Open Access Article
Open Access ArticleProtective effects of SKLB023 on a mouse model of unilateral ureteral obstruction by the modulation of gut microbiota
Yanhuan Feng†
a,
Lingzhi Li†a,
Fan Guoa,
Yanping Lib,
Yan Liangc,
Lin Baid,
Liang Ma *a and
Ping Fu
*a and
Ping Fu *a
*a
aKidney Research Laboratory, Division of Nephrology, National Clinical Research Center for Geriatrics, West China Hospital of Sichuan University, Chengdu 610041, China. E-mail: liang_m@scu.edu.cn; fupinghx@scu.edu.cn
bLaboratory of Clinical Pharmacy and Adverse Drug Reaction, Department of Pharmacy, West China Hospital of Sichuan University, Chengdu 610041, China
cCore Facility of West China Hospital, West China Hospital of Sichuan University, Chengdu 610041, China
dKey Laboratory of Transplant Engineering and Immunology, Ministry of Health, Regenerative Medicine Research Center, West China Hospital of Sichuan University, Chengdu 610041, China
First published on 3rd December 2018
Abstract
Renal interstitial fibrosis is the common pathway underlying the progression of chronic kidney disease (CKD) to end-stage renal disease (ESRD) and the corresponding therapies are limited. Quantitative and qualitative alterations in gut microbiota are noted in patients with CKD and ESRD. In our previous study, SKLB023 exhibited antifibrotic effects by interfering TGF-β1/Smad3 signaling in obstructive nephropathy. However, it remained unclear that oral administration of SKLB023 drives the alteration of gut microbiota to attenuate renal fibrosis. In the study, the marked inflammation and interstitial fibrosis were found in the kidney tissues of unilateral ureteral obstruction (UUO) mice. While treatment with SKLB023 significantly alleviated renal interstitial fibrosis and reduced serum proinflammatory cytokines TNF-α, IL-6 levels. Importantly, SKLB023 derived the modulation of gut microbiota with the increasing similarity between the composition of gut microbiota in the control and UUO. The number of Turicibacter and Candidatus_Arthromitus was significantly decreased following UUO surgery and recovered by SKLB023, which positively correlated with pro-inflammatory cytokine expression. These results indicated the potential relationship between the antifibrotic benefits of SKLB023 and gut microbiota alteration, which provided new insights into drug therapy via gut microbiota modulation in obstructive nephropathy.
Introduction
The gut microbiota play crucial roles in health and diseases.1 Every person's gut harbors a complex community of >100 trillion microbial cells, and >1000 different bacterial species constitute gut microbiota.2 Normal gut microbiota shares a mutually beneficial relationship with the host by contributing to its nutrition, metabolism, physiology and immune function.3,4 Changes in microbial abundance or composition has been implicated in the pathogenesis of diverse illnesses, such as obesity,5,6 diabetes,7–9 inflammatory bowel disease,10–12 cardiovascular disease13–16 and cancer,17,18 etc. In addition, quantitative and qualitative alterations of gut microbiota are also noted in patients with chronic kidney disease (CKD) and end-stage renal disease (ESRD).19–21 Besides gut microbiota themselves, gut microbiota-derived metabolites, both harmful and beneficial, have been shown to influence the development and progression of CKD, particularly, short-chain fatty acids and other uremic toxins, including indoles, ammonia, and trimethylamine N-oxide.22As is known, CKD is a global health problem associated with cardiovascular disease, infectious diseases, and premature death.23 Renal interstitial fibrosis is considered as the final common pathway from CKD to ESRD.24–28 Numerous studies focused on the development of potential drug for renal fibrosis treatment. In a phase I study of patients with treatment-resistant primary focal segmental glomerulosclerosis a single-dose of anti-TGF-β antibody was well-tolerated.29 However, the phase II study of the antibody in patients with diabetic kidney disease was terminated in 2014 due to a lack of efficacy. In total, no approved therapy for renal interstitial fibrosis appears at present.
In recent years, considerable studies have explored a new and exciting area: the interaction between gut microbiome and disease.22 Several studies reported the benefits of gut microbiota in liver fibrosis. The analysis of fecal microbiome in patients with liver fibrosis showed the significant differences with an increase in pathogenic bacterial group, and a decrease of beneficial bacterial group compared with health individuals,30–32 suggesting the gut microbiota played potential role in fibrosis. Although there is no evidence proving the direct relationship between gut microbiota and renal fibrosis, circumstantial evidence supports that they are closely connected. Recently, researchers examined the gut microbiome in progressor vs. nonprogressor immunoglobulin A nephropathy (IgAN) patients and compared them with healthy controls,33 indicated that the gut microbiota significantly varied between the three groups.
(Z)-N-(3-Chlorophenyl)-2-(4-((2,4-dioxothiazolidin-5-ylidene)methyl)phenoxy) acetamide (SKLB023, Fig. 1A) is a synthetic anti-inflammatory compound on the basis of thiazolidine-2,4-dione moiety. SKLB023 exhibited the potent inhibitory effects (IC50 = 8.66 μM) without cytotoxicity on RAW 264.7 microphages shown by MTT assay (without or with LPS, IC50 > 100 μM). The rat acute toxicity and Beagle long-term toxicity test by oral gavage exhibited no marked toxicity.34 In our previous studies, SKLB023 blocked joint inflammation and cartilage destruction in arthritis by the inhibition of pro-inflammatory cytokines expression.35 In the kidney of UUO mice, SKLB023 also could hinder renal interstitial fibrosis by interfering with TGF-β1/Smad3 signaling pathway.36 However, the protective effects of SKLB023 on UUO mice model by modulating gut microbiota remains unclear.
In the study, our results indicated the potential relationship between antifibrotic benefits of SKLB023 and gut microbiota alteration in obstructive nephropathy. Our data showed that SKLB023 exerted an anti-inflammatory property in the regulation of gut microbiota in renal interstitium and the suppressed expression of inflammatory cytokines after UUO, suggesting that the inhibition of inflammatory response through gut microbiota may be one mechanism by which SKLB023 relieved renal interstitial fibrosis.
