade and R. Rivas, Symbiosis, 2016, 67, 1–8 Search PubMed.
- X. Chen, A. Koumoutsi, R. Scholz, A. Eisenreich, K. Schneider, I. Heinemeyer, B. Morgenstern, B. Voss, W. Hess and O. Reva, Nat. Biotechnol., 2007, 25, 1007–1014 CrossRef CAS PubMed.
- A. Sadeghi, E. Karimi, P. A. Dahaji, M. G. Javid, Y. Dalvand and H. Askari, World J. Microbiol. Biotechnol., 2012, 28, 1503–1509 CrossRef CAS PubMed.
- W. A. Chandanie, M. Kubota and M. Hyakumachi, Appl. Soil Ecol., 2009, 41, 336–341 CrossRef.
- S. Compant, B. Duffy, J. Nowak, C. Clément and E. A. Barka, Appl. Environ. Microbiol., 2005, 71, 4951–4959 CrossRef CAS PubMed.
- T. Tsuge, J. Biosci. Bioeng., 2002, 94, 579–584 CrossRef CAS PubMed.
- D. Mattanovich, Trends Biotechnol., 2008, 26, 100–108 CrossRef PubMed.
- A. Drewnowski, Am. J. Prev. Med., 2004, 27, 154–162 CrossRef PubMed.
- J. Fischer, C. Lefèvre, E. Morava, J. M. Mussini, P. Laforêt, A. Negre-Salvayre, M. Lathrop and R. Salvayre, Nat. Genet., 2007, 39, 28–30 CrossRef CAS PubMed.
- S. Goepfert and Y. Poirier, Curr. Opin. Plant Biol., 2007, 10, 245–251 CrossRef CAS PubMed.
- M. J. Mcinerney, M. P. Bryant and N. Pfennig, Arch. Microbiol., 1979, 122, 129–135 CrossRef CAS.
- Y. Li, Y. M. Yin, X. Y. Wang, H. Wu and X. Z. Ge, J. Asian Nat. Prod. Res., 2017, 20, 148–162 CrossRef PubMed.
- L. T. Miller, J. Clin. Microbiol., 1982, 16, 584–586 CAS.
- L. Zelles and Q. Bai, Soil Biol. Biochem., 1993, 25, 495–507 CrossRef CAS.
- S. J. Salipante, D. J. SenGupta, L. A. Cummings, T. A. Land, D. R. Hoogestraat and B. T. Cookson, J. Clin. Microbiol., 2015, 53, 1072–1079 CrossRef CAS PubMed.
- S. Steinigeweg and J. Gmehling, Ind. Eng. Chem. Res., 2002, 41, 5483–5490 CrossRef CAS.
- H. H. Dai, L. T. Yang, B. Lin, C. S. Wang and G. A. Shi, J. Am. Oil Chem. Soc., 2009, 86, 261–267 CrossRef CAS.
- O. Sarkar, A. N. Kumar, S. Dahiya, K. V. Krishna, D. K. Yeruva and S. V. Mohan, RSC Adv., 2016, 6, 18641–18653 RSC.
- L. A. Johnson, I. R. Beacham, I. C. Macrae and M. L. Free, Appl. Environ. Microbiol., 1992, 58, 1776–1779 CAS.
- A. Beopoulos, J. M. Nicaud and C. Gaillardin, Appl. Microbiol. Biotechnol., 2011, 90, 1193–1206 CrossRef CAS PubMed.
- S. L. Cao, Y. M. Huang, X. H. Li, P. Xu, H. Wu, N. Li, W. Y. Lou and M. H. Zong, Sci. Rep., 2016, 6, 20420 CrossRef CAS PubMed.
- C. H. Ali, A. S. Qureshi, S. M. Mbadinga, J. F. Liu, S. Z. Yang and B. Z. Mu, Renew. Energ., 2017, 109, 93–100 CrossRef CAS.
- H. Noureddini, X. Gao and R. S. Philkana, Bioresour. Technol., 2005, 96, 769–777 CrossRef CAS PubMed.
- S. K. Yadav, A. A. Juwarkar, G. P. Kumar, P. R. Thawale, S. K. Singh and T. Chakrabarti, Bioresour. Technol., 2009, 100, 4616–4622 CrossRef CAS PubMed.
- S. C. Wu, Z. H. Cao, Z. G. Li, K. C. Cheung and M. H. Wong, Geoderma, 2005, 125, 155–166 CrossRef.
- M. Lucy, E. Reed and B. R. Glick, Antonie van Leeuwenhoek, 2004, 86, 1–25 CrossRef CAS PubMed.
- S. Mehnaz and G. Lazarovits, Microb. Ecol., 2006, 51, 326–335 CrossRef PubMed.
- B. M. Hoffman, D. Lukoyanov, Z. Y. Yang, D. R. Dean and L. C. Seefeldt, Chem. Rev., 2014, 114, 4041–4062 CrossRef CAS PubMed.
- M. Ongena and P. Jacques, Trends Microbiol., 2008, 16, 115–125 CrossRef CAS.
- E. Montesinos, FEMS Microbiol. Lett., 2007, 270, 1–11 CrossRef CAS.
- E. A. Barka and J. Nowak, Appl. Environ. Microbiol., 2006, 72, 7246–7252 CrossRef CAS.
- H. RodríGuez and R. Fraga, Biotechnol. Adv., 1999, 17, 319–339 CrossRef.
- G. Paiva, P. Abreu, D. N. Proença, S. Santos, M. F. Nobre and P. V. Morais, Int. J. Syst. Evol. Microbiol., 2014, 64, 2223–2228 CrossRef CAS.
- S. Timmusk and E. G. Wagner, Mol. Plant-Microbe Interact., 1999, 12, 951–959 CrossRef CAS PubMed.
- Y. Bai, F. D'Aoust, D. L. Smith and B. T. Driscoll, Can. J. Microbiol., 2002, 48, 230–238 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2018 |