 Open Access Article
Open Access ArticleEffect of Ling-Gui-Zhu-Gan decoction major components on the plasma protein binding of metoprolol using UPLC analysis coupled with ultrafiltration†
Peng Zhou *abc,
Jinling Huang*abc and
Wanxue Dingab
*abc,
Jinling Huang*abc and
Wanxue Dingab
aDepartment of Integrated Traditional Chinese and Western Medicine, Anhui University of Chinese Medicine, Hefei 230012, People's Republic of China. E-mail: zp0108@126.com; jinling6181@126.com; Fax: +86-0551-68129468; Tel: +86-0551-68129468
bResearch Institute of Integrated Traditional Chinese and Western Medicine, Anhui Academy of Chinese Medicine, Hefei 230012, People's Republic of China
cKey Laboratory of Chinese Herbal Compound Formula in Anhui Province, Hefei 230012, People's Republic of China
First published on 22nd October 2018
Abstract
Using traditional Chinese medicine formula Ling-Gui-Zhu-Gan decoction (LGZGD) plus selective β1-adrenergic receptor inhibitor metoprolol to treat arrhythmia of coronary heart disease can significantly improve efficiency with no adverse reactions. However, the effect of major components of LGZGD on the plasma protein binding of metoprolol is unclear. Firstly, this study aimed to computationally predict the molecular interactions between metoprolol, the major components of LGZGD, and bovine serum albumin (BSA). Secondly, the plasma protein binding of metoprolol combined with major components of LGZGD was investigated by UPLC analysis coupled with ultrafiltration. The MOE (2008.10) software package was used to investigate the molecular interactions among metoprolol, the major components of LGZGD, and BSA. Using in vitro experiments, BSA was separately spiked with a mixtures of metoprolol and the major components of LGZGD. The results showed that metoprolol interacted with BSA mainly through arene–arene interactions, as did cinnamic acid and liquiritin. However, the energy scores of cinnamic acid and liquiritin were lower than that of metoprolol. There were no interactions between metoprolol and the major components of LGZGD. Further studies in vitro showed that the presence of the major components of LGZGD did not change the plasma protein binding of metoprolol. We adopted molecular docking to predict the drug–herb plasma protein binding interactions of metoprolol and then used ultrafiltration to verify the docking results. There were no drug–herb interactions between metoprolol and LGZGD in BSA, which indicated that this combination therapy might be safe and feasible.
Introduction
Metoprolol, a class II antiarrhythmic agent, is currently used as a clinical treatment for arrhythmia, hypertension, and chronic heart failure, and to improve cardiac autonomic function. Metoprolol can bind to β-adrenoceptors in cardiac nodule tissue, conduction systems, and contractile cardiomyocytes. The mechanism of action involves blocking the effects of catecholamines at β1-adrenergic receptors, which decreases sympathetic activity on the heart.1,2 Previous reports found that metoprolol has been widely used to treat chronic heart failure (CHF) in several randomized clinical trials, and can reduce hospitalization and improve survival.3 Only free drugs can have a pharmacological effect through the cell membrane and bind to their corresponding targets, meaning that the determination of free drugs is crucial for determining the safety of clinical trials. Drug–protein binding plays an important role in pharmacology and toxicology.4,5 Pharmacokinetic properties, such as distribution volume, hepatic metabolism rate, and renal excretion rate, are functions of the ratio of free components.6 Therefore, it is necessary to determine plasma protein binding to regulate the therapeutic dose and understand the pharmacologic behavior of medicines. Methods for determining the free drug content in plasma include equilibrium dialysis (ED), ultrafiltration (UF), and capillary electrophoresis (CE).7,8 Previous reports found that the mean plasma protein binding (PPB) of metoprolol is 30.70 ± 2.76% in humans.9 Therefore, whether the combination of metoprolol and other drugs will result in adverse reactions also depends on changes in plasma protein binding.Ling-Gui-Zhu-Gan decoction (LGZGD) is a classic traditional Chinese medicine (TCM) formula recorded in Shang Han Lun by Zhongjing Zhang that consists of Poria cocos (Schw.) Wolf (Fu-Ling), Cinnamomi Ramulus (Gui-Zhi), Rhizoma Atractylodis Macrocephalae (Bai-Zhu), and Radix Glycyrrhizae (Gan-Cao).10 LGZGD is often used to treat arrhythmia, cardiac failure, and other cardiovascular diseases in China.11 Presently, many clinical studies have indicated that LGZGD is effective for CHF treatment.12,13 Our previous studies have shown that LGZGD effectively regulates the structure and function of the heart by regulating neuroendocrine cytokines, such as brain natriuretic peptide (BNP), aldosterone (Ald), angiotensin II (Ang II), interleukin-1 beta (IL-1β), interleukin-6 (IL-6), and tumor necrosis factor-alpha (TNF-α) in rat models of CHF.14,15 An in vivo animal study found that LGZGD can improve myocardial tissue injury and inhibit ventricular remodeling by inhibiting excessive activation of the nuclear factor-kappa B (NF-κB) signaling pathway.16 An in vitro study found that LGZGD could regulate protein expressions related to IKK/I-κB/NF-κB signaling pathways, intervene in the transcriptional control of IKK/I-κB/NF-κB downstream target molecules, and effectively inhibit excessive activation of cytokines.17 Our study found that the main characteristic chemical components of LGZGD were cinnamic acid, atractylenolide III, liquiritin, and glycyrrhizic acid. Clinical studies have shown that using LGZGD plus metoprolol to treat arrhythmia in coronary heart disease can significantly improve efficiency, and reduce atrial contraction and ventricular contraction, with no adverse reactions.18,19 Combinations of two drugs can alter drug concentrations and change their clinical effects. In other words, when the free drug concentration of drug A increases due to plasma protein binding displacement by another strongly binding drug, drug B, the increase in free drug A concentration could potentially cause toxicity and may result in a dose adjustment being required.20 However, the effect of major components of LGZGD on the plasma protein binding of metoprolol is not clear.