Experimental section
Animal model and treatment
The UUO model was established in male C57BL/6J mice (8–10 weeks old; 20–25 g) from the Animal Laboratory Center of Sichuan University (Chengdu, China). All animal protocols were approved by the Institute of Animal Care and Use Committee. Briefly, the abdominal cavity was exposed via a midline incision and the left ureter was isolated and ligated. To examine the efficacy of SKLB023 in renal fibrosis after UUO injury, the mice were randomized into three groups (n = 7 in each group): (1) sham operated mice, (2) UUO mice that received daily saline for 7 d by oral gavage (o.g.) after UUO, (3) UUO mice treated with SKLB023 50 mg kg−1 d−1 for 7 d by o.g. after UUO. The mice were sacrificed and the kidneys were removed at day 7 for morphologic analyses. All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Sichuan University and approved by the Animal Ethics Committee of West China Hospital, Sichuan University.Morphologic analyses
Two-micrometer sections of paraffin-embedded kidney tissue were subjected to Masson's or HE staining using commercial kits (Sigma-Aldrich, USA) according to the manufacturer's protocol.37 Heat-induced epitope retrieval was performed on dewaxed slides in citrate buffer (pH 6.0) at 95 °C for 40 min. Then sections were exposed to peroxidase blocking solution (3% H2O2) prior to the addition of primary antibody, anti-α-SMA antibody (Abcam, USA) diluted to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 in PBS. After incubation with primary antibody overnight at 4 °C, the slides were washed three times with PBS, and incubated with horseradish peroxidase (HRP)-incubated secondary antibody (Abcam, USA) for 45 min. The sections were washed again with PBS for three times. Subsequently, the slides were developed by diaminobenzidine (DAB) and counterstained with hematoxylin. Finally, the slides were observed under a light microscope.
500 in PBS. After incubation with primary antibody overnight at 4 °C, the slides were washed three times with PBS, and incubated with horseradish peroxidase (HRP)-incubated secondary antibody (Abcam, USA) for 45 min. The sections were washed again with PBS for three times. Subsequently, the slides were developed by diaminobenzidine (DAB) and counterstained with hematoxylin. Finally, the slides were observed under a light microscope.
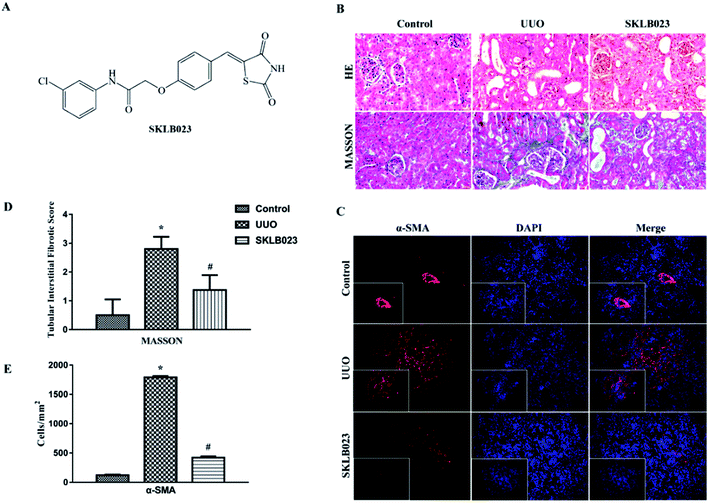
Masson staining as well as immunostaining intensity were scored, and the scoring criteria were as follows: 10 high-power fields (×200) were randomly selected and photographed in each group. None, mild, moderate and severe involvement were scored as 0, 1, 2, or 3 according to the degree and extent of tubular degeneration and necrosis, tubular atrophy, inflammatory cell infiltration and fibrosis.38 The blue area of collagen by Masson staining, which represents the extent of the lesion, was calculated. The dyed area was measured by the average optical density in the immunostaining intensity scores.
Immunofluorescence staining and image analysis
Immunofluorescence staining was performed as previously described.39 Briefly, the tissue sections were fixed, permeabilized with 0.2% Triton X-100, and incubated with the primary antibodies overnight at 4 °C, followed by incubation with secondary antibodies conjugated with Cy3 (Jackson Inc., USA). Cells were counterstained with DAPI to visualize the nuclei. Images were taken by confocal microscopy (Olympus Corporation, Tokyo, Japan). Positive signals were examined using a fluorescent microscope (Axioplan2 imaging; Carl Zeiss, Oberkoche, Germany) and single-positive cells were counted in 10 high-power fields (×40) per section and expressed as cells per square millimeter.DNA extraction and PCR amplification
Microbial DNA was extracted from ileum content samples using the E.Z.N.A.® DNA Kit (Omega Bio-tek, Norcross, GA, U.S.) according to standard protocol. The V3–V4 hypervariable regions of the bacteria 16S rRNA gene were amplified with primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) by thermocycler PCR system. PCR reactions were performed in triplicate 20 μL mixture containing 10 ng template DNA, 2 μL dNTPs (2.5 mmol L−1), 0.8 μL forward primer (5 μmol L−1), 0.8 μL reverse primer (5 μmol L−1), 4 μL 5 × FastPfu Buffer, 0.4 μL FastPfu Polymerase, and ddH2O. Amplicons were extracted from 2% agarose gels, purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA), and quantified using QuantiFluor™-ST (Promega, USA) according to the standard protocols.Processing of sequencing data
