Open Access Article
Open Access ArticleEnhancing the thermostability of Rhizopus oryzae lipase by combined mutation of hot-spots and engineering a disulfide bond†
Jiong-feng Zhaoa,
Zhe Wangb,
Fei-long Gaoa,
Jian-ping Lina,
Li-rong Yanga and
Mian-bin Wu *a
*a
aKey Laboratory of Biomass Chemical Engineering of Ministry of Education, College of Chemical and Biological Engineering, Zhejiang University, Hangzhou 310027, China. E-mail: wumb@zju.edu.cn; Tel: +86-135-88828774 Tel: +86-0571-87952363
bGenclonn Biotech (Hangzhou) Co. Ltd, Hangzhou 310027, China
First published on 11th December 2018
Abstract
Rhizopus oryzae lipase (ROL) is important because of its extreme sn-1,3-regioselectivity, but it shows poor thermostability, which severely restricts its application. In this work, the thermostability of ROL was greatly enhanced by rational design. First, several sites that may affect the thermostability of ROL were identified by multiple-sequence alignment. The half-lives of mutants V209L and D262G at 55 °C were about 4.38- and 4.2-times those of the wild-type, respectively. Then, a disulfide bond was introduced between positions 190 and 238 based on the prediction of Disulfide by Design 2, which greatly improved the thermostability of the protein. The activity of variant E190C/E238C retained about 58.2% after incubation at 55 °C for 720 min, whereas the half-life of wild type ROL was only about 11.7 min. On the basis of the results obtained by the two methods, we carried out a combined mutation. Quadruple mutant V209L/D262G/E190C/E238C was constructed and the thermostability was improved even further. The half-lives at 55 °C and 65 °C were 102.5- and 20-times those of the wild-type ROL. This improvement in thermostability will give ROL wider industrial applicability, especially in the preparation of structured lipids.
Introduction
sn-1,3-Regioselective lipases are excellent tools for the production of triacylglycerols or diglycerides with different fatty acid compositions in the sn-1 and sn-3 positions compared with the sn-2 position.1 They are widely used in industry, especially in the modification of lipids such as 1,3-diglycerides and structurally designed triglycerides (e.g., 1,3-dioleoyl-2-palmitoylglycerol), which have unique nutritional and functional properties.2,3As reported, lipases derived from the genus Rhizopus show high sn-1,3-regioselectivity, for example, the lipases from R. delemar and R. oryzae.4 In previous work, we found that R. oryzae lipase (ROL) showed extreme sn-1,3-regioselectivity in 1,3-diglyceride synthesis. However, the thermostabilities of ROL and other Rhizopus lipases are poor, seriously limiting their industrial application.5–7 Because of the sn-1,3-regioselectivity of ROL and other lipases from Rhizopus, and their potential for unique applications, it is urgently necessary to improve their thermostabilities. However, research into improvement of the thermostabilities of Rhizopus lipases has been limited, and the results have not been satisfactory.6,8
Many methods have been developed for the enhancement of enzyme thermostability, including of lipases.9–12 Among them, point mutations based on multiple-sequence alignments and the introduction of disulfide bonds through in silico design are two common and effective approaches. Based on multiple-sequence alignment results, the thermostabilities of fumarase C from Corynebacterium glutamicum, Candida rugosa lipases and Candida albicans lipase Lip5 were successfully enhanced via molecular modification.12–14 Engineering disulfide bonds in proteins has been applied to improve the thermostabilities of C. antarctica lipase B and α-type carbonic anhydrase.15,16
Disulfide by design 2 (DbD2) is a computational tool used to predict potential disulfide bond sites in proteins, which has been successfully applied to modify the thermostabilities of many proteins.9,17 It uses a disulfide model with fixed bond lengths for Cβ–Sγ (1.81 Å) and Sγ–Sγ (2.04 Å) and a fixed Cβ–Sγ–Sγ bond angle (104.15°). The model is used to predict potential residue pairs that could form disulfide bonds after appropriate mutation of amino acids to cysteine. The predictions are made using the formation energies of disulfide bonds. A residue pair with lower energy has a higher possibility of forming a disulfide bond than that with higher energy.18
The aim of this study was to obtain thermostable mutants of ROL via rational design with a limited screening library. Considering the limited effect of mutation in one method, we combined two mutation methods. First, using sequence alignment, we identified hot-spots that could affect the thermostability of ROL. Then, using the crystal structure of the protein, we engineered a novel disulfide bond into ROL to improve its thermostability. After obtaining positive mutation results, we combined mutation of the hot-spots and introduction of the disulfide bond to further improve the thermostability of ROL. The thermostability improvement was elucidated at the molecular level and by molecular dynamics (MD) simulation.