Molecular docking has become essential for the study of molecular structures and protein interactions, and can predict the most detailed possible view of drug–target interactions.21 Chemical compounds exert their biological activities by binding to one or more targets. Herein, we introduce a new, potentially widely applicable and accurate, drug–plasma protein binding assay that predicts competitive effects.
In this study, the molecular interactions between the major components of LGZGD, metoprolol, and bovine serum albumin (BSA) were predicted using molecular docking. UPLC and ultrafiltration techniques were used to determine the plasma protein binding of metoprolol alone and combined with the major components of LGZGD to verify the docking results.
Materials and methods
Chemicals and materials
Metoprolol was purchased from the China National Institutes for Food and Drug Control (Beijing, China). The purity of the reference compound was greater than 98% by HPLC. PBS buffer (pH 7.4) was composed of sodium chloride (1.35 mol L−1), potassium chloride (27 mmol L−1), sodium phosphate (43 mmol L−1), and potassium phosphate (14 mmol L−1). BSA (purity, ≥98%) was obtained from Sigma-Aldrich. BSA solution was prepared at a concentration of 40 mg mL−1 in PBS buffer. Chromatographic grade acetonitrile and methanol were obtained from Merck company (Germany). Deionized water was purified using a Milli-Q system (Millipore, Bedford, MA, USA). Centrifugal filter units (Centrifree YM-10 regenerated cellulose membrane, MWCO 10K) were obtained from Millipore (Billerica, MA, USA). All other chemicals and solvents used in the present study were of reagent grade or better.Molecular docking between the major components of LGZGD, metoprolol, and BSA
Interactions of the major components of LGZGD, metoprolol, and BSA were obtained using the MOE (2008.10) software package.22,23 The structures of cinnamic acid, atractylenolide III, liquiritin, glycyrrhizic acid, and metoprolol were drawn using ChemDraw software and the three-dimensional crystal protein structure of BSA (PDB ID: 4F5S) was obtained from the Protein Data Bank (PDB) (https://www.rcsb.org/pdb).24 Briefly, 3D structures of cinnamic acid, atractylenolide III, liquiritin, glycyrrhizic acid, and metoprolol were optimized by adding hydrogens and energy minimized.The correct conformations of the ligands were found and then minimum energy of the structures was obtained. After energy optimization, ligands were allowed to be flexible. Water molecules were removed and the 3D protonation of BSA was conducted using the MOE (2008.10) software package. After removing the ligand from BSA, they were minimized using MMFF94 Force field. The predicted molecules were docked with the hydrophobic pocket of the receptor and the values of the combination were given. After docking, the resultant complexes were subjected to hydrogen bonding analysis, and then interactions among the active components and targets were found.
Chromatography conditions
UPLC analyses were performed on a Waters Acquity Ultra-performance Liquid Chromatography (UPLC) H-Class system (Waters, Milford, MA). An Acquity BEH C18 (2.1 mm × 100 mm, 1.7 μm; Waters, Milford, MA) analytical column coupled with a column filter were used to test samples, and column temperature was set at 30 °C. Mobile phase A consisted of 0.05% (v/v) phosphoric acid in distilled water, while mobile phase B was acetonitrile (75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25) at a flow rate of 0.2 mL min−1 for 4 min. Samples were detected at 225 nm. The autosampler temperature was kept at 4 °C and 1 μL of the samples was injected into the UPLC system.