Purified amplicons were pooled and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA) according to the standard protocols. All raw reads were screened according to barcode and primer sequences, using Quantitative Insights Into Microbial Ecology (QIIME, version 1.17), with the following criteria: (1) the reads were truncated at any site receiving an average quality score <20 over a 50 bp sliding window; (2) primers were exactly matched allowing 2 nucleotides mismatching, and reads containing ambiguous bases were removed; (3) sequences whose overlap longer than 10 bp were merged according to their overlap sequence. Operational taxonomic units (OTUs) were clustered with the cut-off of 97% similarity using UPARSE (version 7.1) and UCHIME was utilized to identify and remove chimeric sequences. Alpha and beta diversity analyses (Unweighted UniFrac principal coordinates analysis) were also performed.Measurement of plasma cytokines
Blood samples were centrifuged at 3000 rpm for 15 min and stored at −80 °C until use. Plasma levels of TNF-α and IL-6 were detected by solid-phase sandwich enzyme-linked immunosorbent assay (ELISA) kits (Elabscience Biotechnology Co., Ltd., China) specific for these factors and absorbance was measured at 450 nm using a plate reader (BioTek ELx800, USA).Statistical analysis
Data are expressed as the mean ± SD. Comparisons between groups were made using one-way analysis of variance (ANOVA). In terms of microbiota analysis, the alpha diversity of the microbiota between two groups were performed by the two-tailed Student's t test. The differences of bacterial composition among the multiple group was analyzed by ANOVA. Wilcoxon rank sum test was utilized to evaluate the differences in bacterial relative proportion comparing each group with model. The correlations between relative abundance of certain genus/class/phylum and hyperlipidemia indexes (including physical and biochemical indexes) were performed by Spearman's correlation analyses. P < 0.05 was considered to be statistically significant.Results
SKLB023 ameliorates renal interstitial fibrosis in UUO mice
To confirm antifibrotic effects of SKLB023 on renal fibrosis, the mice were subjected to UUO surgery and treated by oral administration of SKLB023 at a dose of 50 mg kg−1 d−1 for 7 days. As shown in Fig. 1B, UUO mice exhibited marked renal interstitial fibrosis stained by HE and Masson's trichrome. Treatment with SKLB023 significantly reduced interstitial fibrosis in kidneys (Fig. 1D). SKLB023 also inhibited the expression of fibrotic biomarker α-SMA in UUO mice (Fig. 1C and E), suggesting that SKLB023 could ameliorate renal interstitial fibrosis.SKLB023 drives the structural modulation of gut microbiota in UUO mice
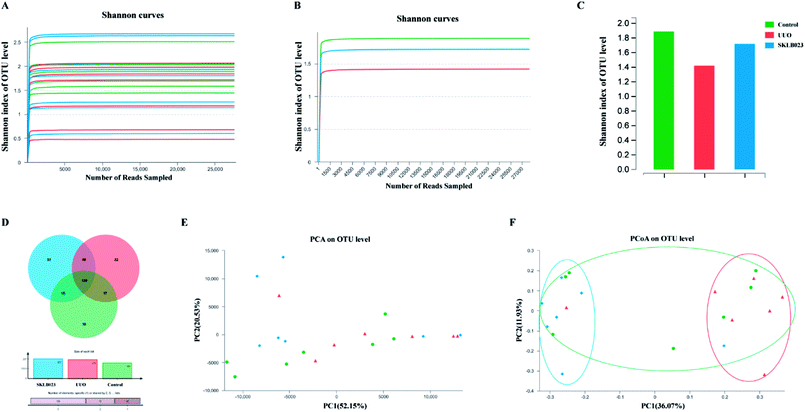
A bar-coded pyrosequencing run was conducted to analyze structural changes of gut microbiota in the three studied groups. In total, 792![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 252 usable reads and 276 OTUs were obtained from the 21 samples. Rarefaction and Shannon diversity curves were exhibited in Fig. 2A–C and Table 1. The rarefaction curves plateau with the current sequencing, indicating that most of the diversity has been captured in all samples. The overlap of OTUs revealed that 139 OTUs coexisted in all three groups. A further 17 OTUs were present in both control and UUO mice, and 40 in the UUO and SKLB023-treated groups, as well as 15 in control and SKLB023 groups. 22 OTUs were only found in UUO and 33 OTUs only found in SKLB023-treated group (Fig. 2D). PCA and PCoA analyses revealed that gut microbiota in the UUO mice deviated from baseline structure, and SKLB023-treated group was approximate to the control level (Fig. 2E and F). As for the PCoA analyses, the changes in the distribution of microbiota were also clear. The balance of gut microbiota was relatively concentrated on the opposite side compared to UUO/control and SKLB023-treated/control mice, respectively (Fig. 2F).