Materials and methods
Materials
A codon optimized ROL gene (GenBank: GQ502721.1, optimized for Pichia pastoris) and PCR primers were synthesized by Sangon Biotech (Shanghai, China). Restriction enzymes (DpnI, EcoR I, Not I, and Sac I), PrimeSTAR Max DNA Polymerase, and T4 DNA ligase were purchased from Takara Biomedical Technology (Beijing, China). Escherichia coli BL21 (DE 3) competent cells were purchased from TransGen Biotech (Beijing, China). The P. pastoris strain GS115 and plasmid pPIC9K were maintained in our laboratory. Kits for plasmid extraction and for extraction of DNA from gels were purchased from Axygen® Brand Products (Hangzhou, China). Olive oil and other analytical reagents were purchased from Aladdin (Shanghai, China).Gene cloning and construction of the recombinant strain
The ROL gene was amplified by PCR using the synthesized gene as the template DNA. The forward and reverse primers were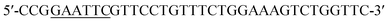 and
and 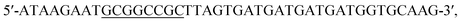 respectively. Cleavage sites for EcoR I and Not I were introduced (underlined). The amplified gene fragment was ligated into plasmid pPIC9K after double digestion, followed by transformation into E. coli BL21 (DE3). The recombinant plasmid was extracted from overnight culture of E. coli BL21 and linearized with Sac I, and then transformed into P. pastoris GS115. Solid medium MD (2% glucose, 0.34% yeast nitrogen base, 1% ammonium sulfate, and 2% agar) was used to screen for recombinant yeast. Point mutations were achieved by whole plasmid PCR.
respectively. Cleavage sites for EcoR I and Not I were introduced (underlined). The amplified gene fragment was ligated into plasmid pPIC9K after double digestion, followed by transformation into E. coli BL21 (DE3). The recombinant plasmid was extracted from overnight culture of E. coli BL21 and linearized with Sac I, and then transformed into P. pastoris GS115. Solid medium MD (2% glucose, 0.34% yeast nitrogen base, 1% ammonium sulfate, and 2% agar) was used to screen for recombinant yeast. Point mutations were achieved by whole plasmid PCR.
Expression and purification of enzyme
Single colonies were inoculated into 50 mL of BMMG medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate buffer, pH 6.0, 1% ammonium sulfate, 0.34% yeast nitrogen base, 4 × 5−10% biotin, and 1% glycerol) and cultured at 30 °C with shaking at 200 rpm until the OD600 reached 2–6. Cells were harvested by centrifugation and resuspended in 50 mL of BMMY medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate buffer, pH 6.0, 1% ammonium sulfate, 0.34% yeast nitrogen base, 4 × 5−10% biotin, and 1% methanol) to an initial OD600 of 1. During the induction period, 0.5 mL of methanol was added every 24 h. After incubation at 30 °C for 96 h, cells were removed and the supernatant was collected.Protein purification was performed as reported previously.19
Multiple-sequence alignment
Multiple-sequence alignment was performed using ClustalW software (http://www.genome.jp/tools-bin/clustalw). Thermomyces lanuginosus lipase and Rhizomucor miehei lipase were used to perform multiple-sequence alignments. The results were uploaded to ESPript 3.0 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) for plotting a more intuitive alignment. On the basis of the sequence alignment results, we used whole-plasmid PCR to make point mutations at potential hot-spots in ROL. The mutated plasmids were transformed into P. pastoris as described above.Engineering disulfide bonds rationally
Disulfide by Design 2 (http://cptweb.cpt.wayne.edu/DbD2/index.php) is an online version of the original DbD software for assessing disulfide bonds. The crystal structure of ROL (PDB ID: 1LGY) was uploaded to DbD2. After preliminary analysis of the protein structure, the software was used to predict residue pairs that were likely to form a disulfide bond if the respective amino acids were mutated to cysteine.20 On the basis of the calculation and scoring results, we performed site-directed mutagenesis of ROL by whole plasmid PCR to introduce cysteines.Homology modeling and MD simulation
Homology modeling of mutants was performed using SWISS-MODEL (http://swissmodel.expasy.org/interacive). All molecular structures were visualized by PyMOL software (http://www.pymol.org). All MD simulations were performed with the Discovery Studio 4.0 package (Accelrys, San Diego, CA, USA). We used an established MD method.15,16 The CHARMm force field and the Momany-Rone partial charge were used to specify the atom type. A solvent environment was added to make the simulation more realistic. MD simulations were performed according to standard dynamic cascade protocols. In the first and second steps, the maximum steps of the steepest descent minimization method and the conjugate gradient minimization method were set to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. The RMS gradients were set to 0.2 and 0.0001, respectively. The temperature of the heating step was set to 300 K for 100 ps. The equilibration and production steps were set to 1 ns and 10 ns, respectively. The tempering temperatures for the corresponding steps were 300 K and 330 K. The type of production was NVT. Using the implicit solvent model, the dielectric constant was set to 1 and the implicit solvent dielectric constant was set to 80. The trajectory of the production step was saved every 2 ps, and 5000 frames were analyzed for comparison.