25) at a flow rate of 0.2 mL min−1 for 4 min. Samples were detected at 225 nm. The autosampler temperature was kept at 4 °C and 1 μL of the samples was injected into the UPLC system.
Calibration curve, precision, stability tests, and extraction recovery
Stock solutions of metoprolol (1 mg mL−1), cinnamic acid (1 mg mL−1), atractylenolide III (1 mg mL−1), liquiritin (1 mg mL−1), and glycyrrhizic acid (1 mg mL−1) were dissolved in methanol. The stock solutions were kept at 4 °C. The stock solutions were successively diluted with methanol to prepare working solutions immediately prior to use.Linearity was assessed by assaying calibration curves at eight concentration levels in the range 0.1–80 μg mL−1 in PBS buffer. The concentrations of metoprolol in samples were calculated from the peak area using a linear regression equation obtained from the calibration curve.
Metoprolol solutions in PBS buffer (0.5 μg mL−1, 5 μg mL−1, and 20 μg mL−1) were prepared as QC samples. For the precision tests, the intra-day variance was determined by assaying each sample in five replicates within one day, while inter-day variance was measured over three consecutive days.
For the stability test, the QC samples of metoprolol were incubated in a water bath at 37 °C for 1, 2, 3, and 4 h, and the incubated fluids were collected and analyzed by UPLC to monitor drug degradation.
The extraction recovery of metoprolol was calculated using the following formula (B/A × 100)%, where A is the metoprolol solution concentration, B is the concentration of the blank BSA samples with added metoprolol. The extraction recovery was also evaluated using the same method.
Nonspecific binding (NSB) of metoprolol
The NSB of metoprolol was determined using the method of Kurlbaum et al. with some modifications.25 Metoprolol was dissolved in PBS buffer to obtain concentrations of 0.5 μg mL−1, 1 μg mL−1, and 2 μg mL−1. Metoprolol solutions (400 μL) with different concentrations were applied to the ultrafiltration unit. After centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 15 min, 1 μL of the filtrate was subjected to UPLC analysis.
000 × g for 15 min, 1 μL of the filtrate was subjected to UPLC analysis.
The nonspecific binding of metoprolol in the ultrafiltration experiments was expressed as NSB [%] = (Cpre − Cpost)/Cpre × 100, where Cpost is the compound concentration recovered after ultrafiltration, and Cpre is the initial compound concentration before filtration.
Plasma protein binding of metoprolol and combined with major components of LGZGD
Plasma protein binding of metoprolol in vitro was studied using an ultrafiltration method.26,27 In brief, metoprolol solutions (30 μL; 10, 20, and 40 μg mL−1) were spiked into BSA (570 μL) and vortexed for 2 min to produce final concentrations of 0.5, 1, and 2 μg mL−1. Three replicates of each concentration were prepared. The spiked BSA samples were incubated at 37 °C for 60 min to achieve equilibrium. Samples (500 μL) were transferred to the centrifugal filter unit and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 15 min. A 1 μL aliquot of the filtrate was subjected to UPLC analysis.
000 × g for 15 min. A 1 μL aliquot of the filtrate was subjected to UPLC analysis.
Metoprolol solutions (30 μL; 10, 20, and 40 μg mL−1) and cinnamic acid solutions (30 μL; 20 μg mL−1) were spiked into BSA (540 μL) in amber vials to yield final solutions of metoprolol (0.5, 1, and 2 μg mL−1) and cinnamic acid (1 μg mL−1). The ultrafiltration steps were conducted as described above.
Metoprolol solutions (30 μL; 10, 20, and 40 μg mL−1) and atractylenolide III solutions (30 μL, 4 μg mL−1) were spiked into BSA (540 μL) in amber vials to yield final solutions of metoprolol (0.5, 1, and 2 μg mL−1) and atractylenolide III (0.2 μg mL−1). The ultrafiltration steps were conducted as described above.
Metoprolol solutions (30 μL; 10, 20, and 40 μg mL−1) and liquiritin or glycyrrhizic acid solutions (30 μL, 40 μg mL−1) were spiked into BSA (540 μL) in amber vials to yield final solutions of metoprolol (0.5, 1, and 2 μg mL−1) and liquiritin or glycyrrhizic acid (2 μg mL−1). The ultrafiltration steps were conducted as described above.
The total free drug concentration in the filtrate in ultrafiltration experiments can be expressed as Cf = Cft/(1 − NSB), where Cft is the actual drug concentration measured in the filtrate and NSB is the nonspecific binding.