252 usable reads and 276 OTUs were obtained from the 21 samples. Rarefaction and Shannon diversity curves were exhibited in Fig. 2A–C and Table 1. The rarefaction curves plateau with the current sequencing, indicating that most of the diversity has been captured in all samples. The overlap of OTUs revealed that 139 OTUs coexisted in all three groups. A further 17 OTUs were present in both control and UUO mice, and 40 in the UUO and SKLB023-treated groups, as well as 15 in control and SKLB023 groups. 22 OTUs were only found in UUO and 33 OTUs only found in SKLB023-treated group (Fig. 2D). PCA and PCoA analyses revealed that gut microbiota in the UUO mice deviated from baseline structure, and SKLB023-treated group was approximate to the control level (Fig. 2E and F). As for the PCoA analyses, the changes in the distribution of microbiota were also clear. The balance of gut microbiota was relatively concentrated on the opposite side compared to UUO/control and SKLB023-treated/control mice, respectively (Fig. 2F).
| OTU | Ace | Chao | Simpson | Shannon | Coverage | |
|---|---|---|---|---|---|---|
| Control | 181 | 102 (100, 123) | 104 (94, 123) | 0.3222 (0.2339, 0.3785) | 1.89(1.63,2.05) | 0.999 361 |
| UUO | 218 | 157 (109, 161) | 140 (109, 149) | 0.3218 (0.2811, 0.6434) | 1.72(0.93,1.90) | 0.999 164 |
| SKLB023 | 227 | 123 (110, 139) | 105 (98, 127) | 0.2979 (0.1914, 0.5022) | 1.80(1.20,2.28) | 0.999 195 |
SKLB023 regulates structural segregation of gut microbiota in UUO mice
At the genus level, the heatmap and circus were created on the basis of the most modified species (Fig. 3). There was a remarkable decrease in Turicibacter, norank_f_Bacteroidales_S24-7_group, Streptococcus, Coriobacteriaceae_UCG-002, Jeotgalicoccus, Haemophilus, Faecalibaculum, unclassified_c_Bacilli, unclassified_p_Firmicutes and increase in Corynebacterium_1, Candidatus_Arthromitus, Rhodocuccus, Lachnospiraceae_NK4A136_group, Lachnospiraceae_UCG-006, Lachnoclostridium, unclassified_f_Lachnospriaceae, norank_f_Lachnospriaceae, Ralstonia, Propionibacterium, Dolosigranulum, unclassified_k_norank in the UUO mice compared with control, and these have all been restored by SKLB023 treatment.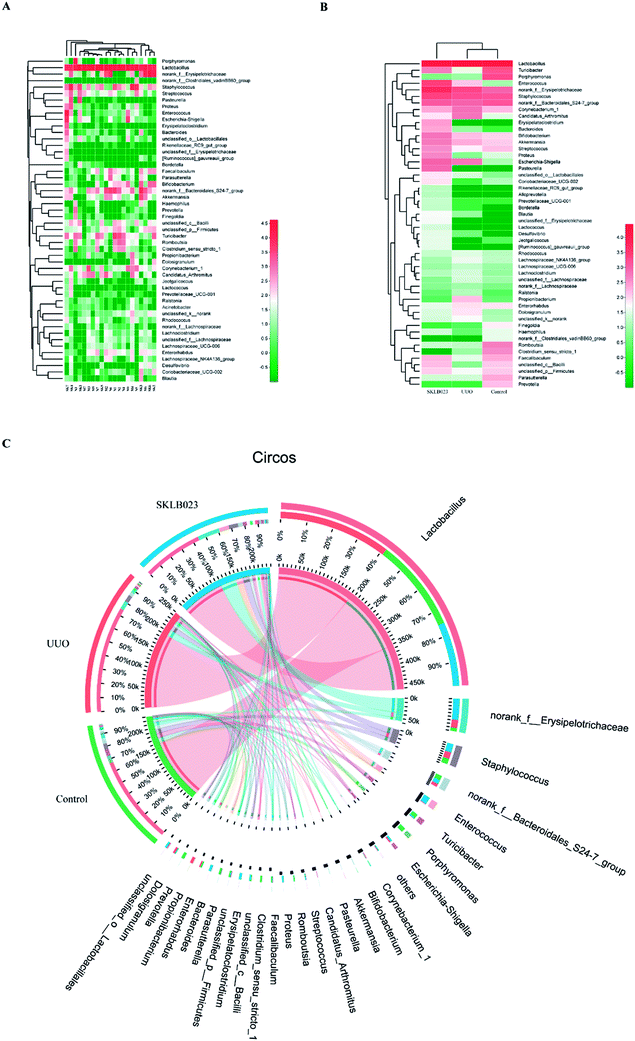 | ||
| Fig. 3 The heatmap of each sample (A), each group (B) and circus (C) on the basis of the most modified species at genus level. | ||
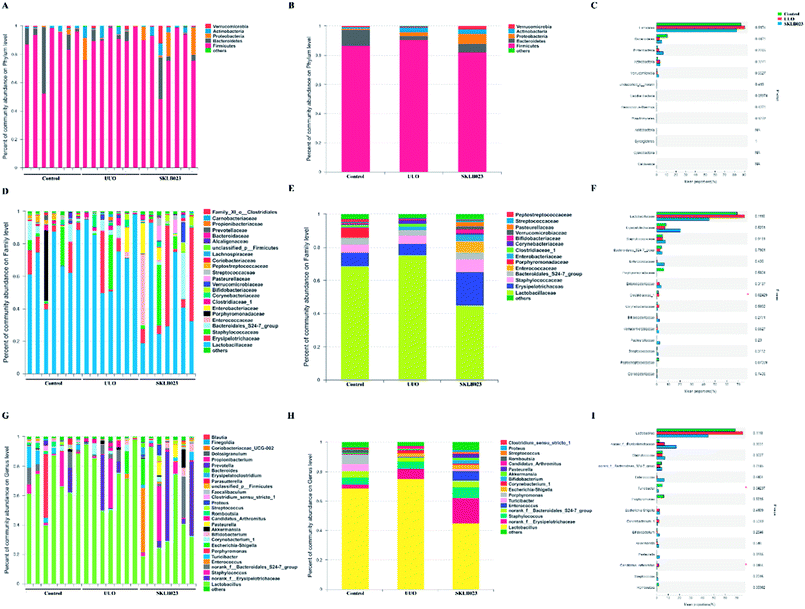
In addition, histograms illustrating the gut microbiota community structure reveal the microbial species and their relative abundance. As exhibited in Fig. 4A–C, the most abundant phyla were Firmicutes in all samples. The relative abundance of microbiota was not significantly different among the three groups at the phylum level. However, sequencing data identified Cyanobacteria of microbial flora were identified in the UUO but were not detected in the control and SKLB023-treated group. Lactobacillaceae accounted for the majority of the families of microflora (Fig. 4D–F). The 111 genera were identified in all samples. Turicibacter and unclassified_c_Bacilli significantly lowered and Candidatus_Arthromitus was increased in the UUO group compared with that of control and reversed by SKLB023 treatment (Fig. 4G–I).
SKLB023 inhibits the growth of certain bacteria in UUO mice
In the study, gut microbiota in the groups was analyzed by LEfSe (LDA > 2, Fig. 5). A histogram of LDA scores was plotted to identify statistically significant biomarkers and to reveal the dominant microorganisms. The results showed that the dominant communities in control were Turicibacter, Staphylococcus, Romboutsia, Clostridium_sensu_stricto_1, Brachybacterium genera, while Streptococcus, Blautia, Desulfovibrio and Jeotgalicoccus were the main genera in SKLB023-treated UUO mice. Among them, Clostridiaceae_1 family, Lachnospiraceae_UCG_001 genus, Lachnospiraceae_bacterium_A2 species, Lachnospiraceae_bacterium_COE1 species and Bacteroides fragilis species exerted crucial influences in the UUO group.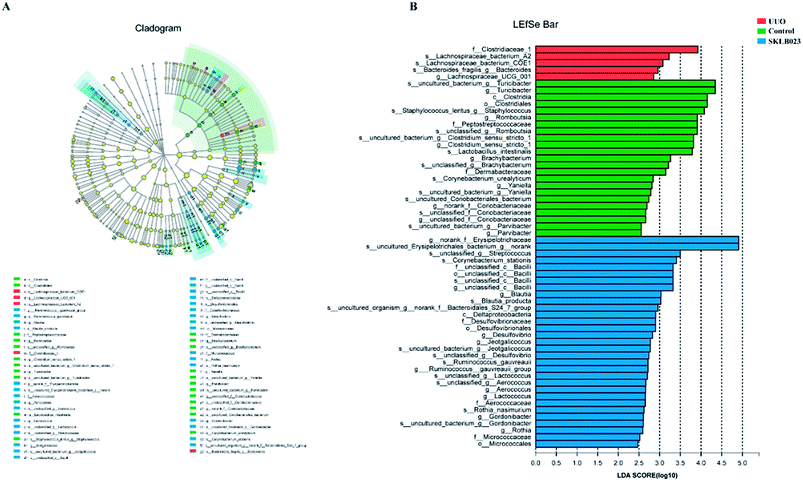 | ||
| Fig. 5 Difference in dominant microorganisms between groups. (A) Cladogram. (B) Distribution histogram based on LDA. | ||
An evolutionary clustering analysis diagram was generated based on LDA score to identify key microflora using taxonomy. As exhibited in Fig. 5, the branches of Clostridia were the major microbiota in the control group. In the SKLB023-treated UUO mice, the predominant intestinal flora, including Deltaproteobacteria, Erysipelotrichaceae, Bacilli and Microbcocales played important roles in the active period of renal fibrosis.