000. The RMS gradients were set to 0.2 and 0.0001, respectively. The temperature of the heating step was set to 300 K for 100 ps. The equilibration and production steps were set to 1 ns and 10 ns, respectively. The tempering temperatures for the corresponding steps were 300 K and 330 K. The type of production was NVT. Using the implicit solvent model, the dielectric constant was set to 1 and the implicit solvent dielectric constant was set to 80. The trajectory of the production step was saved every 2 ps, and 5000 frames were analyzed for comparison.
Analysis of lipase activity
Olive oil titration was used to determine the hydrolysis activity of lipase.21 The olive oil was mixed with a polyvinyl alcohol solution (20 g L−1) in a volume ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 to form an olive oil emulsion, which was used as the substrate. An appropriate amount of the enzyme was added to a mixture of 10 mL of phosphate buffer (25 mM, pH 7.5) and 8 mL of the olive oil emulsion. After reacting at 30 °C for 10 min, 20 mL of ethanol was added to terminate the reaction. The free fatty acid released by the reaction was titrated with a sodium hydroxide solution. We defined enzyme activity as the amount of enzyme required to hydrolyze sufficient olive oil to release 1 μmol of fatty acid per minute.
4 to form an olive oil emulsion, which was used as the substrate. An appropriate amount of the enzyme was added to a mixture of 10 mL of phosphate buffer (25 mM, pH 7.5) and 8 mL of the olive oil emulsion. After reacting at 30 °C for 10 min, 20 mL of ethanol was added to terminate the reaction. The free fatty acid released by the reaction was titrated with a sodium hydroxide solution. We defined enzyme activity as the amount of enzyme required to hydrolyze sufficient olive oil to release 1 μmol of fatty acid per minute.
Analysis of enzyme properties
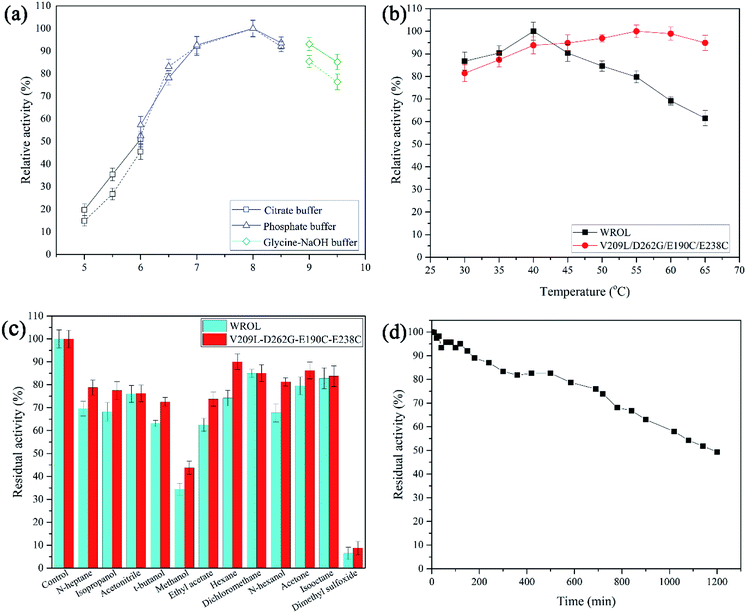
Thermostability was determined by calculating the residual enzyme activity after heat treatment for different times or at different temperatures. The optimum reaction pH was determined by measuring the hydrolysis activity of the enzyme at pH values from 5 to 9.5 at 30 °C. The optimum temperature was determined by measuring hydrolysis activity in the temperature range 30–65 °C at pH 8.0. Solvent tolerance was determined by measuring residual enzyme activity after treatment in different solvents for 12 h. Kinetic parameters of wild-type ROL (WROL) and mutants toward p-nitrophenyl butyrate (10–100 mM) were determined at 30 °C in phosphate buffer (25 mM, pH 8). To evaluate regioselectivity, lipase-catalyzed esterification of oleic acid with glycerol was carried out at 30 °C as described previously.22 We defined regioselectivity as Re = (1,3-diolein% − 1,2-diolein%)/(1,3-diolein% + 1,2-diolein%).Results and discussion
Multiple-sequence alignment
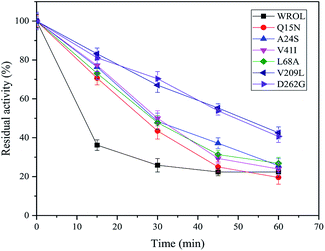
The multiple-sequence alignment method was used to identify amino acid residues that could affect the protein thermostability.12 To improve the thermostability of ROL, we applied a sequence-based rational design approach. Rhizomucor miehei lipase and Thermomyces lanuginosus lipase are more thermo-tolerant than ROL23,24 and share 53.4% and 29.7% sequence identity with ROL, respectively. Thus, these two lipases were selected to perform multiple-sequence alignments to identify sites that could enhance the thermostability of ROL. The amino acid residues that could affect the thermostability of ROL were marked with black asterisks (Fig. 1). Some of these amino acid residues were located on the lid of ROL (Fig. 1, red asterisks), and were not selected for mutation. Point mutations Q15N, A24S, V41I, L68A, Y123L, E129D, T184A, V209L, S223Y, and D262G were performed according to the alignment results.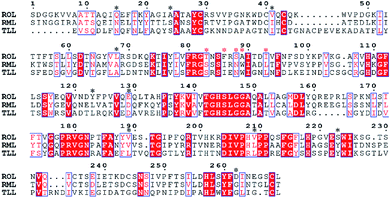 | ||
