Open Access Article
Open Access ArticleGlucuronidation of [6]-shogaol, [8]-shogaol and [10]-shogaol by human tissues and expressed UGT enzymes: identification of UGT2B7 as the major contributor†
Liangliang He‡
a,
Jinjin Xu‡a,
Qi Wangab,
Yezi Zhanga,
Zifei Qin *ac,
Yang Yuad,
Zhengming Qianb,
Zhihong Yao
*ac,
Yang Yuad,
Zhengming Qianb,
Zhihong Yao *ad and
Xinsheng Yao
*ad and
Xinsheng Yao ad
ad
aCollege of Pharmacy, Jinan University, Guangzhou 510632, P. R. China. E-mail: yaozhihong_jnu@163.com; yaozhihong.jnu@gmail.com
bKey Laboratory of State Administration of Traditional Chinese Medicine, Sunshine Lake Pharma Co., LTD, Dongguan, Guangdong 523850, P. R. China
cDepartment of Pharmacy, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, P. R. China. E-mail: qzf1989@163.com
dGuangdong Provincial Key Laboratory of Pharmacodynamic Constituents of TCM and New Drugs Research, College of Pharmacy, Jinan University, Guangzhou 510632, China
First published on 12th December 2018
Abstract
Shogaols, mainly [6]-shogaol (6S), [8]-shogaol (8S) and [10]-shogaol (10S), the predominant and characteristic pungent phytochemicals in ginger, are responsible for most of its beneficial effects. However, poor oral bioavailability owing to extensive glucuronidation limits their application. The present study aimed to characterize the glucuronidation pathways of 6S, 8S and 10S by using pooled human liver microsomes (HLM), human intestine microsomes (HIM) and recombinant human UDP-glucosyltransferases (UGTs). The rates of glucuronidation were determined by incubating shogaols with uridine diphosphate glucuronic acid-supplemented microsomes. Kinetic parameters were derived by appropriate model fitting. Reaction phenotyping assays, activity correlation analyses and relative activity factors were performed to identify the main UGT isoforms. As a result, one mono-4′-O-glucuronide was detected after incubating each shogaol with HLM and HIM. Enzymes kinetic analysis demonstrated that glucuronidation of shogaols consistently displayed the substrate inhibition profile, and the liver showed higher metabolic activity for shogaols (CLint = 1.37–2.87 mL min−1 mg−1) than the intestine (CLint = 0.67–0.85 mL min−1 mg−1). Besides, reaction phenotyping assays revealed that UGT2B7 displayed the highest catalytic ability (CLint = 0.47–1.17 mL min−1 mg−1) among all tested UGTs. In addition, glucuronidation of shogaols was strongly correlated with AZT glucuronidation (r = 0.886, 0.803 and 0.871 for glucuronidation of 6S, 8S and 10S, respectively; p < 0.01) in a bank of individual HLMs (n = 9). Furthermore, UGT2B7 contributed to 40.8%, 34.2% and 36.0% for the glucuronidation of 6S, 8S and 10S in HLM, respectively. Taken altogether, shogaols were efficiently metabolized through the glucuronidation pathway, and UGT2B7 was the main contributor to their glucuronidation.
Introduction
Ginger, the rhizome of Zingiber officinale (ZO), has been widely used throughout the world for centuries.1 As a functional food, ZO is also well known for its nutraceutical and medicinal values, including antioxidation, antitumor, antidiabetic, and anti-inflammatory.2–9 These beneficial effects are believed to be mainly attributed to its major pungent phytochemicals (mainly gingerols and shogaols).10,11 In particular, shogaols (i.e. [6]-shogaol, 6S; [8]-shogaol, 8S and [10]-shogaol, 10S), the predominant and characteristic pungent constituents in ginger, have drawn considerable attention owing to their stronger anti-inflammatory and anticarcinogenic effects than that of the gingerols.12–14 Meanwhile, 6S, 8S and 10S have also been reported to induce G2/M arrest and aberrant mitotic cell death associated with tubulin.15 Furthermore, it is also revealed that 6S, 8S and 10S inhibited the invasion of human-derived breast carcinoma MDA-MB-231 cell with a decrease in matrix metalloproteinase-9 secretion.16 Altogether, shogaols have been proved as potential antimetastatic agents for clinical use.15 These beneficial activities have stimulated increasing interests in their physiological disposal.Previous pharmacokinetics reported that pungent chemicals showed poor oral bioavailability with only small amounts of the prototypes entering the systemic circulation, and they were mainly present in the blood in the form of metabolites (Cmax = 99.99 ng mL−1 at 0.25 h for 6S).17–20 These findings indicated that their metabolic characteristics had an important impact on the bioavailability and even the health-promoting effects. Although, the mercapturic acid and redox pathways were considered as the major metabolic routes for shogaols, whereas these thiol-conjugated and phase I metabolites were mainly excreted in the form of glucuronidated conjugates.21–24 Furthermore, our previous study also showed that shogaols related glucuronides had high exposure in rat plasma after oral administration of ginger.24 Generally, glucuronidation is the primary phase II reaction since it contributes to more than 35% of all phase II metabolites.25 In human, UDP-glucuronosyltransferases (UGTs) are a superfamily of enzymes that mediate the glucuronidation of many endogenous and exogenous compounds to generate more polar and excretable compounds.26 Thus, the glucuronidation of shogaols was probably the main reason affecting their oral bioavailability and functional effects. Nevertheless, so far, only one literature reported the kinetics for glucuronidation of 6S in human liver and intestinal microsomes,27 and the glucuronidation characteristics for 8S and 10S have never been reported. Hence, the UGT enzymes which catalyse the glucuronidation of shogaols and the major contributing enzyme have not been fully characterized, and further research is needed.