Protein binding was calculated using % protein binding = (Ct − Cf)/Ct × 100, where Ct represents the total drug concentration in BSA and Cf is the total free drug concentration in the filtrate.
Statistical analysis
The measured data are presented as mean ± standard deviation (![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) ± s). Multiple group means were compared using one-way analysis of variance and LSD methods. All analyses were performed using SPSS version 23.0 software, with a significance level of p < 0.05.
± s). Multiple group means were compared using one-way analysis of variance and LSD methods. All analyses were performed using SPSS version 23.0 software, with a significance level of p < 0.05.
Results
Molecular interactions between metoprolol, the major components of LGZGD, and BSA
Molecular interactions between metoprolol and BSA are arene–arene interactions. Metoprolol bound to BSA via arene–arene interactions at Phe 550. Cinnamic acid was bound to BSA via hydrogen bond formation at Tyr 400 and arene–arene interactions at Phe 550. Atractylenolide III was bound to BSA via hydrogen bond formation at Tyr 578. Liquiritin was bound to BSA via hydrogen bond formation at Tyr 400 and Phe 506, and arene–arene interactions at Phe 550. Glycyrrhizic acid was bound to BSA via hydrogen bond formation at Tyr 400, Tyr 578, Lys 524, Asn 404, and Asp 561 (Table 1 and Fig. 1).| Chemicals | Interaction energy (kcal mol−1) | Number of binding to residues | Residues involved in H-bond formation | Arene–arene |
|---|---|---|---|---|
| Metoprolol | −10.6796 | 1 | — | Phe 550 |
| Cinnamic acid | −9.5724 | 2 | Tyr 400 | Phe 550 |
| Atractylenolide III | −9.2092 | 1 | Tyr 578 | — |
| Liquiritin | −10.5589 | 3 | Tyr 400, Phe 506 | Phe 550 |
| Glycyrrhizic acid | 1.3939 | 8 | Tyr 400, Tyr 578, Lys 524, Asn 404, Asp 561 | — |
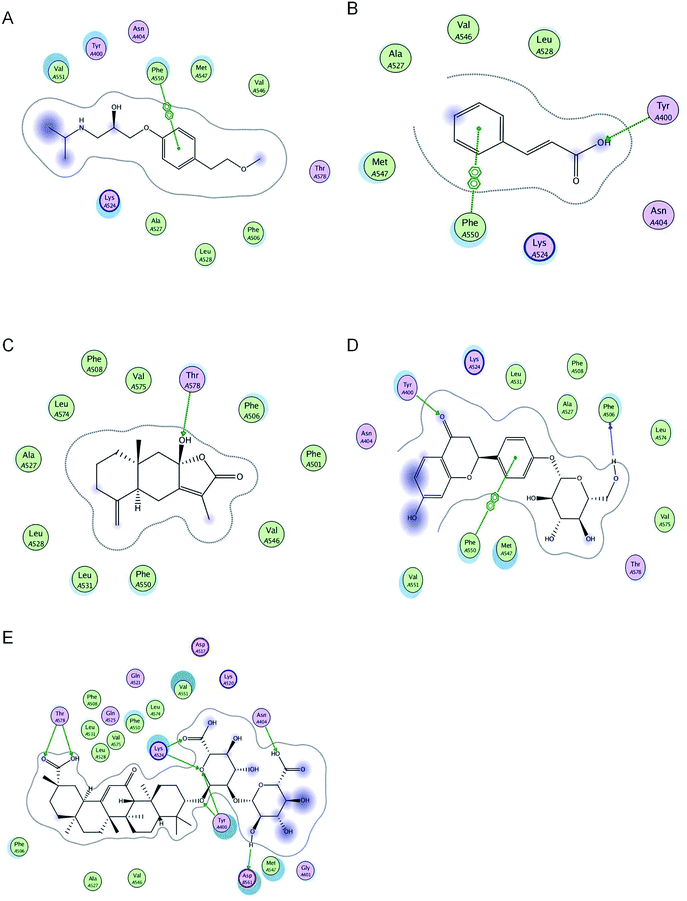 | ||
| Fig. 1 2D pictures of the docked conformations of chemicals and active site of BSA: (A) metoprolol, (B) cinnamic acid, (C) atractylenolide III, (D) liquiritin, and (E) glycyrrhizic acid. | ||
Specificity
The specificity was evaluated by comparing the chromatograms of blank BSA, metoprolol, and blank BSA spiked with metoprolol. The retention time of metoprolol was approximately 2.1 min. No significant endogenous peaks interfering with metoprolol were obtained in BSA (Fig. S1†).Standard curve, precision, and stability
The calibration curves of metoprolol were linear over the range 0.1–80 μg mL−1 (0.1, 0.5, 1, 5, 10, 20, 40, and 80 μg mL−1). The calibration curve for metoprolol was prepared with the equation y = 4931.5x − 61.152, r = 0.9999 (n = 3). The intra-day and inter-day analyses met the requirements of quantitative analysis (Table 2). Consequently, these results indicated that the analytical method was suitable for the present study.![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) ± s)
± s)
| Drug | Concentration (μg mL−1) | Intra-day (n = 5) | Inter-day (n = 15) | ||
|---|---|---|---|---|---|
| Concentration (μg mL−1) | RSD (%) | Concentration (μg mL−1) | RSD (%) | ||
| Metoprolol | 0.5 | 0.51 ± 0.02 | 3.92 | 0.52 ± 0.03 | 5.77 |
| 5 | 4.03 ± 0.13 | 3.23 | 4.12 ± 0.18 | 4.37 | |
| 20 | 20.43 ± 1.07 | 5.24 | 20.86 ± 1.22 | 5.85 | |
Extraction recovery
The extraction recovery of metoprolol in BSA at each QC level was 84.87 ± 3.54%, 86.45 ± 4.18%, and 86.95 ± 4.42% (Table 3).![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) ± s, n = 3)
± s, n = 3)
| Drug | Concentration (μg mL−1) | Extraction recovery (%) | RSD (%) |
|---|---|---|---|
| Metoprolol | 0.5 | 84.87 ± 3.54 | 4.17 |
| 5 | 86.45 ± 4.18 | 4.84 | |
| 20 | 86.95 ± 4.42 | 5.08 |
NSB of metoprolol