| Fig. 1 Multiple-sequence alignment of ROL with Thermomyces lanuginosus (TLL) lipase and Rhizomucor miehei (RML) lipase. | ||
The thermostabilities of the mutants and WROL were determined at 55 °C by olive oil titration. The probability of obtaining a thermostatically enhanced mutant by a multi-sequence alignment method is large, but it does not ensure that all mutations will exhibit positive results. The structures and locations of the amino acid residues before and after the mutation will affect the results of the mutation. Mutants Y123L, E129D, T184A, and S223Y showed no enhancement of thermostability compared with the wild-type (data not shown). However, mutants Q15N, A24S, V41I, L68A, V209L, and D262G showed varying degrees of improvement in thermostability (Fig. 2). Among these mutants, V209L and D262G showed the best thermostabilities. The half-lives of V209L and D262G at 55 °C were 51.25 and 49.37 min, respectively, whereas that of WROL was 11.7 min.
Postulations based on molecular modeling
Compared with WROL, the mutants V209L and D262G were much more thermostable. To explain this phenomenon, molecular models of these two mutants were built using SWISS-MODEL and the crystal structure of ROL (PDB ID: 1LGY) (Fig. 3).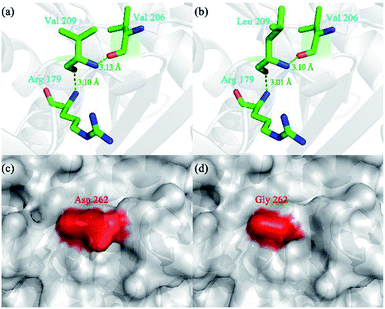 | ||
| Fig. 3 Three-dimensional structure of wild-type ROL (WROL) and structural models of its mutants. Interactions of V209 in WROL (a) and L209 in mutant V209L (b). The green dashed line represents a hydrogen bond and the bond length is indicated in green text. Surface representation of three-dimensional homology model of WROL (c) and mutant D262G (d). The red regions are the surfaces of D262 and G262. The surfaces of other areas are shown in gray. The structure of WROL was obtained from the Protein Data Bank (PDB ID: 1LGY). Molecular models of the mutants were built using SWISS-MODEL. | ||
Hydrogen bonds play an important role in stabilizing proteins.25 When the valine at position 209 of ROL was mutated to leucine, no new bond was formed (Fig. 3). However, the length of the hydrogen bond between this Leu and the adjacent amino acids (R179 and V206) decreased, which rendered the protein structure more resistant to unfolding and resulted in a more stable protein structure.26 When the aspartic acid at position 262 was replaced by glycine, the thermostability of the mutant also greatly improved compared to the wild-type. The mutation D262G was found in a surface loop and no new bond or interaction was predicted to form after this mutation (Fig. 3). Similar reports have shown that mutations to glycine at specific sites led to an increase in protein thermostability.27–29 This could be because the surface loop of ROL containing G262 plays an important role in the thermostability of the enzyme. Glycine is the simplest amino acid, and the introduction of a mutation (glycine) to this loop may alleviate conformational strain and eliminate unfavorable steric interactions, thereby enhancing the thermostability of the protein.30