In this study, the metabolism of shogaols (6S, 8S and 10S) via the glucuronidation pathway was characterized in vitro, and the main UGT enzymes involved in the glucuronidation of shogaols were identified. The uridine diphosphate glucuronic acid (UDPGA)-supplemented microsomes were incubated with shogaols to determine the rates of glucuronidation. Kinetic parameters were derived by fitting an appropriate model to the data. The reaction phenotyping, relative activity factors and activity correlation analyses were performed to identify the main UGT enzymes contributing to hepatic metabolism of shogaols. With these methods, this study was of great value for a better predicting the disposal of shogaols and understanding of their mechanism action in human tissues, which also helped to illustrate the variation in their beneficial effects and potential drug–herbal interactions.
Experimental
Materials
Alamethicin, D-saccharic-1,4-lactone, magnesium chloride, and uridine diphosphate glucuronic acid (UDPGA), were provided from Sigma-Aldrich (St Louis, MO). Pooled human liver microsomes (HLM), human intestine microsomes (HIM) and recombinant expressed human UGT Supersomes™ (UGT1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B10, 2B15 and 2B17) were obtained from Corning Biosciences (New York, USA). Zidovudine (AZT) were purchased from Aladdin Chemicals (Shanghai, China). [6]-shogaol (6S), [8]-shogaol (8S), [10]-shogaol (10S) with purity over 99% were isolated and identified in our laboratory, and their 13C-NMR and HRMS data were listed in the ESI.† Other chemicals and materials were all analytical grade.Glucuronidation assay
The test compound (6S, 8S and 10S) was incubated with HLM, HIM and expressed UGTs enzymes, respectively, to determine the rates of glucuronidation as published previously.28–30 Briefly, the incubation mixture (200 μL, 37 °C) mainly contained 50 mM Tris–hydrochloric acid buffer (pH 7.4), 22 μg mL−1 alamethicin, 0.88 mM MgCl2, 4.4 mM saccharolactone and 3.5 mM UDPGA. The reaction was terminated by adding ice-cold acetonitrile (containing the internal standard N-vanillylnonanamide for 6S and 10S, or [6]-gingerol for 8S), and the samples were vortexed and centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800g for 10 min. Then, the supernatant was subjected to UPLC-Q/TOF-MS analysis. Incubation without UDPGA served as negative control to confirm the metabolites produced were UDPGA-dependent. All experiments were performed in triplicate. Preliminary experiments were performed to ensure that the rates of glucuronidation were determined under linear conditions with respect to the incubation time and protein concentration.
800g for 10 min. Then, the supernatant was subjected to UPLC-Q/TOF-MS analysis. Incubation without UDPGA served as negative control to confirm the metabolites produced were UDPGA-dependent. All experiments were performed in triplicate. Preliminary experiments were performed to ensure that the rates of glucuronidation were determined under linear conditions with respect to the incubation time and protein concentration.
UPLC-Q/TOF-MS analytical condition
The UPLC analysis was performed on an Acquity UPLC 1-Class system equipped with a binary solvent system, an automatic sample manager, and photodiode array detector (Waters Corporation). A BEH RP C18 column (2.1 × 50 mm, 1.7 μm) was used for separation of shogaols (6S and 10S) and their glucuronides, whereas a BEH C18 column (2.1 × 50 mm, 1.7 μm) was selected to separate 8S and its glucuronide. The mobile phases consisted of water (A) and acetonitrile (B), both including 0.1% formic acid (v/v), which were delivered at a flow rate of 0.3 mL min−1 using a gradient elution program as follow: 20% B from 0–0.2 min, 20–90% B from 0.2–3.0 min, 90–90% B from 3.0–4.0 min, 90–20% B from 4.0–4.5 min. 20–20% B from 4.5–5.0 min. The detection wavelength was set at 280 nm and the injection volume was 4 μL. Quantitation of the shogaol glucuronide was based on the standard curve of the parent compound using the same method as described by UPLC analysis.29,30 The limit of quantity for 6S, 8S and 10S was 0.098 μM. Calibration curves were constructed by plotting shogaol (analyte)/quercetin (internal standard) peak area ratios (Y) versus shogaol concentrations (X) using a 1/X2 weighting factor. Acceptable linear correlation for 6S, 8S and 10S (Y6s = 0.0380X6s, Y8s = 0.0535X8s, Y10s = 0.0389X10s) was confirmed by correlation coefficients (r2) of 0.9900, 0.9940 and 0.9993, respectively. The linear range was 0.098–50 μM.The UPLC system was coupled to a hybrid quadrupole orthogonal time-of-flight (Q-TOF) tandem mass spectrometer equipped with electrospray ionization (SYNAPT™ G2 HDMS, Waters, Manchester, U.K.). The operating parameters were as follows: capillary voltage of 3 kV (ESI+), sample cone voltage of 30 V (ESI+), ramp trap collision energy of 30–50 V (ESI+), extraction cone voltage of 4 V, source temperature of 100 °C, desolvation temperature 300 °C, cone gas flow of 50 L h−1 and desolvation gas flow of 800 L h−1. Argon was used as collision gas for CID in MSE mode. Leucine enkephalin was used as an external reference of LockSpray™ infused at a constant flow of 5 μL min−1 and data were centroided during acquisition.