The NSB between metoprolol and the ultrafiltration membrane was approximately 22% (Table 4). As the results showed that the ultrafiltration membrane had a certain adsorption effect on metoprolol, the formula of plasma protein binding should be corrected.| Drug | Added concentration (μg mL−1) | Cpre (μg mL−1) | Cpost (μg mL−1) | NSB (%) |
|---|---|---|---|---|
| Metoprolol | 0.5 | 0.51 ± 0.02 | 0.40 ± 0.02 | 22.47 ± 1.79 |
| 1.0 | 0.99 ± 0.04 | 0.78 ± 0.03 | 21.94 ± 2.50 | |
| 2.0 | 2.00 ± 0.05 | 1.55 ± 0.05 | 22.55 ± 2.14 |
Plasma protein binding rate of metoprolol in BSA
As shown in Table 5, the plasma protein binding rate of metoprolol in BSA at 37 °C through the ultracentrifugation process was consistently 28% over three concentrations of 0.5, 1, and 5 μg mL−1.![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) ± s, n = 3)
± s, n = 3)
| Drug | Ct (μg mL−1) | Cf (μg mL−1) | Plasma protein binding (%) |
|---|---|---|---|
| Metoprolol | 0.50 ± 0.02 | 0.36 ± 0.01 | 28.15 ± 2.02 |
| 0.99 ± 0.04 | 0.70 ± 0.02 | 29.67 ± 2.42 | |
| 2.02 ± 0.06 | 1.45 ± 0.03 | 28.36 ± 1.65 |
Effects of cinnamic acid on plasma protein binding of metoprolol
Table 6 shows that the plasma protein binding of metoprolol (0.5 μg mL−1) might be influenced by cinnamic acid (1 μg mL−1), while the plasma protein binding of metoprolol at concentrations of 1 and 2 μg mL−1 was not be affected. The statistical results showed that cinnamic acid was unlikely to affect the plasma protein binding of metoprolol (Table 6).![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) ± s, n = 3)
± s, n = 3)
| Drug | Ct (μg mL−1) | Cf (μg mL−1) | Plasma protein binding (%) (with cinnamic acid) | Plasma protein binding (%) (without cinnamic acid) |
|---|---|---|---|---|
| a p > 0.05, vs. metoprolol without cinnamic acid. | ||||
| Metoprolol | 0.50 ± 0.03 | 0.37 ± 0.01 | 25.94 ± 2.43a | 28.15 ± 2.02 |
| 1.00 ± 0.04 | 0.69 ± 0.02 | 30.72 ± 1.87a | 29.67 ± 2.42 | |
| 2.01 ± 0.06 | 1.44 ± 0.03 | 28.43 ± 1.53a | 28.36 ± 1.65 | |
Effects of atractylenolide III on plasma protein binding of metoprolol
The statistical results showed that atractylenolide III was unlikely to affect the plasma protein binding of metoprolol (Table 7).![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) ± s, n = 3)
± s, n = 3)
| Drug | Ctotal (μg mL−1) | Cfree (μg mL−1) | Plasma protein binding (%) (with atractylenolide III) | Plasma protein binding (%) (without atractylenolide III) |
|---|---|---|---|---|
| a p > 0.05, vs. metoprolol without atractylenolide III. | ||||
| Metoprolol | 0.51 ± 0.02 | 0.37 ± 0.02 | 27.19 ± 2.07a | 28.15 ± 2.02 |
| 1.00 ± 0.03 | 0.70 ± 0.02 | 30.96 ± 1.84a | 29.67 ± 2.42 | |
| 2.01 ± 0.05 | 1.40 ± 0.08 | 30.22 ± 4.14a | 28.36 ± 1.65 | |
Effects of liquiritin on plasma protein binding of metoprolol
Table 8 shows that the plasma protein binding of metoprolol (0.5 μg mL−1) might be influenced by liquiritin (2 μg mL−1), while the plasma protein binding of metoprolol at concentrations of 1 and 2 μg mL−1 was not affected. The statistical results showed that liquiritin was unlikely to affect the plasma protein binding of metoprolol (Table 8).![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) ± s, n = 3)
± s, n = 3)
| Drug | Ctotal (μg mL−1) | Cfree (μg mL−1) | Plasma protein binding (%) (with liquiritin) | Plasma protein binding (%) (without liquiritin) |
|---|---|---|---|---|
| a p > 0.05, vs. metoprolol without liquiritin. | ||||
| Metoprolol | 0.50 ± 0.02 | 0.37 ± 0.01 | 26.01 ± 2.37a | 28.15 ± 2.02 |
| 1.00 ± 0.03 | 0.71 ± 0.03 | 29.19 ± 2.66a | 29.67 ± 2.42 | |
| 1.99 ± 0.04 | 1.39 ± 0.03 | 29.95 ± 1.60a | 28.36 ± 1.65 | |
Effects of glycyrrhizic acid on plasma protein binding of metoprolol