Engineering disulfide bonds rationally
All residue pairs that may form disulfide bonds if mutated to cysteine were identified using the online prediction server DbD2. Our results indicated that 32 residue pairs may form disulfide bonds after mutation to cysteines (ESI, Fig. s1†). The prediction results were scored and sorted in terms of energy, bad contacts, thermal mobility, and sequence separation. We selected those that scored >90 and mutated the corresponding amino acid residue pairs to cysteines. Several of these variants showed an increase in thermostability (Fig. 4). Among them, variant E190C/E238C showed great improvement of thermostability with about 93% of its initial activity retained after heating at 55 °C for 15 min, whereas WROL retained only about 37% of its initial activity after the same treatment. However, the introduced pair of cysteines do not definitely form disulfide bonds. In addition, the introduction of disulfide bonds into proteins does not always enhance stability, and it is still difficult to predict the stabilizing effects of disulfide bonds. This is because original favorable interactions may be lost after the introduction of the disulfide, and unfavorable contacts may be formed by amino acid residues around the introduced disulfide.31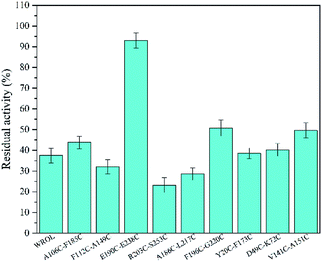 | ||
| Fig. 4 Effect of preheating at 55 °C for 15 min on the enzyme activity of WROL and variants containing an introduced disulfide bond. | ||
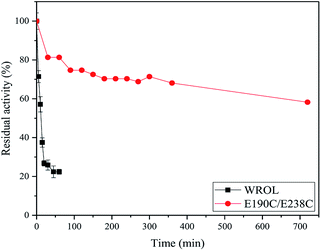
The deactivation of variant E190C/E238C and WROL at 55 °C were measured (Fig. 5). The activity of variant E190C/E238C remained at about 58.2% after incubated at 55 °C for 720 min, whereas the half-life of WROL was only about 11.7 min. The introduction of a disulfide bond between sites 190 and 238 greatly improved the rigidity of the enzyme.
Conformational changes at the molecular level for WROL and E190C/E238C
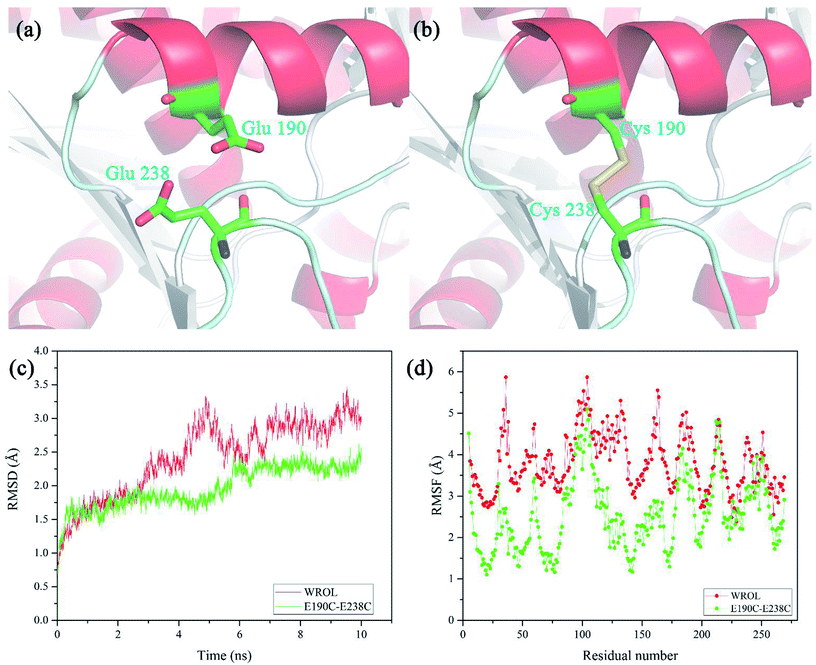
The newly formed disulfide bond C190–C238 that improved the thermostability of ROL was located on the protein surface and distant from the catalytic center of the enzyme (Fig. 6a and b). Positions 190 and 238 are located in the alpha helix and random coil of the protein, respectively. The restriction of random coil or free loop structures by disulfide bond crosslinking between these two sites reduces the conformational entropy of the unfolded state, thereby contributing to enhanced protein stability.32 The structural properties of the disulfide-containing variant of ROL were assessed using MD simulations at 330 K for 10 ns. The overall molecular root-mean-square deviation (RMSD) of the E190C/E238C variant was much lower than that of WROL (Fig. 6c). Therefore, during heat treatment, the structure of WROL becomes looser than that of E190C/E238C and the introduction of the C190–C238 disulfide improves the overall structural rigidity of the protein. The overall molecular root-mean-square fluctuations also indicate that the overall structure of E190C/E238C is more rigid than that of WROL, resulting in the observed enhancement of the thermostability of the enzyme.Improving thermostability by combined mutation of hot-spots and introduction of disulfide bond
By combining individual point mutations that lead to improved thermostability, it is highly probable that variants with further improvements in thermostability will be obtained.12 On the basis of our results, we produced ROL with combinations of the mutations V209L and D262G with E190C and E238C. The thermostabilities of these mutants were tested at 65 °C because of the good thermostability of E190C/E238C variant at 55 °C (Fig. 7). All the variants showed enhancement of thermostability compared with the mutant E190C/E238C. The quadruple variant V209L/D262G/E190C/E238C showed the best thermostability because of the positive superposition effect of multi-sequence mutation with the introduced disulfide bond. The half-life of this variant at 65 °C was 37.93 min, which was about 20 times that of the wild-type at this temperature. However, the double mutant V209L/D262G showed poor thermostability at 65 °C (data not shown). This could be caused by the limited thermostabilities of these two mutation sites compared with the introduction of disulfide bonds.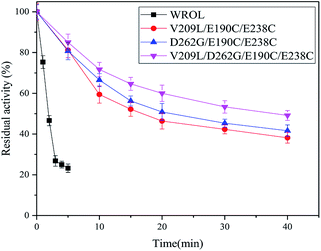 | ||
| Fig. 7 Inactivation curves of WROL and its variants V209L/E190C/E238C, D262G/E190C/E238C, and V209L/D262G/E190C/E238C at 65 °C. | ||
Conformational change of WROL and the V209L/D262G/E190C/E238C mutant
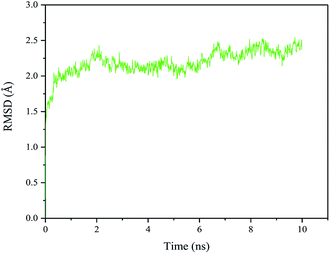
The RMSD of the quadruple variant was analyzed as shown in Fig. 8. Compared with the wild type, the RMSD of the quadruple variant V209L/D262G/E190C/E238C was reduced, but was higher than that of E190C/E238C (Fig. 6c). This could be because the glycine introduced at position 262 increased the flexibility of the entire structure. However, the thermostability of the quadruple variant V209L/D262G/E190C/E238C increased drastically because of the cumulative effect of multiple positive mutations.Enzymatic properties of WROL and the V209L/D262G/E190C/E238C mutant
The properties of WROL and the mutant V209L/D262G/E190C/E238C were compared (Fig. 9). The pH of the reaction system is an important factor affecting the catalytic performance of an enzyme. The optimum pH values for the reaction of WROL and the quadruple mutant were determined at 30 °C. The two forms of the enzyme exhibited similar properties with respect to pH, and maximum activity occurred at pH 8.0. This can be attributed to the location of the mutated sites far from the active center, where they have little effect on its catalytic performance. The optimum reaction temperature shifted from 40 °C for the wild-type to 55 °C for the variant because of the substantial increase in the thermostability of the latter. WROL rapidly lost its activity when incubated at temperatures above 40 °C because of its poor thermostability. However, the quadruple mutant had higher heat tolerance than WROL because it was more rigid, which increased the optimum temperature. Solvent tolerance is an important property of lipases. With the enhancement of thermostability, the solvent tolerance of ROL was also enhanced for most of the solvents tested because the increased rigidity of the enzyme made it more stable in organic solvents. The thermostability of this quadruple variant was also tested at 55 °C. When incubated at 55 °C for 1200 min, the quadruple variant retained about 49.3% of its initial activity. The half-life of this variant at 55 °C was about 102.5-times that of WROL.Kinetic parameters of WROL and mutant lipase