Enzymes kinetic evaluation
According to the glucuronidation assay protocol as described previously, the rates of glucuronidation were determined for 6S, 8S and 10S at a series of concentrations. The kinetic model of substrate inhibition (eqn (1)) was fitted to the data of metabolic rates versus substrate concentrations. Appropriate models were selected based on visual inspection of the Eadie–Hofstee plot.31–33 Model fitting and parameter estimation were performed using the GraphPad Prism 6 software (San Diego, CA, USA).
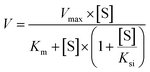 | (1) |
Activity correlation analysis
The metabolic activities of individual HLMs (n = 9) towards 6S, 8S, 10S and AZT (a probe substrate for UGT2B7) were determined, according to the glucuronidation assay protocol as described previously.30 6S (10 μM), 8S (20 μM) and 10S (10 μM) were incubated with UDPGA-supplemented individual HLM (1.0 mg mL−1) for 15 min, whereas AZT (1.25 mM) was treated with UDPGA-supplemented individual HLM (1.0 mg mL−1) for 120 min. Correlation analysis was performed between 4′-O-glucuronidated shogaol and AZT glucuronidation. Correlation analysis was performed using GraphPad Prism 6 software.Contribution of UGT2B7
The relative activity factor (RAF) approach was applied to estimate the contributing of individual expressed UGT enzymes to glucuronidated shogaols in HLM as described in previous studies.26,30 The value of RAF was calculated by the activity ratio of pooled HLM to UGT2B7 enzyme toward a probe substrate (AZT) for this enzyme (eqn (2)). The relative amount of shogaol glucuronidation in HLM attributed to UGT2B7 enzyme was estimated by multiplying the glucuronidation activity derived with this enzyme by the corresponding RAF. The contribution of individual UGT enzyme were calculated according to eqn (3).
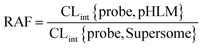 | (2) |
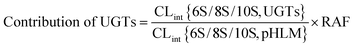 | (3) |
Statistical analysis
Data are expressed as mean ± standard deviation (SD). The two-tailed Student's t-test was used to compare the mean differences, and the prior level of significance was set at p < 0.05 (*), p < 0.01 (**) or p < 0.001 (***).Results
Structural identification of shogaols metabolites
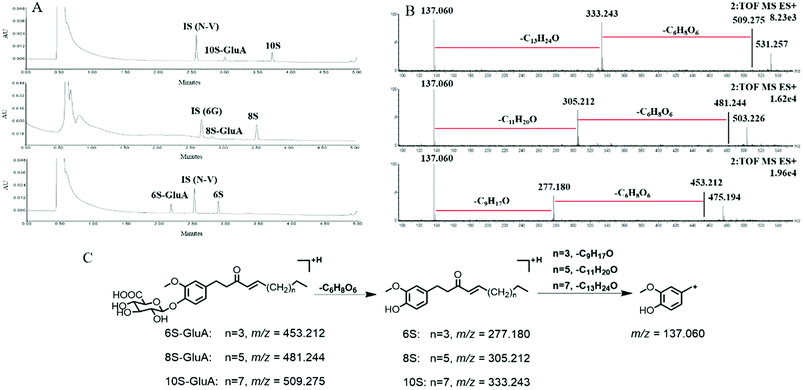
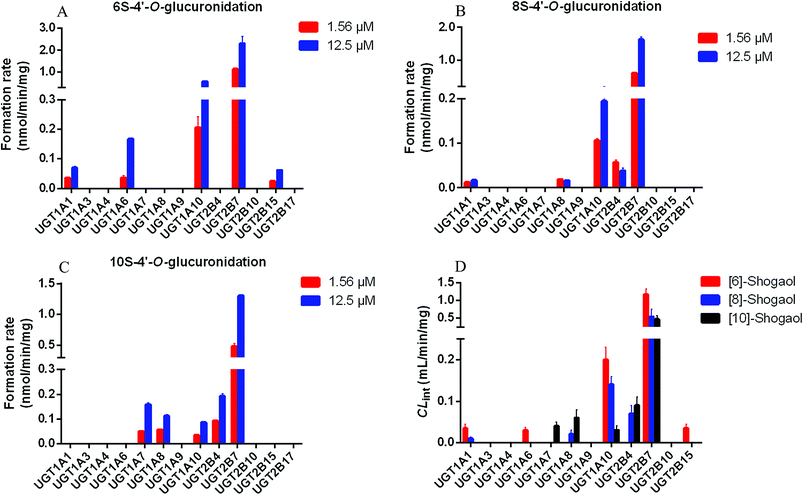
After incubation of 6S, 8S and 10S with UDPGA-supplemented various enzyme sources, respectively, one additional peak, which showed a similar ultraviolet absorption profile (λmax: 280 nm) with the parent shogaol, was detected by UPLC-PDA analysis (Fig. 1). In (ESI+) MSE analysis, an obvious neural loss of 176.032 Da was detected from the parent ions of the corresponding metabolites (i.e. m/z 453.212, 481.244 and 509.275 for the metabolites of 6S, 8S and 10S, respectively), which suggested that these metabolites were glucuronide conjugation products of 6S, 8S and 10S, respectively. The site of glucuronidation was assigned on the C4′ position of the aromatic ring since there was only one hydroxyl group (at the C4′ position) for each parent shogaol. Hence, for 6S, 8S and 10S, their related glucuronidated metabolites were characterized as 6S-4′-O-glucuronide, 8S-4′-O-glucuronide and 10S-4′-O-glucuronide, respectively.Glucuronidation of shogaols in HLM and HIM
Kinetic profiling revealed that glucuronidation of shogaols (6S, 8S and 10S) in HLM and HIM both followed the inhibition kinetics (Fig. S1†). The glucuronide formation for 6S, 8S and 10S in HLM was 20.43, 28.55 and 14.29 nmol min−1 mg−1, respectively, which showed a remarkable statistical difference (p < 0.001) compared with that in HIM (Vmax = 4.57 nmol min−1 mg−1 for 6S, Table 1; 2.62 nmol min−1 mg−1 for 8S, Table 2; 1.27 nmol min−1 mg−1 for 10S, Table 3). This phenomenon indicated that glucuronidation of shogaols was much more efficient in HLM than in HIM. Furthermore, in HLM, the total CLint values were 2.87, 1.66 and 1.37 mL min−1 mg−1 for glucuronidation of 6S, 8S and 10S (Tables 1–3), respectively. Meanwhile, the corresponding total CLint values were 0.85 mL min−1 mg−1 for 6S glucuronidation (Table 1), 0.77 mL min−1 mg−1 for 8S glucuronidation (Table 2) and 0.67 mL min−1 mg−1 for 10S glucuronidation (Table 3) in HIM. This suggested that the glucuronidation activity for shogaols in HLM and HIM might decrease as the length of their alkyl chains increased.| Protein source | Vmax (nmol min−1 mg−1) | Km (μM) | Ksi (μM) | CLint (mL min−1 mg−1) | Model |