The alteration of gut microbiota in UUO mice is associated with inflammatory events
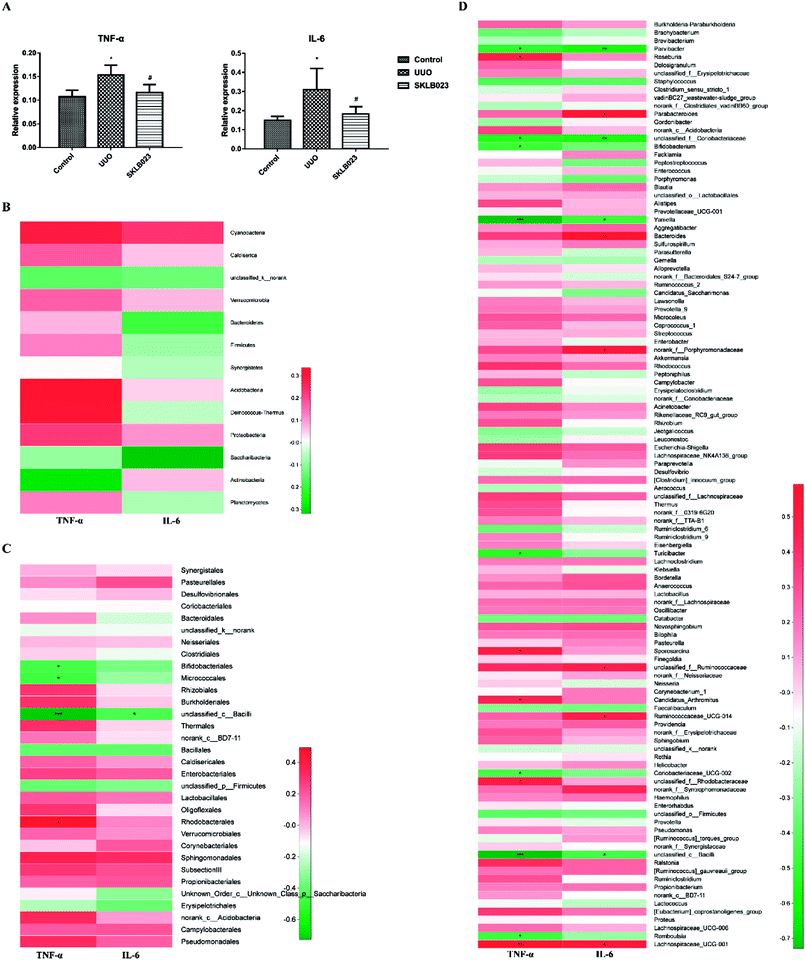
Pro-inflammatory cytokines tumor necrosis factor alpha (TNF-α) and interleukin 6 (IL-6) play a pivotal role in the process of renal fibrosis. Compared with control, the levels of TNF-α and IL-6 were significantly increased in the UUO. Oral administration of SKLB023 suppressed the accumulation of TNF-α and IL-6 in the UUO mice (Fig. 6A). Although serum cytokines were not significantly correlated with the concentration of gut microbiota in phylum level (Fig. 6B), when down to the order level, Bifidobacteriales, Micrococcales and unclassified_c_Bacilli exhibited robust negative correlations (Fig. 6C). As far as genus level was concerned, Turicibacter (r = −0.533, p < 0.05 of TNF-α), Candidatus_Arthromitus (r = 0.455, p < 0.05 of TNF-α) and unclassified_c_Bacilli (r = −0.727, p < 0.001 of TNF-α and r = −0.455, p < 0.05 of IL-6), as was observed in Fig. 6D. These findings suggested that SKLB023 prevented renal fibrosis by anti-inflammatory action through regulating gut microbiota in obstructive nephropathy.Discussion
SKLB023 is a synthetic, potent, and orally available drug candidate based on thiazolidine-2,4-dione moiety.34 In the study, we investigated the therapeutic role of SKLB023 by the regulation of gut microbiota in a mice model of renal interstitial fibrosis by UUO surgery. The results indicated that SKLB023 significantly attenuated renal fibrosis and drive the alteration of gut microbiota in obstructive nephropathy.The underlying mechanisms of renal interstitial fibrosis are not completely understood and antifibrotic therapies are lacking. The host–microbiota interaction has been the focus of increasing interest in recent years. It has become clear that host–microbe interactions are essential to many aspects of “Bmammalian” physiology. Previously, serum samples of IgAN patients showed altered levels of some metabolites.40,41 And recently, a marked damage of the colonic epithelial barrier structure and an alteration of the colonic microbiota and intestinal microbiota were found in humans and animals with CKD and IgAN.33,42 Targeting the large intestine and understanding the composition of gut microbial communities might be a promising approach to tackle the high morbidity and mortality of CKD. Therefore, an intimate connection between the gut and kidney has been proposed, which is called the gut–kidney axis.43,44 However, supportive data in patients or experimental animals were limited.
Microbiological composition was determined by high-throughput sequencing. As shown in Table 1, the results of Simpson and Shannon indices indicated that the microbial diversity in control and SKLB023-treated groups was greater than that of UUO mice. It is noteworthy that UUO and SKLB023 changed the concentration of gut microbiota toward the opposite sides (Fig. 2F). The gut microbiota community in all samples was evaluated on the basis of the following criteria: phylum, class, order, family, and genus. In the UUO group, Firmicutes was the dominant phyla in the gut, while Bacteroidetes occupied a second tier. Although there was no difference of microbiological composition in phylum level, our study revealed in the first time that there were significantly different between the groups in Clostridiaceae_1 and unclassified_c_Bacilli in family level and Turicibacter, Candidatus_Arthromitus, unclassified_c_Bacilli in genus level.
One bacterial taxon in the study was found to be significantly decreased following UUO surgery and treatment of SKLB023 could recover the level of Turicibacter. Turicibacter is a bacterium that considered to be beneficial to the host and associated with inflammatory diseases.45–49 Studies showed that Turicibacter markedly decreased in mice access to HFD and reported that the prevalence of Turicibacter infection was lower in patients with inflammatory bowel disease,45–48 that was consistent with our findings in UUO. In contrast, the UUO group increased the abundance of Candidatus_Arthromitus which belonged to Clostridiaceae_1 family in the ileum and SKLB023-treated group decreased it. Candidatus_Arthromitus is a commensal bacterium with an important role in the post-natal development of the gut immune functions by increasing the number of lymphoid cells in the lamina propria of the ileal and cecal mucosa.50,51 It also plays an key role in modulating host immune function.52,53 It induces IgA secreting B-cells and steers the development of the T-cell repertoire.54,55 In the previous studies, the higher level of Candidatus_Arthromitus was observed in central nervous system and played immune functions.56,57 However, these genera have not been reported in the relationship with renal interstitial fibrosis whether in humans or animal models. Therefore, further studies are warranted to confirm the mechanisms that link gut microbiome and metabolic alterations in the development of renal interstitial fibrosis.
The gastrointestinal tract has emerged as a major instigator of systemic inflammation associated with kidney injury.58 The single layer of intestinal epithelial cells not only constitute a physical barrier against the entrance of microorganisms, but also serve as active component of the host defense to prevent the systemic translocation of pathogenic microorganisms or their molecules into the portal circulation by sensing and responding to them.59,60 Commensal bacteria are important in maintaining the intestinal epithelial barrier by enhancing the tight junction between epithelial cells and suppressing intestinal inflammation, and the intestinal immune system becomes largely tolerant to resident commensals.61,62 Exposure to the gut-derived endotoxin will initiate a systemic inflammatory response and oxidative stress, which will further induce and accelerate the kidney injury.63,64
On the other hand, inflammation plays crucial roles in renal interstitial fibrosis. Renal inflammation is induced as a protective response to injuries to eliminate the cause and promote repair, but ongoing inflammation promotes progressive renal fibrosis.65 The discovery of “gut-derived inflammation” strongly suggested that interventions aimed at establishing gut symbiosis and blocking microbiome-related pathogenic biochemical pathways should be explored to ameliorate kidney inflammation and uremic syndrome. In the present study, SKLB023 significantly inhibit renal interstitial fibrosis in morphology and α-SMA expression in tissue section of immunofluorescence staining. And SKLB023 could block joint inflammation and cartilage destruction in arthritis by the inhibition of pro-inflammatory cytokines TNF-α and IL-6 expression.34,35 Considered studies revealed that gut microbiota was involved in progressive disease, and we also speculated the SKLB023 exhibited antifibrotic effects by inhibiting inflammation to recover the balance of gut microbiota. In agreement with that, pro-inflammatory cytokines TNF-α and IL-6 were significantly increased in serum of UUO mice and decreased after SKLB023 treatment, with positively correlations with the expression of numbers of Turicibacter and Candidatus_Arthromitus. Besides, several gut microbiotas were in close with cytokine factors (Fig. 6D), which consistent with the previous studies, such as unclassified_Coriobacteriaceae66,67 Coriobacteriaceae_UCG-002 (ref. 67) and Lachnospiraceae_UCG-001.67,68
Conclusion
Consistent with these findings, our data showed that SKLB023 exerted an anti-inflammatory property in the regulation of gut microbiota in renal interstitium and the suppressed expression of inflammatory cytokines, such as TNF-α and IL-6 after UUO surgery. These results suggested that the inhibition of inflammatory response through gut microbiota may be one mechanism by which SKLB023 relieved renal interstitial fibrosis. To the best of our knowledge, the present study was the first to survey the dynamic shifts of colonic mucosal microbiomes in renal interstitial fibrosis and the therapeutic effects by SKLB023 in UUO. Our data revealed that UUO reduced the diversity and richness of colonic mucosa associated microbiota, with significantly-dynamic shifts in the abundance of Clostridiaceae_1 and unclassified_c_Bacilli in family level and Turicibacte, Candidatus_Arthromitus, unclassified_c_Bacilli in genus level. Oral administration of SKLB023 inhibited renal interstitial fibrosis and reversed the change of gut microbiota. These results indicated that the process of renal fibrosis and treatment of SKLB023 altered or recovered the microbial fermentation and the composition and potential function of the mucosa-associated bacterial communities in the colon. In summary, our data highlighted the potential relationship between antifibrotic benefits of SKLB023 and gut microbiota alteration, which provided new insights into drug therapy via gut microbiota modulation in obstructive nephropathy.Conflicts of interest
There are no conflicts to declare. All authors declare no competing interests.Acknowledgements
The work was supported by grants from the National Key Research and Development Program (2016YFC1305403) and National Postdoctoral Program for Innovative Talents of China (BX201600112).References
- L. V. Hooper and J. I. Gordon, Science, 2001, 292, 1115–1118 CrossRef CAS.
- R. E. Ley, D. A. Peterson and J. I. Gordon, Cell, 2006, 124, 837–848 CrossRef CAS PubMed.
- I. Solt, M. J. Kim and C. Offer, Harefuah, 2011, 150, 484–488 Search PubMed.
- K. J. Pflughoeft and J. Versalovic, Annu. Rev. Pathol.: Mech. Dis., 2012, 7, 99–122 CrossRef CAS PubMed.
- R. E. Ley, P. J. Turnbaugh, S. Klein and J. I. Gordon, Nature, 2006, 444, 1022–1023 CrossRef CAS PubMed.
- H. L. Bradlow, Horm. Mol. Biol. Clin. Invest., 2014, 17, 53–61 CAS.
- J. Qin, Y. Li, Z. Cai, S. Li, J. Zhu, F. Zhang, S. Liang, W. Zhang, Y. Guan, D. Shen, Y. Peng, D. Zhang, Z. Jie, W. Wu, Y. Qin, W. Xue, J. Li, L. Han, D. Lu, P. Wu, Y. Dai, X. Sun, Z. Li, A. Tang, S. Zhong, X. Li, W. Chen, R. Xu, M. Wang, Q. Feng, M. Gong, J. Yu, Y. Zhang, M. Zhang, T. Hansen, G. Sanchez, J. Raes, G. Falony, S. Okuda, M. Almeida, E. LeChatelier, P. Renault, N. Pons, J. M. Batto, Z. Zhang, H. Chen, R. Yang, W. Zheng, S. Li, H. Yang, J. Wang, S. D. Ehrlich, R. Nielsen, O. Pedersen, K. Kristiansen and J. Wang, Nature, 2012, 490, 55–60 CrossRef CAS PubMed.
- J. L. Dunne, E. W. Triplett, D. Gevers, R. Xavier, R. Insel, J. Danska and M. A. Atkinson, Clin. Exp. Immunol., 2014, 177, 30–37 CrossRef CAS PubMed.
- M. Gross, Curr. Biol., 2013, 23, R359–R362 CrossRef CAS PubMed.
- D. N. Frank, A. L. St Amand, R. A. Feldman, E. C. Boedeker, N. Harpaz and N. R. Pace, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 13780–13785 CrossRef CAS PubMed.
- G. Major and R. Spiller, Curr. Opin. Endocrinol., Diabetes Obes., 2014, 21, 15–21 CrossRef CAS PubMed.
- A. D. Kostic, R. J. Xavier and D. Gevers, Gastroenterology, 2014, 146, 1489–1499 CrossRef CAS.
- V. Lam, J. Su, S. Koprowski, A. Hsu, J. S. Tweddell, P. Rafiee, G. J. Gross, N. H. Salzman and J. E. Baker, FASEB J., 2012, 26, 1727–1735 CrossRef CAS PubMed.
- Z. Wang, E. Klipfell, B. J. Bennett, R. Koeth, B. S. Levison, B. Dugar, A. E. Feldstein, E. B. Britt, X. Fu, Y. M. Chung, Y. Wu, P. Schauer, J. D. Smith, H. Allayee, W. H. Tang, J. A. DiDonato, A. J. Lusis and S. L. Hazen, Nature, 2011, 472, 57–63 CrossRef CAS PubMed.
- V. Dinakaran, A. Rathinavel, M. Pushpanathan, R. Sivakumar, P. Gunasekaran and J. Rajendhran, PLoS One, 2014, 9, e105221 CrossRef PubMed.
- J. Rajendhran, M. Shankar, V. Dinakaran, A. Rathinavel and P. Gunasekaran, Int. J. Cardiol., 2013, 168, 5118–5120 CrossRef PubMed.
- C. J. Rogers, K. S. Prabhu and M. Vijay-Kumar, Cancer J., 2014, 20, 176–180 CrossRef CAS PubMed.
- R. Francescone, V. Hou and S. I. Grivennikov, Cancer J., 2014, 20, 181–189 CrossRef CAS PubMed.
- M. Hida, Y. Aiba, S. Sawamura, N. Suzuki, T. Satoh and Y. Koga, Nephron, 1996, 74, 349–355 CrossRef CAS PubMed.
- M. L. Simenhoff, S. R. Dunn, G. P. Zollner, M. E. Fitzpatrick, S. M. Emery, W. E. Sandine and J. W. Ayres, Miner. Electrolyte Metab., 1996, 22, 92–96 CAS.
- N. D. Vaziri, J. Wong, M. Pahl, Y. M. Piceno, J. Yuan, T. Z. DeSantis, Z. Ni, T. H. Nguyen and G. L. Andersen, Kidney Int., 2013, 83, 308–315 CrossRef PubMed.
- L. L. Li, L. Ma and P. Fu, Drug Des., Dev. Ther., 2017, 11, 3531–3542 CrossRef PubMed.
- A. S. Levey, K. U. Eckardt, Y. Tsukamoto, A. Levin, J. Coresh, J. Rossert, D. De Zeeuw, T. H. Hostetter, N. Lameire and G. Eknoyan, Kidney Int., 2005, 67, 2089–2100 CrossRef PubMed.
- A. Bohle, R. Bader, K. E. Grund, S. Mackensen and J. Neunhoeffer, Virchows Arch. A: Pathol. Anat. Histol., 1977, 375, 87–96 CrossRef CAS.
- M. Iwano, D. Plieth, T. M. Danoff, C. Xue, H. Okada and E. G. Neilson, J. Clin. Invest., 2002, 110, 341–350 CrossRef CAS.
- P. Boor, T. Ostendorf and J. Floege, Nat. Rev. Nephrol., 2010, 6, 643–656 CrossRef PubMed.
- M. Zeisberg and E. G. Neilson, J. Am. Soc. Nephrol., 2010, 21, 1819–1834 CrossRef CAS PubMed.
- T. D. Hewitson, Am. J. Physiol. Renal. Physiol., 2009, 296, F1239–F1244 CrossRef CAS PubMed.
- H. Trachtman, F. C. Fervenza, D. S. Gipson, P. Heering, D. R. Jayne, H. Peters, S. Rota, G. Remuzzi, L. C. Rump, L. K. Sellin, J. P. Heaton, J. B. Streisand, M. L. Hard, S. R. Ledbetter and F. Vincenti, Kidney Int., 2011, 79, 1236–1243 CrossRef CAS PubMed.
- Y. Chen, F. Yang, H. Lu, B. Wang, D. Lei, Y. Wang, D. Lei, Y. Wang, B. Zhu and L. Li, Hepatology, 2011, 54, 562–572 CrossRef.
- H. Lu, Z. Wu, W. Xu, J. Yang, Y. Chen and L. Li, Microb. Ecol., 2011, 61, 693–703 CrossRef.
- I. Gomez-Hurtado, A. Santacruz, G. Peiro, P. Zapater, A. Gutierrez, M. Perez-Mateo, Y. Sanz and R. Francés, PLoS One, 2011, 6, e23037 CrossRef CAS PubMed.
- M. De Angelis, E. Montemurno, M. Piccolo, L. Vannini, G. Lauriero, V. Maranzano, G. Gozzi, D. Serrazanetti, G. Dalfino, M. Gobbetti and L. Gesualdo, PLoS One, 2014, 9, e99006 CrossRef PubMed.
- L. Ma, C. Xie, Y. Ma, J. Liu, M. Xiang, X. Ye, H. Zheng, Z. Chen, Q. Xu, T. Chen, J. Chen, J. Yang, N. Qiu, G. Wang, X. Liang, A. Peng, S. Yang, Y. Wei and L. Chen, J. Med. Chem., 2011, 54, 2060–2068 CrossRef CAS PubMed.
- C. Xie, L. Ma, J. Liu, X. Li, H. Pei, M. Xiang and L. Chen, PLoS One, 2013, 8, e56349 CrossRef CAS.
- Y. H. Feng, J. Xu, F. Guo, R. S. Huang, M. Shi, L. Z. Li, L. Ma and P. Fu, RSC Adv., 2018, 8, 5891–5896 RSC.
- L. Wang and T. Lyerla, Histochem. Cell Biol., 2010, 134, 205–213 CrossRef CAS.
- H. Kikuchi, T. Katsuramaki, K. Kukita, S. Taketani, M. Meguro, M. Nagayama, M. Isobe, T. Mizuguchi and K. Hirata, Wound Repair Regen., 2007, 15, 881–888 CrossRef PubMed.
- M. Pang, J. Kothapally, H. Mao, E. Tolbert, M. Ponnusamy, Y. E. Chin and S. Zhuang, Am. J. Physiol. Renal. Physiol., 2009, 297, F996–F1005 CrossRef CAS PubMed.
- W. Sui, L. Li, W. Che, G. Zuo, J. Chen, W. Li and Y. Dai, Clinics, 2012, 67, 363–373 CrossRef.
- N. Ranganathan, E. A. Friedman, P. Tam, V. Rao, P. Ranganathan and R. Dheer, Curr. Med. Res. Opin., 2009, 25, 1919–1930 CrossRef CAS.
- I. W. Wu, K. H. Hsu, C. C. Lee, C. Y. Sun, H. J. Hsu, C. J. Tsai, C. Y. Tzen, Y. C. Wang, C. Y. Lin and M. S. Wu, Nephrol., Dial., Transplant., 2011, 26, 938–947 CrossRef CAS PubMed.
- E. J. Hoorn and R. Zietse, Gut-kidney kaliuretic signaling: looking forward to feeding, Kidney Int., 2015, 88, 1230–1232 CrossRef CAS PubMed.
- R. Inagi, The gut-kidney connection in advanced chronic kidney disease, Kidney Res. Clin. Pract., 2015, 34, 191–193 CrossRef PubMed.
- J. S. Suchodolski, M. E. Markel, J. F. Garcia-Mazcorro, S. Unterer, R. M. Heilmann, S. E. Dowd, P. Kachroo, I. Ivanov, Y. Minamoto, E. M. Dillman, J. M. Steiner, A. K. Cook and L. Toresson, PLoS One, 2012, 7, e51907 CrossRef CAS PubMed.
- G. Rossi, G. Pengo, M. Caldin, A. Palumbo Piccionello, J. M. Steiner, N. D. Cohen, A. E. Jergens and J. S. Suchodolski, PLoS One, 2014, 9, e94699 CrossRef PubMed.
- Y. Minamoto, C. C. Otoni, S. M. Steelman, O. Büyükleblebici, J. M. Steiner, A. E. Jergens and J. S. Suchodolski, Gut Microbes, 2015, 6, 33–47 CrossRef CAS PubMed.
- Y. Vazquez-Baeza, E. R. Hyde, J. S. Suchodolski and R. Knight, Nat. Microbiol., 2016, 1, 16177 CrossRef CAS PubMed.
- Y. L. Jones-Hall, A. Kozik and C. Nakatsu, PLoS One, 2015, 10, e0119441 CrossRef.
- H. L. Klaasen, P. J. Van der Heijden, W. Stok, F. G. Poelma, J. P. Koopman, M. E. Van den Brink, M. H. Bakker, W. M. Eling and A. C. Beynen, Infect. Immun., 1993, 61, 303–306 CAS.
- A. Bolotin, T. de Wouters, P. Schnupf, C. Bouchier, V. Loux, M. Rhimi, A. Jamet, R. Dervyn, S. Boudebbouze, H. M. Blottière, A. Sorokin, J. Snel, N. Cerf-Bensussan, V. Gaboriau-Routhiau, M. van de Guchte and E. Maguin, Genome Announc., 2014, 2, e00705-–14 Search PubMed.
- J. Snel, P. P. Heinen, H. J. Blok, R. J. Carman, A. J. Duncan, P. C. Allen and M. D. Collins, Int. J. Syst. Bacteriol., 1995, 45, 780–782 CrossRef CAS PubMed.
- C. L. Thompson, A. Mikaelyan and A. Brune, Mucosal Immunol., 2013, 6, 200–201 CrossRef CAS PubMed.
- C. L. Maynard, C. O. Elson, R. D. Hatton and C. T. Weaver, Nature, 2012, 489, 231–241 CrossRef CAS.
- A. C. Ericsson, C. E. Hagan, D. J. Davis and C. L. Franklin, Comp. Med., 2014, 64, 90–98 CAS.
- N. Yacoubi, L. Saulnier, E. Bonnin, E. Devillard, V. Eeckhaut, L. Rhayat, R. Ducatelle and F. Van Immerseel, Poult. Sci., 2018, 97, 412–424 CrossRef CAS PubMed.
- M. Yu, H. Jia, C. Zhou, Y. Yang, Y. Zhao, M. Yang and Z. Zou, J. Pharm. Biomed. Anal., 2017, 138, 231–239 CrossRef CAS PubMed.
- L. Vitetta, A. W. Linnane and G. C. Gobe, Toxins, 2013, 5, 2042–2057 CrossRef CAS PubMed.
- S. Robijn, B. Hoppe, B. A. Vervaet, P. C. D'Haese and A. Verhulst, Kidney Int., 2011, 80, 1146–1158 CrossRef CAS PubMed.
- M. E. Andrade, R. S. Araújo, P. A. de Barros, A. D. Soares, F. A. Abrantes, V. Generoso Sde, S. O. Fernandes and V. N. Cardoso, Clin. Nutr., 2015, 34, 1080–1087 CrossRef CAS PubMed.
- Y. J. Sun, H. J. Cao, D. D. Song, Y. G. Diao, J. Zhou and T. Z. Zhang, Dig. Dis. Sci., 2013, 58, 1528–1536 CrossRef PubMed.
- C. Barrios, M. Beaumont, T. Pallister, J. Villar, J. K. Goodrich, A. Clark, J. Pascual, R. E. Ley, T. D. Spector, J. T. Bell and C. Menni, PLoS One, 2015, 10, e0134311 CrossRef.
- N. D. Vaziri, Clin. J. Am. Soc. Nephrol., 2016, 11, 199–201 CrossRef CAS PubMed.
- A. R. Aroor, S. McKarns, V. G. Demarco, G. Jia and J. R. Sowers, Metabolism, 2013, 62, 1543–1552 CrossRef CAS.
- X. M. Meng, D. J. Nikolic-Paterson and H. Y. Lan, Nat. Rev. Nephrol., 2014, 10, 493–503 CrossRef CAS PubMed.
- J. Wang, Y. Wang, X. Zhang, J. Liu, Q. Zhang, Y. Zhang, J. Peng, Q. Feng, J. Dai, S. Sun, Y. Zhao, L. Zhao, Y. Zhang, Y. Hu and M. Zhang, Front. Microbiol., 2017, 8, 2222 CrossRef PubMed.
- L. Zhao, Q. Zhang, W. Ma, F. Tian, H. Shen and M. Zhou, Food Funct., 2017, 8, 4644–4656 RSC.
- R. Song, J. Yao, Q. Shi and R. Wei, Mar. Drugs, 2018, 16(pii), E23 CrossRef PubMed.
Footnote |
| † The authors contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2018 |