Kinetic constants were calculated from the initial rate of reaction of WROL and the mutant V209L/D262G/E190C/E238C using p-nitrophenyl butyrate as the substrate (Table 1). The kinetic parameters of the two lipases were similar, indicating that the difference in catalytic ability before and after the mutation was small. When these two lipases were used to catalyze the esterification reaction of the model substrate oleic acid with glycerol, both of them exhibited excellent 1,3-regioselectivity. Therefore, the mutant V209L/D262G/E190C/E238C with much better thermostability than WROL has wider applicability for preparation of structured lipids. The similar properties of these two lipases may be attributed to the locations of the mutation sites far from the active site, where they had little effect on the enzyme activities and selectivity (Fig. 10).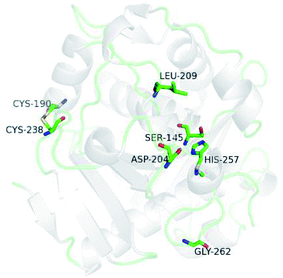 | ||
| Fig. 10 Three-dimensional model of the mutant V209L/D262G/E190C/E238C. Residues S145, D204, and H257 are the catalytic triad. | ||
Conclusions
In this work, we modified the thermostability of ROL because it has high sn-1,3-regioselectivity but poor thermostability. Ten variants with one point-mutant were constructed by multiple-sequence alignment. The most thermotolerant were variants V209L and D262G, with half-lives at 55 °C of 51.25 and 49.37 min, respectively. By contrast, the half-life of WROL was only 11.7 min. This improvement could be caused by enhancement of the strength of hydrogen bond and alleviation of conformational strain, respectively. The enzyme became more rigid after introduction of a disulfide bond (E190C/E238C). The activity of this variant remained at about 58.2% after incubation at 55 °C for 720 min. The thermostability of the enzyme was further enhanced by combined introduction of two point-mutations and the disulfide bond. When incubated at 55 °C for 1200 min, the quadruple variant retained about 49.3% of its initial activity. The half-lives of this variant at 55 °C and 65 °C were about 102.5- and 20-times that of WROL, respectively. The enhancement of thermostability for this quadruple variant could be attributed to the positive superposition effect of multi-sequence mutation with the introduced disulfide bonds. The mutant V209L/D262G/E190C/E238C showed only a small difference in catalytic performance compared with WROL. This improvement in thermostability will give ROL wider industrial applicability, especially in the preparation of structured lipids.Conflicts of interest
None.Acknowledgements
This work was supported by the National Natural Science Foundation of China [Grant No. 21376215], National Basic Research Program of China [973, 2011CB710803], and the National High-Tech Research and Development Program of China [863, 2012AA022302].Notes and references
- P. Adlercreutz, Chem. Soc. Rev., 2013, 42, 6406–6436 RSC.
- X. Wang, W. Zou, X. Sun, Y. Zhang, L. Wei, Q. Jin and X. Wang, Biotechnol. Lett., 2015, 37, 691–696 CrossRef CAS PubMed.
- R. Dhara, P. Dhar and M. Ghosh, J. Food Sci. Technol., 2013, 50, 678–686 CrossRef CAS PubMed.
- M. M. Soumanou, M. Pérignon and P. Villeneuve, Eur. J. Lipid Sci. Technol., 2013, 115, 270–285 CrossRef CAS.
- R. B. Salah, H. Mosbah, A. Fendri, A. Gargouri, Y. Gargouri and H. Mejdoub, FEMS Microbiol. Lett., 2006, 260, 241–248 CrossRef PubMed.
- X. W. Yu, W. Rui, Z. Meng, X. Yan and X. Rong, Microb. Cell Fact., 2012, 11, 102–112 CrossRef CAS PubMed.
- W.-n. Niu, Z.-p. Li and T. Tan, Mol. Biotechnol., 2006, 32, 73–81 CrossRef CAS.
- X. W. Yu, N. J. Tan, R. Xiao and Y. Xu, PLoS One, 2012, 7, e46388 CrossRef CAS PubMed.
- Z. L. Han, S. Y. Han, S. P. Zheng and Y. Lin, Appl. Microbiol. Biotechnol., 2009, 85, 117–126 CrossRef CAS.
- H. Yu and H. Huang, Biotechnol. Adv., 2014, 32, 308–315 CrossRef CAS PubMed.
- J. P. Wu, M. Li, Y. Zhou, L. R. Yang and G. Xu, Biotechnol. Lett., 2015, 37, 403–407 CrossRef CAS PubMed.
- L. Lin, Y. Wang, M. Wu, L. Zhu, L. Yang and J. Lin, Enzyme Microb. Technol., 2018, 115, 45–51 CrossRef CAS PubMed.
- D. Yuan, Z. Zhao, X. Wang, S. Guo, B. Yang and Y. Wang, Eur. J. Lipid Sci. Technol., 2016, 118, 821–826 CrossRef CAS.
- G. Li, Y. Chen, X. Fang, F. Su, L. Xu and Y. Yan, RSC Adv., 2018, 8, 1948–1957 RSC.
- Q. A. Le, J. C. Joo, Y. J. Yoo and Y. H. Kim, Biotechnol. Bioeng., 2012, 109, 867–876 CrossRef CAS PubMed.
- B. H. Jo, T. Y. Park, H. J. Park, Y. J. Yeon, Y. J. Yoo and H. J. Cha, Sci. Rep., 2016, 6, 29322 CrossRef PubMed.
- G. Li, X. Fang, F. Su, Y. Chen, L. Xu and Y. Yan, Appl. Environ. Microbiol., 2017, 84, 1–16 Search PubMed.
- A. A. Dombkowski, Bioinformatics, 2003, 19, 1852–1853 CrossRef CAS.
- X. F. Zhang, G. Y. Yang, Y. Zhang, Y. Xie, S. G. Withers and Y. Feng, Sci. Rep., 2016, 6, 33797 CrossRef CAS PubMed.
- A. A. Dombkowski, Bioinformatics, 2003, 19, 1852–1853 CrossRef CAS.
- D. Y. Kwon and J. S. Rhee, J. Am. Oil Chem. Soc., 1986, 63, 89–92 CrossRef CAS.
- X. Meng, G. Xu, Q. L. Zhou, J. P. Wu and L. R. Yang, Food Chem., 2014, 143, 319–324 CrossRef CAS PubMed.
- M. L. M. Fernandes, N. Krieger, A. M. Baron, P. P. Zamora, L. P. Ramos and D. A. Mitchell, J. Mol. Catal. B: Enzym., 2004, 30, 43–49 CrossRef CAS.
- M. Noel and D. Combes, J. Biotechnol., 2003, 102, 23–32 CrossRef CAS PubMed.
- L. Zhang and Y. Ding, Protein Pept. Lett., 2017, 24, 643–648 CAS.
- F. I. Khan, B. Nizami, R. Anwer, K. R. Gu, K. Bisetty, M. I. Hassan and D. Q. Wei, J. Biomol. Struct. Dyn., 2017, 35, 2123–2135 CrossRef CAS PubMed.
- B. Madan and P. Mishra, Biochem. Eng. J., 2014, 91, 276–282 CrossRef CAS.
- M. Anbar, O. Gul, R. Lamed, U. O. Sezerman and E. A. Bayer, Appl. Environ. Microbiol., 2012, 78, 3458–3464 CrossRef CAS PubMed.
- S. R. Trevino, S. Schaefer, J. M. Scholtz and C. N. Pace, J. Mol. Biol., 2007, 373, 211–218 CrossRef CAS PubMed.
- M. Anbar, R. Lamed and E. A. Bayer, ChemCatChem, 2010, 2, 997–1003 CrossRef CAS.
- T. Hibi, A. Kume, A. Kawamura, T. Itoh, H. Fukada and Y. Nishiya, Biochemistry, 2016, 55, 724–732 CrossRef CAS PubMed.
- C. N. Pace, G. R. Grimsley, J. A. Thomson and B. J. Barnett, J. Biol. Chem., 1988, 263, 11820–11825 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra07767c |
| This journal is © The Royal Society of Chemistry 2018 |