|---|---|---|---|---|---|
| a UGT, UDP-glucuronosyltransferase; HLM, human liver microsomes; HIM, human intestine microsomes; SI, substrate inhibition model. | |||||
| HLM | 20.43 ± 1.11 | 7.12 ± 0.82 | 183.20 ± 36.10 | 2.87 ± 0.36 | SI |
| HIM | 4.57 ± 0.36 | 5.37 ± 0.88 | 78.91 ± 15.60 | 0.85 ± 0.15 | SI |
| UGT1A1 | 0.11 ± 0.01 | 2.96 ± 0.79 | 33.97 ± 9.60 | 0.04 ± 0.01 | SI |
| UGT1A6 | 0.36 ± 0.05 | 13.81 ± 3.29 | 64.76 ± 18.61 | 0.03 ± 0.007 | SI |
| UGT1A10 | 0.86 ± 0.06 | 4.28 ± 0.66 | 86.74 ± 16.34 | 0.20 ± 0.03 | SI |
| UGT2B7 | 3.21 ± 0.17 | 2.75 ± 0.35 | 91.79 ± 14.92 | 1.17 ± 0.16 | SI |
| UGT2B15 | 0.17 ± 0.04 | 10.24 ± 3.63 | 10.78 ± 3.91 | 0.02 ± 0.007 | SI |
| Protein source | Vmax (nmol min−1 mg−1) | Km (μM) | Ksi (μM) | CLint (mL min−1 mg−1) | Model |
|---|---|---|---|---|---|
| a UGT, UDP-glucuronosyltransferase; HLM, human liver microsomes; HIM, human intestine microsomes; SI, substrate inhibition model. | |||||
| HLM | 28.55 ± 3.78 | 17.18 ± 2.96 | 14.08 ± 2.63 | 1.66 ± 0.36 | SI |
| HIM | 2.62 ± 0.26 | 3.40 ± 0.64 | 32.26 ± 7.26 | 0.77 ± 0.17 | SI |
| UGT1A1 | 0.04 ± 0.007 | 3.49 ± 1.11 | 13.46 ± 3.99 | 0.01 ± 0.004 | SI |
| UGT1A8 | 0.08 ± 0.03 | 3.81 ± 1.83 | 4.01 ± 1.92 | 0.02 ± 0.01 | SI |
| UGT1A10 | 0.29 ± 0.02 | 2.15 ± 0.35 | 49.85 ± 10.49 | 0.14 ± 0.02 | SI |
| UGT2B4 | 0.15 ± 0.02 | 2.11 ± 0.51 | 5.22 ± 1.23 | 0.07 ± 0.02 | SI |
| UGT2B7 | 4.88 ± 1.10 | 9.01 ± 2.83 | 11.59 ± 3.84 | 0.54 ± 0.21 | SI |
| Protein source | Vmax (nmol min−1 mg−1) | Km (μM) | Ksi (μM) | CLint (mL min−1 mg−1) | Model |
|---|---|---|---|---|---|
| a UGT, UDP-glucuronosyltransferase; HLM, human liver microsomes; HIM, human intestine microsomes; SI, substrate inhibition model. | |||||
| HLM | 14.29 ± 1.60 | 10.43 ± 1.69 | 19.50 ± 3.51 | 1.37 ± 0.27 | SI |
| HIM | 1.27 ± 0.06 | 1.90 ± 0.20 | 39.73 ± 5.12 | 0.67 ± 0.08 | SI |
| UGT1A7 | 0.34 ± 0.06 | 9.50 ± 2.35 | 22.24 ± 6.11 | 0.04 ± 0.01 | SI |
| UGT1A8 | 0.25 ± 0.03 | 4.27 ± 0.93 | 17.19 ± 3.96 | 0.06 ± 0.02 | SI |
| UGT1A10 | 0.29 ± 0.05 | 10.11 ± 2.35 | 8.21 ± 1.96 | 0.03 ± 0.008 | SI |
| UGT2B4 | 0.26 ± 0.02 | 2.93 ± 0.56 | 47.70 ± 11.56 | 0.09 ± 0.02 | SI |
| UGT2B7 | 2.31 ± 0.27 | 4.88 ± 0.99 | 30.12 ± 7.27 | 0.47 ± 0.11 | SI |
Reaction kinetics for regioselective glucuronidation of [6]-shogaol by expressed UGT enzymes
To identify the enzymes involving in the glucuronidation of 6S, various UGT enzymes (UGT1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B10, 2B15 and 2B17) were tested for their catalysis activities. The results showed that UGT1A1, 1A6, 1A10, 2B7 and 2B15 were the main enzymes to catalyse the formation of 6S-4′-O-glucuronide (Fig. 2A). Furthermore, glucuronidation of 6S by these active UGT enzymes followed the substrate inhibition kinetics (Fig. S2†). According to the CLint values (Table 1), the most efficient enzyme in glucuronidating 6S was UGT2B7 (1.17 mL min−1 mg−1), followed by UGT1A10 (0.20 mL min−1 mg−1), 1A1 (0.04 mL min−1 mg−1), 1A6 (0.03 mL min−1 mg−1) and 2B15 (0.02 mL min−1 mg−1). It was known that UGT1A10 was mainly expressed in the small intestine, and hardly expressed in the liver, which indicated that hepatic and intestinal glucuronidation were important clearance mechanisms for 6S in vivo.Reaction kinetics for regioselective glucuronidation of [8]-shogaol by expressed UGT enzymes
Of the 13 tested UGT enzymes, five of them, including UGT1A1, 1A8, 1A10, 2B4 and 2B7, generated 4′-O-glucuronide from 8S (Fig. 2B). Enzymes kinetic studies showed that glucuronidation of 8S by the five UGT enzymes consistently displayed the substrate inhibition profile (Fig. S3†). Furthermore, on the basis of a comparison of CLint values (Table 2), it was found that UGT1A10 and 2B7 were the two most active enzymes in glucuronidating 8S, which was similar to the main enzymes to catalyse the formation of 6S-4′-O-glucuronide. Of note, UGT2B7 showed the highest CLint value of 0.54 mL min−1 mg−1, which was 4 times of that for UGT1A10 mediated glucuronidation. The results suggested that UGT2B7 was important for the bioavailability of 8S.Reaction kinetics for regioselective glucuronidation of [10]-shogaol by expressed UGT enzymes