The statistical results showed that glycyrrhizic acid was unlikely to affect the plasma protein binding of metoprolol (Table 9).![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) ± s, n = 3)
± s, n = 3)
| Drug | Ctotal (μg mL−1) | Cfree (μg mL−1) | Plasma protein binding (%) (with glycyrrhizic acid) | Plasma protein binding (%) (without glycyrrhizic acid) |
|---|---|---|---|---|
| a p > 0.05, vs. metoprolol without glycyrrhizic acid. | ||||
| Metoprolol | 0.49 ± 0.02 | 0.35 ± 0.02 | 27.86 ± 2.53a | 28.15 ± 2.02 |
| 1.00 ± 0.03 | 0.72 ± 0.02 | 28.34 ± 1.74a | 29.67 ± 2.42 | |
| 1.99 ± 0.03 | 1.40 ± 0.03 | 29.61 ± 1.44a | 28.36 ± 1.65 | |
Discussion
Prediction of drug interaction in BSA by molecular docking
Molecular docking is essential for studying drug–target interactions, providing information on possible drug–target interactions and predicting drug–herb protein binding interactions.28 The binding of drugs to plasma proteins can also be assessed using this method. For the first time, we have adopted molecular docking to predict the effect of LGZGD on the plasma protein binding of metoprolol in BSA. The molecular docking results showed that metoprolol, cinnamic acid, and liquiritin can combine with the same amino acid residue, Phe 550, which means that metoprolol and cinnamic acid or liquiritin will produce competitive replacement. However, the interaction energy scores of cinnamic acid and liquiritin were lower than that of metoprolol, indicating that cinnamic acid and liquiritin had lower binding abilities than metoprolol. Therefore, the major components of LGZGD might not affect the plasma protein binding rate of metoprolol, meaning that the major components of LGZGD and metoprolol might have no competitive inhibition in clinical treatment.Reasons for selecting bovine serum albumin
Drugs need to be distributed through plasma to target tissues and play a pharmacological role after entering blood circulation.29 Serum albumin is the most abundant carrier protein in plasma and plays a vital role in the transport of various exogenous and endogenous substances, such as fatty acids, amino acids, drugs, and nutrients.30,31 BSA is used as a model protein that can bind a variety of biologically active ingredients, including drugs, natural polyphenols, and vitamins.32 BSA has an amino acid sequence very similar to that of human serum albumin (HSA), and a high homology with HSA (approximately 76%).33 BSA can better simulate the binding of drugs to plasma proteins. Therefore, BSA was used instead of HSA to study the interactions between serum albumin and drugs in this experiment.Selection of ultrafiltration technique conditions
Optimization of the ultrafiltration conditions can affect the plasma protein binding rate results. A disadvantage of ultrafiltration is that it can be susceptible to the non-specific binding (NSB) of test compounds to the polymer-constructed components of the devices.34 In this study, the NSB of metoprolol was detected and then the corrected formula of plasma protein binding was used for calculations. The incubation temperature and time can influence the speed and processes of drug–protein binding equilibrium. Therefore, the plasma protein binding of metoprolol was performed with incubation times of 15, 30, 45, and 60 min at 37 °C. The results showed that 30 min was the optimal incubation time for subsequent experiments. The rate and time of centrifugation also had an important influence on the ultrafiltration performance. Centrifugal rates of 8000, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, and 12
000, and 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g and times of 10, 15, and 20 min were tested. The results showed that 10
000 × g and times of 10, 15, and 20 min were tested. The results showed that 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g and 15 min were the optimal centrifugal conditions.35
000 × g and 15 min were the optimal centrifugal conditions.35
Plasma protein binding of metoprolol combined with the major components of LGZGD
The existence of the major components of LGZGD did not change the plasma protein binding of metoprolol. Furthermore, there were no significant differences in plasma protein binding between metoprolol and the major components of LGZGD. Molecular docking results showed that the binding sites of cinnamic acid and liquiritin were the same as that of metoprolol, but with interaction energy scores lower than that of metoprolol, indicating that there was no competitive inhibition. Glycyrrhizic acid and atractylenolide III bound to BSA through hydrogen bond formation rather than arene–arene interactions, and the interaction energy scores were also lower than that of metoprolol, indicating that they were less able to compete with metoprolol. The results of in vitro experiments further verified this molecular docking prediction.Conclusion
In conclusion, molecular docking was adopted to predict the drug–herb plasma protein binding interactions of metoprolol, and ultrafiltration was used to verify these docking results. The factors influencing ultrafiltration efficiency were exhaustively optimized. The results showed that the presence of the major components of LGZGD did not affect the plasma protein binding of metoprolol. No drug–herb interactions were observed in vitro between metoprolol and LGZGD in BSA, which indicated that this combination therapy might be safe and feasible. Further in-depth studies are needed to improve this experimental technology and enrich research indicators.Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
This work was financially supported by the Key Project Foundation of Natural Science Research in Universities of Anhui Province in China (No. KJ2017A303), the National Natural Science Foundation of China (No. 81373533), and the 2017 Anhui University of Traditional Chinese Medicine Postgraduate Science and Technology Innovation Fund (2017YB12).References
- W. R. Chen, X. M. Shi, T. S. Yang, L. C. Zhao and L. G. Gao, J. Interv. Card. Electrophysiol., 2013, 36, 267–272 CrossRef PubMed.
- H. Fröhlich, L. Torres, T. Täger, D. Schellberg, A. Corletto, S. Kazmi, K. Goode, M. Grundtvig, T. Hole and H. A. Katus, Clin. Res. Cardiol., 2017, 106, 711–721 CrossRef PubMed.
- P. A. Heidenrich, T. T. Lee and B. M. Massie, J. Am. Coll. Cardiol., 1997, 30, 27–34 CrossRef.
- P. Li, Y. Fan, Y. Wang, Y. Lu and Z. Yin, Sci. Rep., 2015, 5, 14866 CrossRef CAS PubMed.
- K. Vuignier, J. Schappler, J. L. Veuthey, P. A. Carrupt and S. Martel, Anal. Bioanal. Chem., 2010, 398, 53–66 CrossRef CAS PubMed.
- J. Ghuman, P. A. Zunszain, I. Petitpas, A. A. Bhattacharya, M. Otagiri and S. Curry, J. Mol. Biol., 2005, 353, 38–52 CrossRef CAS PubMed.
- M. Li, X. Chen, S. Hu, R. Wang, X. Peng and X. Bai, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2016, 1031, 139–145 CrossRef PubMed.
- Y. Wang, G. Xie, Q. Liu, X. Duan, Z. Liu and X. Liu, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2018, 1072, 355–361 CrossRef PubMed.
- J. H. Zhang, R. Wang, H. Xie, Q. Yin, Z. P. Jia and W. B. Li, J. South. Med. Univ., 2014, 34, 1616–1620 CAS.
- M. L. Qiu, J. Y. Mao, J. Y. Wang, X. L. Wang and Y. Z. Hou, Chin. J. Exp. Tradit. Med. Formulae, 2011, 17, 243–247 Search PubMed.
- Z. Song, K. Bi and X. Luo, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2003, 788, 387–391 CrossRef CAS.
- X. Q. Li, X. Y. Gen, X. Y. Wang, H. Y. Jiang and Z. Lai, Zhonghua Zhongyiyao Zazhi, 2005, 20, 220–222 Search PubMed.
- Y. L. Liu and G. X. Xu, Zhonghua Zhongyiyao Zazhi, 2012, 36, 600–602 Search PubMed.
- H. Y. Fang, J. L. Huang, F. F. Sang, T. S. Wang, L. Wang, X. Y. Cheng, J. Yuan, Z. J. Long and X. H. Liu, Anhui Zhongyi Xueyuan Xuebao, 2010, 29, 53–55 Search PubMed.
- J. L. Huang, L. Wang, H. Shi and X. Y. Hou, Effect of Lingguizhugan decoction on myocardial nuclear factor kappa B protein expression in rats with chronic heart failure, J. Tradit. Chin. Med., 2011, 33, 343–348 CrossRef.
- H. Shi, S. Xu, L. Wang, J. L. Huang, L. Liu, Y. Y. Liu and J. Peng, J. Chin. Med. Mater., 2017, 40, 680–683 Search PubMed.
- H. Shi, L. Wang, J. L. Huang, S. Xu, L. Liu, Y. Y. Liu and J. Peng, Zhongguo Zhongxiyi Jiehe Zazhi, 2017, 37, 1215–1219 Search PubMed.
- S. F. Liu, Clin. J. Chin. Med., 2017, 20, 35–36 Search PubMed.
- H. H. Zheng, Q. Fang and J. Y. Dong, Chin. Modern. Doct., 2013, 51, 58–60 Search PubMed.
- T. Bohnert and L. S. Gan, J. Pharm. Sci., 2013, 102, 2953–2994 CrossRef CAS PubMed.
- N. M. Mostafa, M. L. Ashour, O. A. Eldahhan and A. N. Singab AN, Nat. Prod. Res., 2015, 30, 2093–3000 CrossRef PubMed.
- A. Ali, Z. Ashraf, N. Kumar, M. Rafiq, F. Jabeen, J. H. Park, K. H. Choi, S. Lee, S. Y. Seo, E. H. Choi and P. Attri, Sci. Rep., 2016, 6, 21779 CrossRef CAS PubMed.
- J. K. Maier and P. Labute, Proteins, 2014, 82, 1599–1610 CrossRef CAS PubMed.
- J. L. Liu, Y. L. He, D. Liu, Y. He, Z. P. Tang, H. Lou, Y. P. Huo and X. Y. Cao, RSC Adv., 2018, 8, 7280–7286 RSC.
- M. Kurlbaum and P. Högger, J. Pharm. Biomed. Anal., 2011, 54, 127–132 CrossRef CAS PubMed.
- J. Chen, H. Wu, G. B. Xu, M. M. Dai, S. L. Hu, L. L. Sun, W. Wang, R. Wang, S. P. Li and G. Q. Li, J. Pharm. Biomed. Anal., 2015, 108, 122–128 CrossRef CAS PubMed.
- D. Q. Tang, Y. J. Li, Z. Li, T. T. Bian, K. Chen, X. X. Zheng, Y. Y. Yu and S. S. Jiang, Biomed. Chromatogr., 2015, 29, 1137–1145 CrossRef CAS PubMed.
- E. D. Coelho, J. P. Arrais and J. L. Oliveira, PLoS Comput. Biol., 2016, 12, e1005219 CrossRef PubMed.
- R. Muslim, A. Aftab, Y. Feng, K. Zahid, J. Yang, W. Yun, R. Saleem, W. Wang, U. Faheem and Q. Yuan, J. Photochem. Photobiol., B, 2017, 170, 6–15 CrossRef PubMed.
- X. Y. Cao, Y. L. He, D. Liu, Y. He, X. Hou, Y. Cheng and J. L. Liu, RSC Adv., 2018, 8, 25519 RSC.
- X. Pan, P. Qin, R. Liu and J. Wang, J. Agric. Food Chem., 2011, 59, 6650–6656 CrossRef CAS PubMed.
- X. Peng, X. Wang, W. Qi, R. Huang, R. Su and Z. He, Food Funct., 2015, 6, 2712–2726 RSC.
- Y. Wu, H. Cheng, Y. Chen, L. Chen, Z. Fang and L. Liang, J. Agric. Food Chem., 2017, 65, 3019–3030 CrossRef CAS PubMed.
- S. Taylor and A. Harker, J. Pharm. Biomed. Anal., 2006, 41, 299–303 CrossRef CAS PubMed.
- H. S. Larsen, P. K. Chin, E. J. Begg and B. P. Jensen, Bioanalysis, 2011, 3, 843–852 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra07153e |
| This journal is © The Royal Society of Chemistry 2018 |