Formation of 10S-4′-O-glucuronide was mainly observed with UGT1A7, 1A8, 1A10, 2B4 and 2B7 among the 13 expressed UGT enzymes (Fig. 2C). The enzymes kinetics for glucuronidation of 10S by the five UGT enzymes consistently displayed the substrate inhibition profile (Fig. S4†). As showed in Table 3, UGT2B7 showed the highest Vmax and CLint value of 2.31 nmol min−1 mg−1 and 0.47 mL min−1 mg−1 with a lower Km at 4.88 μM, respectively. In addition, based on a comparison of the apparent CLint values (Table 3), UGT2B7 was the most active enzyme in glucuronidation of 10S. By contrast, glucuronidation of 10S by UGT1A7, 1A8, 1A10 and 2B4 was less efficient with much lower CLint values compared to that of UGT2B7. Of note, UGT1A10 did not exhibit higher catalytic activity to 10S compare to other four UGT enzymes, which was different from the importance of UGT1A10 in catalysing the formation of 6S and 8S-4′-O-glucuronide. The results suggested that UGT2B7 may be a main enzyme contributing to glucuronidation of 10S.Activity correlation analysis for UGT2B7
As mentioned, glucuronidation activity of AZT in pooled HLM is a well-accepted functional maker for UGT2B7.26,30 Hence, the formation rates of individual HLMs (n = 9) toward shogaol-4′-O-glucuronidate and AZT glucuronidation were measured. The results showed that 6S-4′-O-glucuronidate exhibited the strongest correlation with AZT glucuronidation (r = 0.886, p = 0.002), followed by 10S-4′-O-glucuronidate (r = 0.871, p = 0.002) and 8S-4′-O-glucuronidate (r = 0.803, p = 0.009) (Table 4 and Fig. S5†). The results indicated that the contribution of UGT2B7 to glucuronidation of shogaols in the liver was appreciable, and UGT2B7 played a critical role in glucuronidation of shogaols.| Compound | Metabolite | r value | p value |
|---|---|---|---|
| a AZT, zidovudine; 6S, [6]-shogaol; 8S, [8]-shogaol; 10S, [10]-shogaol; GluA: glucuronic acid. (**p < 0.01). | |||
| 6S | 6S-GluA | 0.886 | 0.002, ** |
| 8S | 8S-GluA | 0.803 | 0.009, ** |
| 10S | 10S-GluA | 0.871 | 0.002, ** |
Contribution of UGT2B7 to shogaols glucuronidation in HLM
As presented in previous study, the RAF value for UGT2B7 was 1.05.26,30 Hence, the scaled CLint value of 6S-4′-O-glucuronide was 2.46 (=1.17 × 1.05) mL min−1 mg−1 for UGT2B7, representing 40.8% of the CLint value in HLM (2.87 mL min−1 mg−1). The scaled CLint value of 8S-4′-O-glucuronide was 1.13 (=0.54 × 1.05) mL min−1 mg−1 for UGT2B7, which was 34.2% of total glucuronidation activity in HLM (1.66 mL min−1 mg−1). The scaled CLint value of 10S-4′-O-glucuronide was 2.46 (=0.47 × 1.05) mL min−1 mg−1 for UGT2B7, which represented 36.0% of the CLint value in HLM (1.37 mL min−1 mg−1). Clearly, the results indicated that the glucuronidation of shogaols in HLM was essentially contributed by UGT2B7.Discussion
As the functional and characteristic pungent chemicals in ginger, shogaols has drawn increasing attentions in recent years due to their stronger anti-inflammatory and anticancer effects than that of the gingerols.12–14 However, their metabolic properties have not been fully characterized. It was known that glucuronidation accounted for an important part of the metabolic pathways of pungent chemicals of ginger (mainly gingerols and shogaols) in animals and humans.17–20 Therefore, the potency of glucuronidation would be an important factor controlling the plasma concentration of gingerols, shogaols and their metabolites, and thereby affecting the beneficial effects on the bodies. Previous study has reported that UGT2B7 (mainly formation of 4′-O-glucuronides of gingerols) and UGT1A9 (mainly formation of 5-O-glucuronides of gingerols) were the main enzymes responsible for regioselective glucuronidation of gingerols.34 Thus far, the metabolism of shogaols (6S, 8S and 10S) via the glucuronidation pathway has not been revealed well.27 Hence, in this study, the glucuronidation pathways of shogaols were characterized by using HLM, HIM, and expressed UGT enzymes. It was reported for the first time that shogaols was efficiently metabolized via the glucuronidation pathway, indicating that the glucuronidation was an important pathway for the clearance of shogaols.In this study, determination of glucuronidation activity was based on the intrinsic clearance values (CLint) obtained by kinetic profiling and modeling over a wide range of substrate concentrations.35 The advantages of using CLint (=Vmax/Km) as an indicator of UGT activity were as follow. Firstly, CLint was independent of substrate concentration and represented the catalytic efficiency of UGT enzymes. Secondly, when compared with other kinetic parameters, such as Km or Vmax, CLint was more relevant in predicting clearance in vivo. Accordingly, in this study, the liver showed a high CLint values (1.37–2.87 mL min−1 mg−1, Tables 1–3) for the glucuronidation of shogaols, suggesting the hepatic glucuronidation should lead to a rapid elimination of shogaols from human body, which was consistent with the lower oral bioavailability of shogaols.
In addition, we provided strong evidences to propose that UGT2B7 played a predominant role in 4′-O-glucuronidation of shogaols in the liver. First, as Tables 1–3 showed, UGT2B7 showed predominant activity toward shogaols of all expressed UGT enzymes (CLint = 1.17, 0.54 and 0.47 mL min−1 mg−1 for glucuronidation of 6S, 8S and 10S, respectively). Second, 4′-O-glucuronidation of shogaols was strongly correlated (r = 0.886, 0.803 and 0.871 for glucuronidation of 6S, 8S and 10S, respectively; p < 0.01) with zidovudine glucuronidation (Table 4 and Fig. S5†). Third, the contribution of UGT2B7 to 6S, 8S and 10S glucuronidation in HLM was calculated as 40.8%, 34.2% and 36.0%, respectively, based on the RAF approach. As mentioned, UGT2B7 played a predominant role in glucuronidation of gingerols at C4′ position. And it was worth noting that the effect of UGT1A9 on the glucuronidation of gingerols at C5 position, whereas UGT1A9 did not show a contribution to glucuronidation of shogaols in the present study. It was known that shogaols could be regarded as the dehydrated products of gingerols at C5 position. These results further demonstrated the regioselective action of the UGT enzymes on glucuronidation of shogaols or gingerols, which would contribute to a better understanding of the biological effects of shogaols and gingerols in vivo.
Furthermore, the results also showed that as the length of the alkyl chain increased, the glucuronidation efficiency of shogaols in HLM and HIM decreased, which suggested that the glucuronidation activity of shogaols by HLM and HIM varied with the chemical structure of the substrate. In addition, the intestinal glucuronidation clearance values of shogaols was approximately 2.0–3.4 fold lower than that of glucuronidation clearance in liver (Tables 1–3), which demonstrated that most of the substrates in the liver have a higher glucuronidation activity than those in intestine.36 However, the overall contribution of intestinal glucuronidation should not be ignored as the higher exposure and longer retention time in the intestine than that in liver after oral administration of shogaols.37 Of note, UGT2B7 was expressed in both the intestine and the liver,27 which indicated that the glucuronidation of shogaols in intestine by UGT2B7 could not be ignored. Hence, the contribution of UGT2B7 to shogaols glucuronidation in HIM need further evaluated. Besides UGT2B7, it was also worthy to mention that UGT1A10 which were abundantly expressed in HIM played an important role in the glucuronidation of 6S (CLint = 0.20 mL min−1 mg−1) and 8S (CLint = 0.14 mL min−1 mg−1) based on their kinetic values. However, UGT1A7 (CLint = 0.04 mL min−1 mg−1) and UGT1A8 (CLint = 0.06 mL min−1 mg−1) were the main enzymes for the glucuronidation of 10S in the intestinal tract compared to that of UGT1A10 (CLint = 0.03 mL min−1 mg−1). These results further indicated that the glucuronidation of shogaols was affected by their structures. It was known that the glucuronidation activity could be associated with the biological effects for chemicals.38 Hence, the modulation of glucuronidation liability might help to optimize their efficacy. However, further studies about their rigorous quantitative–activity relationships and predictive models for glucuronidation were necessary.
Normally, glucuronidation was considered as a detoxification mechanism as glucuronides often had no pharmacological bioactive due to the high extremely polarity and rapid excretion from the body.34 Previous study revealed that the glucuronidation of 6S greatly reduced its cytotoxicity on human colon cancer cells.27 Hence, for 6S and other shogaols, the inhibition of UGT2B7 enzyme was of some significance to the exertion of their anticancer efficacy. Furthermore, it was also reported that 6S was a significant inhibitor of UGT2B7 (Ki = 3.4 μM).39 These results indicated that there was a high possibility of drug–drug interaction between shogaols and the drugs such as zidovudine, naloxone, morphine, and others whose main metabolic pathways was catalysed by UGT2B7.
Of note, the UGT2B7 gene was highly polymorphic with three nonsynonymous coding single nucleotide polymorphisms (SNPs), including exon 1-211G > T (UGT2B7*71S), exon 2-802C > T (UGT2B7*2), and exon 5 1192G > A (UGT2B7*5). Furthermore, evidences have been shown that the genetic variations of UGT2B7 gene were different in different populations.40 For example, the allelic frequency of UGT2B7*2 in the Korean population was 0.39, which was lower than that in the Caucasian (0.49), but higher than that in the Japanese (0.27).41 Therefore, after oral administration of ginger or the shogaols, their metabolism via UGT2B7 could be affected owing to the genetic polymorphisms of UGT2B7, which could further increase or decrease their plasma concentrations, causing enhanced or limited beneficial effects. In addition, there was also a need to consider ethnic variability in assessing the beneficial effects of ginger or shogaols.
Conclusion
In summary, shogaols, the functional compounds in ginger, was subjected to efficient glucuronidation in HLM and HIM. However, the liver displayed a higher glucuronidation activity with the CLint values of 1.37–2.87 mL min−1 mg−1 than that of the intestine (CLint values of 0.67–0.85 mL min−1 mg−1). Of note, UGT2B7 showed the highest metabolic activity toward shogaols compared to other expressed UGT enzymes used in this study. And the shogaols glucuronidation was strongly correlated with AZT glucuronidation (r = 0.886, 0.803 and 0.871 for glucuronidation of 6S, 8S and 10S, respectively; p < 0.01) in a bank of individual HLMs (n = 9). Also, based on the RAF approach, the contribution of UGT2B7 was estimated as 40.8%, 34.2% and 36.0% for the glucuronidation of 6S, 8S and 10S in HLM. Overall, this study provided a better insight of the impact of glucuronidation on the functional effects of shogaols, which also gave an in-depth data on understanding of their mechanism action in human tissues and potential drug–herbal interactions.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by National Natural Science Foundation of China (81774219), State Key Program of National Natural Science Foundation of China (81630097), and National Major Scientific and Program of Introducing Talents of Discipline to Universities (B13038).References
- M. Afzal, D. Al-Hadidi, M. Menon, J. Pesek and M. S. Dhami, Drug Metab. Drug Interact., 2001, 18, 159–190 CAS.
- K. R. Shanmugam, K. Mallikarjuna, N. Kesireddy and K. S. Reddy, Food Chem. Toxicol., 2011, 49, 893–897 CrossRef CAS PubMed.
- Z. Ghlissi, R. Atheymen, M. A. Boujbiha, Z. Sahnoun, F. M. Ayedi, K. Zeghal, A. El Feki and A. Hakim, Int. J. Food Sci. Nutr., 2013, 64, 974–978 CrossRef CAS PubMed.
- M. Akimoto, M. Iizuka, R. Kanematsu, M. Yoshida and K. Takenaga, PLoS One, 2015, 10, e0126605 CrossRef PubMed.
- L. T. Bidinotto, A. L. T. Spinardi-Barbisan, N. S. Rocha, D. M. Favero Salvadori and L. F. Barbisan, Environ. Mol. Mutagen., 2006, 47, 624–630 CrossRef CAS PubMed.
- S. Nammi, S. Sreemantula and B. D. Roufogalis, Basic Clin. Pharmacol. Toxicol., 2009, 104, 366–373 CrossRef CAS PubMed.
- Y. Li, V. H. Tran, B. P. Kota, S. Nammi, C. C. Duke and B. D. Roufogalis, Basic Clin. Pharmacol. Toxicol., 2014, 115, 209–215 CrossRef CAS PubMed.
- X.-H. Li, K. C. Y. McGrath, S. Nammi, A. K. Heather and B. D. Roufogalis, Basic Clin. Pharmacol. Toxicol., 2012, 110, 238–244 CrossRef CAS.
- A. J. Akinyemi, G. R. Thome, V. M. Morsch, N. B. Bottari, J. Baldissarelli, L. S. de Oliveira, J. F. Goularte, A. Bello-Klein, T. Duarte, M. Duarte, A. A. Boligon, M. L. Athayde, A. A. Akindahunsi, G. Oboh and M. R. Chitolina Schetinger, Planta Med., 2016, 82, 612–620 CrossRef CAS PubMed.
- I. R. Kubra and L. J. M. Rao, Crit. Rev. Food Sci. Nutr., 2012, 52, 651–688 CrossRef CAS PubMed.
- M. S. Baliga, R. Haniadka, M. M. Pereira, J. J. D'Souza, P. L. Pallaty, H. P. Bhat and S. Popuri, Crit. Rev. Food Sci. Nutr., 2011, 51, 499–523 CrossRef CAS PubMed.
- C.-Z. Wang, L.-W. Qi and C.-S. Yuan, Am. J. Chin. Med., 2015, 43, 1351–1363 CrossRef CAS PubMed.
- S. Sang, J. Hong, H. Wu, J. Liu, C. S. Yang, M.-H. Pan, V. Badmaev and C.-T. Ho, J. Agric. Food Chem., 2009, 57, 10645–10650 CrossRef CAS PubMed.
- P. Wang, R. Wang, Y. Zhu and S. Sang, J. Agric. Food Chem., 2017, 65, 9618–9625 CrossRef CAS PubMed.
- F.-F. Gan, A. A. Nagle, X. Ang, O. H. Ho, S.-H. Tan, H. Yang, W.-K. Chui and E.-H. Chew, Apoptosis, 2011, 16, 856–867 CrossRef CAS PubMed.
- H. Ling, H. Yang, S. H. Tan, W. K. Chui and E. H. Chew, Br. J. Pharmacol., 2010, 161, 1763–1777 CrossRef CAS PubMed.
- S. M. Zick, Z. Djuric, M. T. Ruffin, A. J. Litzinger, D. P. Normolle, S. Alrawi, M. R. Feng and D. E. Brenner, Cancer Epidemiol., Biomarkers Prev., 2008, 17, 1930–1936 CrossRef CAS PubMed.
- Y. Yu, S. Zick, X. Li, P. Zou, B. Wright and D. Sun, AAPS J., 2011, 13, 417–426 CrossRef CAS PubMed.
- J. Iwabu, J. Watanabe, K. Hirakura, Y. Ozaki and K. Hanazaki, Drug Metab. Dispos., 2010, 38, 2040–2048 CrossRef CAS PubMed.
- A. Asami, T. Shimada, Y. Mizuhara, T. Asano, S. Takeda, T. Aburada, K.-i. Miyamoto and M. Aburada, J. Nat. Med., 2010, 64, 281–287 CrossRef CAS PubMed.
- H. Chen, L. Lv, D. Soroka, R. F. Warin, T. A. Parks, Y. Hu, Y. Zhu, X. Chen and S. Sang, Drug Metab. Dispos., 2012, 40, 742–753 CrossRef CAS PubMed.
- H. Chen, D. N. Soroka, Y. Hu, X. Chen and S. Sang, Mol. Nutr. Food Res., 2013, 57, 447–458 CrossRef CAS PubMed.
- H. Chen and S. Sang, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2012, 907, 126–139 CrossRef CAS PubMed.
- L. He, Z. Qin, M. Li, Z. Chen, C. Zeng, Z. Yao, Y. Yu, Y. Dai and X. Yao, J. Agric. Food Chem., 2018, 66, 9010–9033 CrossRef CAS PubMed.
- W. E. Evans and M. V. Relling, Sciences, 1999, 286, 487–491 CrossRef CAS.
- X. Hong, Y. Zheng, Z. Qin, B. Wu, Y. Dai, H. Gao, Z. Yao, F. J. Gonzalez and X. Yao, Int. J. Mol. Sci., 2017, 18, 1983 CrossRef PubMed.
- P. Wang, Y. Zhao, Y. Zhu and S. Sang, Mol. Nutr. Food Res., 2017, 61 Search PubMed.
- H. Sun, H. Wang, H. Liu, X. Zhang and B. Wu, Expert Opin. Drug Metab. Toxicol., 2014, 10, 1325–1336 CrossRef CAS PubMed.
- Z. Yao, S. Li, Z. Qin, X. Hong, Y. Dai, B. Wu, W. Ye, F. J. Gonzalez and X. Yao, RSC Adv., 2017, 7, 52661–52671 RSC.
- L. Wang, X. Hong, Z. Yao, Y. Dai, G. Zhao, Z. Qin, B. Wu, F. J. Gonzalez and X. Yao, Xenobiotica, 2018, 48, 357–367 CrossRef CAS PubMed.
- J. M. Hutzler and T. S. Tracy, Drug Metab. Dispos., 2002, 30, 355–362 CrossRef CAS.
- D. Hallifax and J. B. Houston, Drug Metab. Dispos., 2006, 34, 724–726 CrossRef CAS PubMed.
- J. Zhou, T. S. Tracy and R. P. Remmel, Drug Metab. Dispos., 2010, 38, 431–440 CrossRef CAS PubMed.
- Z. Wu, H. Liu and B. Wu, J. Pharm. Pharmacol., 2015, 67, 583–596 CrossRef CAS PubMed.
- B. Wu, D. Dong, M. Hu and S. Zhang, Curr. Top. Med. Chem., 2013, 13, 1343–1352 CrossRef CAS PubMed.
- H. Shiratani, M. Katoh, M. Nakajima and T. Yokoi, Drug Metab. Dispos., 2008, 36, 1745–1752 CrossRef CAS PubMed.
- H. Hao, G. Wang, N. Cui, J. Li, L. Xie and Z. Ding, Curr. Drug Metab., 2007, 8, 137–149 CrossRef CAS PubMed.
- G. J. Kilpatrick and T. W. Smith, Med. Res. Rev., 2005, 25, 521–544 CrossRef CAS PubMed.
- Y. Ji and Y. Yu, Lat. Am. J. Pharm., 2016, 35, 1686–1691 CAS.
- M.-S. Hwang, S.-J. Lee, H.-E. Jeong, S. Lee, M.-A. Yoo and J.-G. Shin, Drug Metab. Pharmacokinet., 2010, 25, 398–402 CrossRef CAS PubMed.
- C. R. Bhasker, W. McKinnon, A. Stone, A. C. T. Lo, T. Kubota, T. Ishizaki and J. O. Miners, Pharmacogenetics, 2000, 10, 679–685 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra08466a |
| ‡ These two authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |