Open Access Article
Open Access ArticleChalcopyrite bioleaching of an in situ leaching system by introducing different functional oxidizers
Caoming Huangab,
Chong Qinac,
Xue Fenga,
Xueduan Liuac,
Huaqun Yinac,
Luhua Jiangac,
Yili Liangac,
Hongwei Liuac and
Jiemeng Tao *ac
*ac
aSchool of Minerals Processing and Bioengineering, Central South University, Changsha, 410083, China. E-mail: 175601003@csu.edu.cn
bChina Nonferrous Metal Mining (Group) Co., Ltd., Beijing, China
cKey Laboratory of Biometallurgy, Ministry of Education, Changsha, 410083, China
First published on 2nd November 2018
Abstract
Introducing exogenous species to an indigenous microbial community is an effective way to reveal the connections between metabolic processes, ecological function and microbial community structure. Herein, three different functional consortia (ferrous oxidizers, sulfur oxidizers and ferrous/sulfur oxidizers) were added to a natural leaching solution system derived from Zijin copper mine, China. The leaching experiment showed that the copper extraction rate of the community invaded by a sulfur-oxidizing consortium was 50.40% higher than that of the indigenous leachate at the endpoint of bioleaching. The variations of ferrous content, total iron, pH and redox potential in leachates also provided evidence that the community with exogenous sulfur oxidizers was more efficient. XRD analysis demonstrated that a proper addition of the sulfur-oxidizing consortium could eliminate sulfur passivation, promote production of chalcocite and enhance leaching. Furthermore, an exogenous ferrous-oxidizing consortium and a sulfur-oxidizing consortium greatly changed the community structure and microbial succession and promoted the cell growth rate during the bioleaching process, while ferrous/sulfur oxidizers showed no obvious effects on the indigenous community. Exogenous ferrous oxidizers, mainly L. ferriphilum, and sulfur oxidizers, mainly A. thiooxidans, successfully established and colonized in the indigenous community. However, only colonized A. thiooxidans, rather than L. ferriphilum, showed advantageous enhancement in the dissolution of chalcopyrite. Results indicated that exogenous sulfur oxidizer A. thiooxidans, which was scarce in the indigenous community, could easily colonize in the indigenous community, significantly change the community structure, sufficiently execute its function, and greatly enhance copper dissolution.
Introduction
With the continuous consumption of high-grade minerals, the demand for exploitation of low-grade ores has been increasingly urgent. In recent decades, bioleaching has been considered as a successful metallurgical technology to extract precious and base metals from low-grade ores,1 such as secondary copper sulfides, pyritic gold ores or nickel-copper sulfide ores.2,3 Bioleaching systems constitute suitable habitats and niches for the colonization of a diversity of microorganisms. To date, more than 40 types of acidophilic species have been detected in bioleaching systems.4 Based on different functional performances, bioleaching microorganisms can be assigned as ferrous-oxidizing acidophiles (mainly Leptospirillum ferriphilum and Ferroplasma thermophilum), sulfur-oxidizing acidophiles (mainly Acidithiobacillus caldus and Acidithiobacillus thiooxidans) and ferrous/sulfur-oxidizing acidophiles (mainly Acidithiobacillus ferrooxidans and Sulfobacillus thermosulfidooxidans).5 Ferrous-oxidizing acidophiles can dissolve sulfide ores to produce ferric iron, which further attack minerals to generate sulfur or polysulfide on the ore surface. Complementary to this, sulfur-oxidizing acidophiles can consume elemental sulfur or polysulfide to drive the bioleaching process.6 Currently, many researchers have turned their attention on microbial communities owing to the close relationships between functional oxidizers, environmental factors and bioleaching performances.7,8 Sulfur and iron oxidizers are often jointly used to improve the bioleaching efficiency.9,10Due to the ever-changing physical and chemical parameters in leachates, the types of microorganisms in different bioleaching systems or in different phases under the same bioleaching condition are always differential.8,11 Under two subsystems in the Zijin Shan copper mine (Fujian province, China), Acidithiobacillus was the dominant genus in the leaching heap system, while Leptospirillum owned the competitive advantage in the leaching solution system.12 In a successive process of chalcopyrite bioleaching, At. caldus, Sulfobacillus acidophilus and F. thermophilum sequentially dominated the system from the initial to the end phase.13 The dynamic changes in different microorganisms played different roles in chalcopyrite dissolution and their synergistic functions were tightly related to the efficiency of chalcopyrite bioleaching.10 However, in a natural ecosystem, microbial invasion is an extremely common scenario. As a dynamic and open environment, such a bioleaching system could be more frequently affected by exotic species. Alien or exotic species can in some cases change the structure and functioning of an entire ecosystem.14 Our previous study has demonstrated that when an exogenous strain At. thiooxidans A01 was introduced to a consortium of pyrite bioleaching, the functional gene expression, structure and function of the indigenous consortium changed, and 10.7% increase in bioleaching rate was observed.15 It has also been reported that the introduction of F. thermophilum into the defined microbial consortium (At. caldus and L. ferriphilum) accelerated the dissolution of the mineral and caused significant differences in the planktonic and attached population dynamics.16 These examples provided more information for the study of exogenous acidophiles on artificial microbial communities in a bioleaching system. However, to date, only a few studies have reported the effects of exogenous acidophiles on the community structure and function in an in situ copper mine system. A study on an in situ copper mine system could be more practical and applicable for understanding natural bioleaching systems.17
In our previous study, the differences in microbial structure and function between leaching heap (LH) and leaching solution (LS) in Zijin Shan copper mine (Fujian province, China) were investigated. Bioleaching experiment showed that microbial communities in LH had stronger leaching ability, while mineral extraction efficiency was significantly positively correlated with sulfur-oxidizing microbes.12 Therefore, we suspected that additional oxidizing microbes could enhance the bioleaching efficiency of LS. To explore the effects of exogenous microorganisms on chalcopyrite bioleaching, acidophiles with different oxidative functions (ferrous oxidizers, sulfur oxidizers and ferrous/sulfur oxidizers) were introduced into in situ microbial communities of LS. Through observing the changes in environmental properties and microbial communities, this study can further reveal the mechanism of chalcopyrite bioleaching upon coupling with exogenous functional oxidizers.
Materials and methods
Chalcopyrite samples
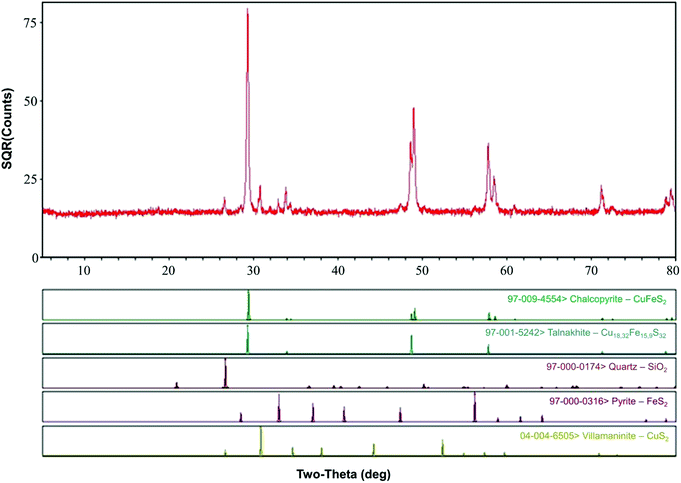
Chalcopyrite samples used in this study were obtained from Guangxi, China. The ore samples were prepared by crushing, hand-sorting to remove the gangue minerals, grinding and dry-sieving to 0.037–0.074 mm (200–400 mesh grid, Tyler series). After washing with 2 M HCl, distilled water and pure ethanol, the ore samples were dried and placed in a 125 mL vacuum desiccator at room temperature for further study. Chemical analysis by inductively coupled plasma-atomic emission spectrometry (ICP-AES, PS-6, Baird, USA) showed that the samples contained Fe (28.70%), Cu (33.10%) and S (35.40%). X-ray diffraction (XRD, D/Max 2500, Rigaku, Japan) pattern (Fig. 1) showed that the main mineral phase was chalcopyrite.Microorganisms and culture media
Six typical acidophilic strains used in this study were provided by the Key Lab of Biometallurgy of Ministry of Education, Central South University. The strains include two ferrous-oxidizers (L. ferriphilum DX2012 and F. thermophilum DX2012), two sulfur-oxidizers (A. caldus DX2012 and A. thiooxidans DX2012) and two ferrous/sulfur-oxidizers (A. ferrooxidans DX2012 and S. thermosulfidooxidans DX2012). All strains were cultured aerobically in sterile 9 K medium (autoclaved at 121 °C for 20 min) comprising (NH4)2SO4 (3.0 g L−1), K2HPO4 (0.5 g L−1), KCl (0.1 g L−1), Ca(NO3)2 (0.01 g L−1), and MgSO4·7H2O (0.5 g L−1). The energy sources were FeSO4·7H2O (44.7 g L−1) for ferrous-oxidizers, S0 (10 g L−1) for sulfur-oxidizers and FeSO4·7H2O (22.4 g L−1) and S0 (5 g L−1) for ferrous/sulfur-oxidizers. In addition, 0.02% (w/v) yeast extract was added for S. thermosulfidooxidans and 0.01% (w/v) yeast extract was added for F. thermophilum. Detailed culture conditions of acidophiles were referenced from a previous study.15Preparation of indigenous leaching communities
Original leaching samples were collected from a leaching solution (LS) of the Zijin Shan copper mine in Fujian province, China. First, microorganisms from LS were collected by filtering the solution with 0.22 μm-filter paper. Then, the microorganisms were cultivated in 9K medium at pH 2.0 and 30 °C for three successive generations with three different energy substrates: (i) FeSO4·7H2O (44.7 g L−1); (ii) S0 (10 g L−1); and (iii) FeSO4·7H2O (22.4 g L−1) + S0 (5 g L−1) + yeast extract (0.2 g L−1).18 Finally, microbial cultures from different media were mixed in a flask. The mixed cultures were inoculated into 500 mL-shake flasks with 250 mL 9K medium and 2% chalcopyrite (w/v) for 30 days for three successive generations. Then, the indigenous leaching microbial communities were obtained by filtering, centrifuging and washing mineral residues from the third leachate.Bioleaching experiments
Bioleaching experiments were performed in 500 mL-shake flasks, each containing 250 mL sterilized 9K medium and 2% pulp density (w/v); the initial pH was adjusted to 2.0 by adding 18 M sulfuric acid. The initial cell density of indigenous microorganisms was 1.0 × 107 cells per mL for inoculated systems. The cell density of each exogenous consortium was 1.0 × 108 cells per mL (e.g., 0.5 × 108 cells per mL for L. ferriphilum and 0.5 × 108 cells per mL for F. thermophilum). These consortiums were introduced to the indigenous leaching systems prior to the log phase (at 6th day). Experimental systems were classified into four series: indigenous community (system A), indigenous community + ferrous-oxidizers of L. ferriphilum and F. thermophilum (system B), indigenous community + sulfur-oxidizers of A. caldus and A. thiooxidans (system C) and indigenous community + ferrous/sulfur-oxidizers of A. ferrooxidans and S. thermosulfidooxidans (system D). Furthermore, one abiotic control was set up for each system. Rotating speed and temperature of the rotary shaker were set at 175 rpm and 30 °C, respectively. During the leaching process, evaporation loss was compensated by adding water of pH = 2.0 and sampling loss was compensated by periodically supplementing with sterile 9K medium to maintain the volume of the system.Analytical methods
Physicochemical parameters, such as pH, oxidation-redox potential (ORP) and the concentration of dissolved metal ions in bioleaching systems, were measured every three days. pH value was measured by a digital pH meter (PHSJ-4A, Leici, Shanghai, China). ORP was monitored by a Pt meter (213-01, Leici, Shanghai, China) using an Ag/AgCl reference electrode. The supernatant of each treatment was collected to detect cell density and the concentrations of dissolved copper ions and ferrous ions in solution. Cell density was measured using blood cell counting chambers (XB-K-25, QiuJing, Shanghai, China). Copper concentration was assayed based on the color reaction with bis(cyclohexanone) oxalyldihydrazone.19 Ferrous iron concentration was analyzed using the 1,10-phenanthroline spectrophotometry assay.20In the bioleaching process, 5 mL of bioleaching solution was collected from each flask on the 0th, 6th, 12th, 18th, 24th, 30th and 36th day for each system. Samples were centrifuged at 2000 × g for 2 min to separate the supernatant from the bottom sediment, following which the supernatant was collected. Then, the sedimented ore samples were re-suspended and shaken on a vortexer by adding 5.0 mL of sterile basal media several times until no cells could be detected under the microscope. Finally, the cells were collected by centrifuging the mixed supernatants at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min. Subsequently, total DNA from each sample was extracted with a TIANamp® Bacteria DNA kit (Tiangen Biotech, Co. Ltd., Beijing, China) and analyzed by 1% (w/v) agarose gel electrophoresis. Independent PCR reactions were conducted to amplify the V4 hyper variable region of the 16S rDNA (∼250 nucleotides) with the primer set 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′).21 Sequencing was performed on the Illumina MiSeq platform (Illumina, San Diego, CA, USA) using a 500 cycles kit (2 × 250 bp paired-ends) at the Key Laboratory of Biometallurgy of Ministry of Education in Central South University. Raw sequence data were processed on an in-house galaxy pipeline developed by Prof. Zhou's lab (http://zhoulab5.rccc.ou.edu/), similarly to our previous studies.22 Through quality trimming (using Btrim to remove sequencing adaptors and low quality regions), merging and clustering (using CD-HIT algorithm), the samples were rarefied to a depth of 10
000 × g for 10 min. Subsequently, total DNA from each sample was extracted with a TIANamp® Bacteria DNA kit (Tiangen Biotech, Co. Ltd., Beijing, China) and analyzed by 1% (w/v) agarose gel electrophoresis. Independent PCR reactions were conducted to amplify the V4 hyper variable region of the 16S rDNA (∼250 nucleotides) with the primer set 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′).21 Sequencing was performed on the Illumina MiSeq platform (Illumina, San Diego, CA, USA) using a 500 cycles kit (2 × 250 bp paired-ends) at the Key Laboratory of Biometallurgy of Ministry of Education in Central South University. Raw sequence data were processed on an in-house galaxy pipeline developed by Prof. Zhou's lab (http://zhoulab5.rccc.ou.edu/), similarly to our previous studies.22 Through quality trimming (using Btrim to remove sequencing adaptors and low quality regions), merging and clustering (using CD-HIT algorithm), the samples were rarefied to a depth of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 sequences per sample and clustered into 289 operational taxonomic units (OTUs) by a 97% identity cutoff using UCLUST.23 Taxonomic assignment was conducted through the RDP Classifier24 with a minimal 50% confidence estimate. Raw data of the high-throughput sequencing used to characterize community variations have been submitted to the Sequence Read Archive (SRA) of NCBI, and the project ID is SRP 141342.
000 sequences per sample and clustered into 289 operational taxonomic units (OTUs) by a 97% identity cutoff using UCLUST.23 Taxonomic assignment was conducted through the RDP Classifier24 with a minimal 50% confidence estimate. Raw data of the high-throughput sequencing used to characterize community variations have been submitted to the Sequence Read Archive (SRA) of NCBI, and the project ID is SRP 141342.
Furthermore, real-time quantification polymerase chain reaction (RT-qPCR) was introduced to clarify the different strains of Acidithiobacillus spp. and further reveal community succession during the bioleaching process. RT-qPCR primers for A. caldus, A. thiooxidans and A. ferrooxidans were designed by Primer Premier 5.0 and synthesized by BioSune (BioSune Biological Engineering Technology & Services Co. Ltd., Shanghai, China). Before RT-qPCR, conventional PCR was performed and the products were diluted serially from 104 to 1010 copies per mL and amplified by RT-qPCR to construct standard curves. The detailed procedure for RT-qPCR was performed on an iCycler iQ Real-time PCR detection system (Bio-Rad Laboratories Inc., Hercules, USA) as per the method described in a previous study.10
Data analysis
All graphs and charts were generated using Origin 9.0 or R v. 3.4.0. Statistical differences in experimental data were determined by a one-way analysis of variance (ANOVA), followed by the Least Significant Difference (LSD) test in SPSS 22.0 (SPSS Inc., Chicago, USA). Detrended correspondence analysis (DCA) and heat maps were used to compare the different community structures. DCA was performed in R v. 3.4.0 (ref. 25) with the vegan package.26 Heat mapping was performed in the environment of statistical analysis of metagenomic profiles (STAMP).27Results and discussion
Variation in main physicochemical parameters in different leachates
As shown in Fig. 2, exogenous acidophiles had a great influence on the variation in physicochemical parameters in indigenous leachate. The whole course in leachate could be divided into three phases: strain-adaptive growing phase (SAG, 0–9th d), rapidly increasing phase (RIC, 9–18th d) and stationary phase (STA, 18–36th d). During the bioleaching process, ferrous concentrations increased rapidly in the early stage for all four treatments, and then decreased sharply until zero (Fig. 2a). Compared with the abiotic control, system D first reached to its peak ferrous concentration (549 ± 24 mg L−1) at day 9, followed by system B (602 ± 24 mg L−1) and system C (675 ± 26 mg L−1), and system A was the last one reach its highest ferrous concentration (603 ± 21 mg L−1) at day 15. It has been reported that a high content of introduced ferrous oxidizers could enhance the iron metabolism during bioleaching and yield more Fe from the mineral.10 However, the ferrous concentration in system D decreased first, followed by systems B, C and A, which indicated that the ferrous oxidative ability of system D was much stronger than that of the other systems. Herein, the stronger ferrous oxidative ability of ferrous/sulfur oxidizers (A. ferrooxidans and S. thermosulfidooxidans) might be attributed to the fact that the ferrous oxidative genes were stimulated and sufficiently activated by long-term ferrous substrate domestication in our laboratory. Moreover, it is generally accepted that iron metabolism could have been enhanced via the synergistic effect with sulfur metabolism.28,29 Therefore, the ferrous-oxidizing ability of ferrous/sulfur oxidizers could be more efficient under the stimulation of their sulfur oxidation. Ferrous oxidative ability has been related to several factors, such as microbial types, cell growth rate and environmental parameters, rather than a single variable.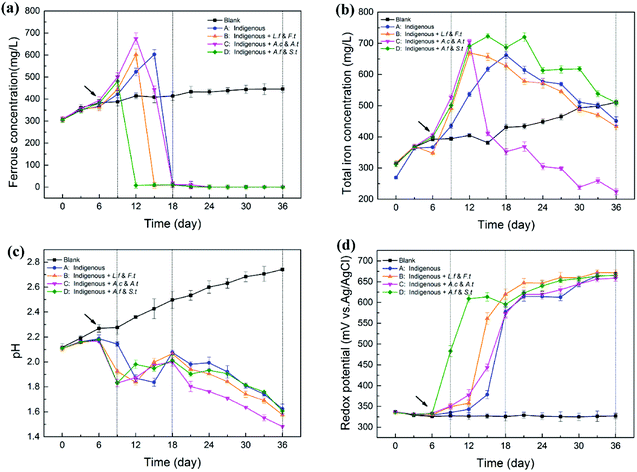 | ||
| Fig. 2 Variation (mean ± SD) in physicochemical parameters in leachates by introducing different functional consortia ((a) ferrous ions, (b) total iron, (c) pH, (d) ORP). | ||
During the whole process, total iron concentration in abiotic control slowly and gradually increased, but the variation in the experimental treatments was high, particularly for system C (Fig. 2b). Total iron concentration in the four experimental systems increased rapidly from day 6, suggesting that the dissolution of chalcopyrite was significantly accelerated with the introduction of exogenous cells.30 Differences among the four systems appeared from the RIC phase onwards. In this period, total iron concentration increased rapidly to a maximum value and then decreased. Remarkably, the concentration of total iron in system C decreased at a much faster rate than that in systems B, A and D from the 12th day. Finally, the concentration of total iron in system C declined to 224 ± 9 mg L−1, while that in systems B, A and D declined to 433 ± 11 mg L−1, 451 ± 14 mg L−1 and 509 ± 11 mg L−1, respectively. In the leaching solution, a decrease in total iron was mainly due to the transformation of Fe3+ to jarosite. System C holds more sulfur-oxidizing bacteria and produced more SO42−, which might drive the reaction between Fe3+ and SO42−.
pH of the abiotic control increased gradually with the leaching process. However, in the four experimental leachates with inoculated microorganisms, pH declined in general, although with fluctuations (Fig. 2c). In the early stage, pH slightly increased before the 6th day, which might be caused by the consumption of protons through the direct chalcopyrite bio-oxidation during initial bioleaching. pH of systems B, C and D appeared to rapidly decrease from the 6th day, while the pH of system A began to decrease rapidly from 9th day, which might be attributed to the addition of bacteria that accelerated the dissolving process of the mineral and produced more acid products. Among the introduced systems, C and D showed lower pH than B because the exogenous cells in C and D exhibited a sulfur-oxidizing function. This observation was in accordance with the consideration that more active sulfur oxidizers can produce more acid and cause a decrease in pH by enhancing the oxidation of elemental sulfur or sulfide ores.31 In the STA phase, the pH value maintained a modest downtrend and the value in system C was always significantly lower than that in other systems . At the endpoint of bioleaching, pH in systems A, B, C and D dropped to 1.62, 1.58, 1.48 and 1.60, respectively.
Compared with the abiotic control, ORP of the four experimental groups increased continuously and then reached a plateau (about 660 mV) in the later stage of the bioleaching process (Fig. 2d). The solution potential is a determinant of the chalcopyrite biohydrometallurgy system.32 The variation of ORP in different leaching systems was closely related to the increase in ratio of Fe3+/Fe2+ during the process.33 Ferrous concentration of system D decreased from 549 ± 24 mg L−1 to 8 mg L−1 from day 9 to day 12 and finally to 0 mg L−1 at day 24, during which ORP of system D increased from 483 ± 13 mV to 609 ± 9 mV and finally to 666 ± 7 mV. Similarly, the variation of ORP in the other three systems also showed a steadily inverse correlation with ferrous concentration, but with changed times, exhibiting varying degrees of latency: system B from day 12 to day 15, system C from day 12 to day 18 and system A from day 15 to day 18. Many studies have shown that the passivation of chalcopyrite occurred quickly if the initial solution potential was too high.32 Similarly, in the STA phase, ORP of the four treatments reached a stable value of about 670 mV, which indicated that iron started to precipitate and consequently inhibit copper dissolution.
Bioleaching efficiency of different leachates and XRD analysis of bioleaching residues
As shown in Fig. 3, copper extraction by all the four treatments continually increased during the entire leaching time. In the SGA phase, there was no significant difference among the four treatments. After exogenous consortia were introduced, chalcopyrite in the introduced systems showed a faster dissolution rate, particularly for system C. Over the course of bioleaching, the differences between systems A, B and D diminished gradually, while system C always maintained a higher dissolution rate. In the latter stage, the extraction rate of copper ions became slower owing to higher jarosite yield, which further formed a passivation layer on the ore surface to block the continuous copper extraction.29,34 After leaching for 36 days, a maximum of 49.48% of copper was released from chalcopyrite in system C, followed by system B (34.94%), system A (32.90%) and system D (30.81%). These results indicated that the oxidative dissolution of chalcopyrite was more extensive with exogenous sulfur-oxidizing bacteria Acidithiobacillus. In our previous studies, it has been demonstrated that the ability of pyrite bioleaching was significantly and positively correlated with Acidithiobacillus,12 which suggested that higher abundance of Acidithiobacillus could enhanced mineral bioleaching, which is in line with our results.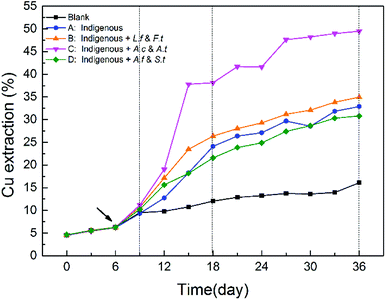 | ||
| Fig. 3 Differences in copper extraction efficiency in different systems during the whole bioleaching process. | ||
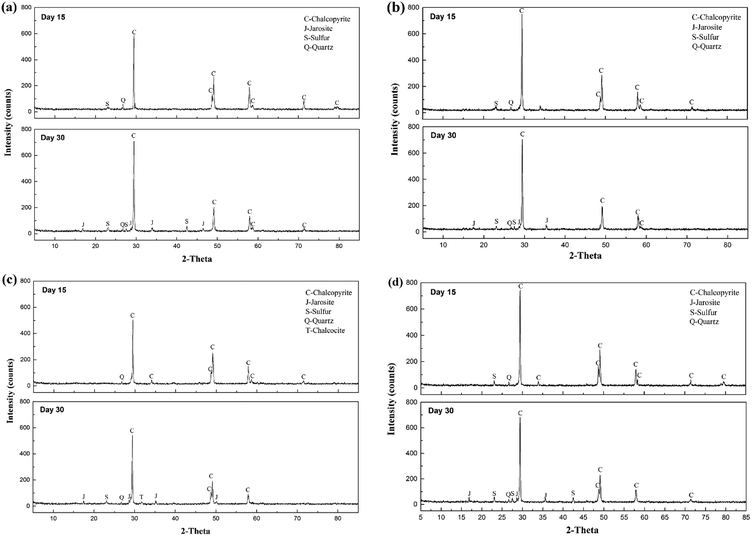
XRD analysis was performed to investigate the composition of the ore residues after bioleaching for 15 and 30 days (Fig. 4). At day 15, more distinct diffraction peaks of chalcopyrite were detected in systems A and D, confirming that chalcopyrite was leached faster in systems B and C. In addition, the peak ascribed to elemental sulfur was observed in the XRD patterns of residues of systems A, B and D, but no such peak appeared in the XRD pattern of the residue of system C, which indicated that most of the sulfur was consumed by the diverse sulfur-oxidizing bacteria in system C. At day 30, jarosite was observed in the residues of all four systems, which was the main factor that inhibited further copper dissolution. Moreover, fewer peaks of elemental sulfur and chalcopyrite were detected in the XRD pattern of the residue of system C, demonstrating its high ability for mineral dissolution and sulfur oxidization. Interestingly, an inconspicuous peak of chalcocite was also observed in system C residue at day 30, implying the reduction of chalcopyrite. A two-step model of chalcopyrite reduction has been proposed in the past.35 This model suggested that chalcopyrite could be reduced by ferrous ions in the presence of cupric ions to form chalcocite at relatively low redox potentials. Then, chalcocite, which was more amenable to leaching than chalcopyrite, was oxidized to cupric ions and elemental sulfur by ferric ions or oxygen. Combined with the redox potential variation, systems A and C seemed to be more liable to the reduction of chalcopyrite. However, no chalcocite was observed in system A residue, which might be owing to small concentration in the feed or fast oxidation by cupric ions.36
Microbial community succession in leachates
As important members of the bioleaching systems, the dynamics of acidophilic microorganisms in a community were closely related to the dissolution process of minerals. Therefore, microbial community succession was investigated by 16S rDNA sequencing and RT-qPCR. A total of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rarified 16S rRNA gene sequences were obtained and clustered into 289 operational taxonomic units (OTUs) at a 97% similarity threshold. On this basis, the effects of exogenous species on microbial structure and function and the relationships between microbial structure and bioleaching performance were analyzed.
000 rarified 16S rRNA gene sequences were obtained and clustered into 289 operational taxonomic units (OTUs) at a 97% similarity threshold. On this basis, the effects of exogenous species on microbial structure and function and the relationships between microbial structure and bioleaching performance were analyzed.
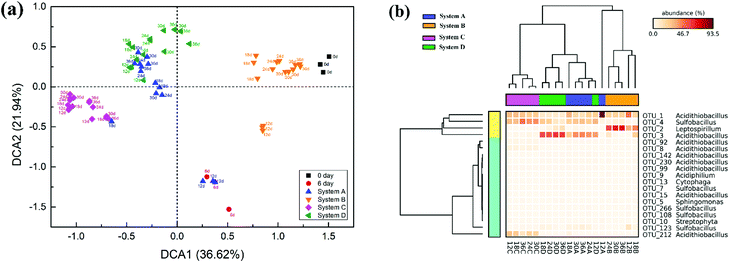
Detrended correspondence analysis (DCA) was performed to portray the overall succession of microbial communities in response to different exogenous oxidizing bacteria (Fig. 5a). At the beginning, the distribution of communities at day 6 showed noticeable deviation from the original communities (day 0) due to intensified competition among the bacteria. After the exogenous consortia were introduced, the samples at different time points in the same system clustered together neatly, but the distance between the samples in system A and system D was less pronounced. Compared with system B, the samples in system C clustered more closely during the whole bioleaching process, indicating a more stable community in system C. Furthermore, cluster analysis was conducted for the 19 abundant OTUs (relative abundance >1.0%) in leachates (Fig. 5b). The obtained results also indicated that exogenous ferrous/sulfur oxidizers did not cause significant changes to the microbial structure, but the structures of microbial communities invaded by sulfur or ferrous oxidizers were significantly different from that of the indigenous leachate. It was noticeable that there exists unique abundant OTUs in different systems; e.g., OTU_3 was more abundant in system A and D, OTU_2 was more abundant in system B and OTU_212 was more abundant in system C. These unique species might have contributed to the dissolution of mineral by different mechanisms.
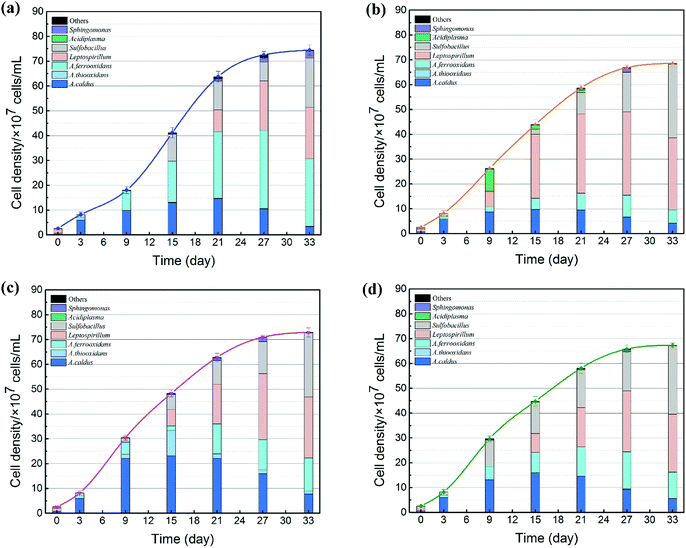
The succession of microbial community is shown in Fig. 6. On the one hand, the cell growth rate showed significant differences due to different exogenous species. During the SAG phase, minor amounts of nutrient substrates were released from the ore for cell growth.37 Therefore, cell growth rate was still weak. With the production of ferrous ions, elemental sulfur and other metabolic intermediates (such as CuS, CuS2, polysulfide and extracellular secretions),38 bacteria with different functions were stimulated to accelerate the bioleaching process; thus, microbial growth increased significantly in the RIC phase. As expected, the introduction of exogenous consortia accelerated the processes of cell growth and mineral dissolution. System C maintained the maximum cell number during the whole bioleaching process. At the endpoint of bioleaching, cell density of the four systems reached 7.1 × 108, 7.9 × 108, 8.2 × 108 and 7.6 × 108 cells per mL. The cell density of system C was approximately 15% higher than that in the indigenous community. These results indicated that exogenous species, particularly sulfur-oxidizing species, had stronger growth activity and produced a competitive advantage to occupy their living niche.
On the other hand, the proportion of the microbial sequences could be assigned to different taxonomic ranks with distinct taxonomic resolution (Fig. 6). The introduction of exogenous species caused drastic changes in the microbial composition. During the first 6 days, the predominant genus in leachates changed from Leptospirillum (72.63%) to A. caldus (85.33%). Compared with the indigenous community, the microbial compositions of systems B and C changed significantly after the exogenous oxidizers were introduced. In system B, the proportion of Leptospirillum increased rapidly and finally became the dominant species. In contrast, a higher amount of sulfur oxidizers (A. caldus and A. thiooxidans) and Sulfobacillus were detected in system C. Different community succession for systems B and C indicated that exogenous species performed successful colonization in the indigenous leaching system. During the STA phase, the amount of Leptospirillum began to increase by different degrees in all the four treatments till the end of the bioleaching process, which was consistent with the previous studies which showed that L. ferriphilum had higher tolerance to high ORP and low pH conditions.39 Moreover, due to the accumulation of organics from microbial death and extracellular metabolites with time, facultative chemolithoautotroph Sulfobacillus became an important genus in the latter stage to accelerate the consumption of organics and thus lessen the toxic effect of organics on Acidithiobacillus and Leptospirillum.40 Furthermore, the variation in community succession was also related to copper recovery efficiency. Copper extraction was much higher in system C, which might be related to its special community. Similar to other systems, A. caldus played an important role in sulfur oxidization. In addition to A. caldus, exogenous A. thiooxidans performed its oxidizing function in system C. Sulfur oxidizers diminished the accumulation of sulfur on the surfaces of mineral particles and provided sulfuric acid for proton attack and maintenance of low pH.41,42 Before introducing the sulfur consortium (A. caldus and A. thiooxidans), the leachate was mainly composed of A. caldus. Therefore, it was difficult for exogenous A. caldus to settle in the indigenous community because of a high niche overlap and repulsive interaction. In contrast, A. thiooxidans seemed to present stronger ability to establish its presence in the indigenous community.
In summary, microbial community structure and composition were greatly altered by exogenous acidophiles. High amounts of sulfur oxidizing bacteria, particularly A. thiooxidans, could enhance the leaching efficiency.
Mechanism of different exogenous consortia on chalcopyrite bioleaching
A mechanism model coupling community succession and main biochemical reactions (eqn (1)–(10)) is proposed according to the different effects of the exogenous consortia on the indigenous leachate (Fig. 7). As shown in Fig. 7, the intensity of the colour (aquamarine blue) filled in the leaching systems represented the concentration of copper ions. In other words, darker the colour, higher would be the copper ion concentration. In the SAG phase, both the sulfur and iron metabolisms were still weak due to low number of cells (L. ferriphilum and A. caldus) and chemical ions. Due to limited substrates, competition among bacteria was intensified. Thus, the structure of communities significantly changed in the beginning (Fig. 5a). When microorganisms colonized on the mineral surface, chalcopyrite was dissolved through a direct mechanism, as shown in eqn (1), which was in line with increasing trend of pH during the initial bioleaching process (Fig. 2c). After a short adaptive phase, exogenous consortia with different oxidizing ability were introduced to the indigenous leachate. During the RIC phase, with the accumulation of ferric ions (according to eqn (1)), chalcopyrite was dissolved rapidly by the ferric ions (eqn (2)) and proton attack (eqn (3)). With the production of more ferrous ions and elemental sulfur (Fig. 4), iron and sulphur metabolisms were greatly enhanced in the presence of oxidizing bacteria to produce more ferric ions (eqn (4)) and sulfuric acid (eqn (6)), respectively. More detailed reaction processes for iron and sulfur metabolisms are shown below.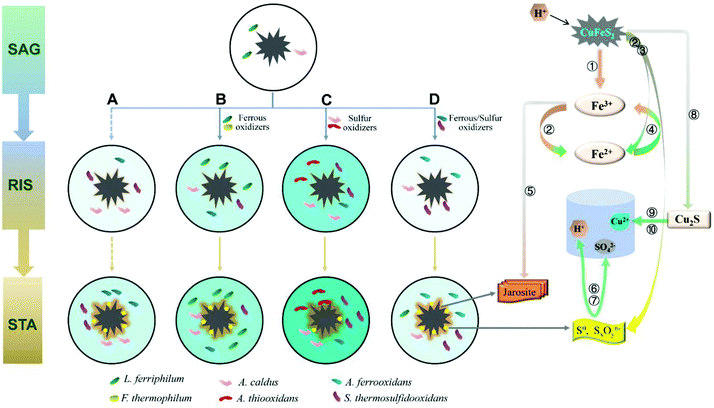 | ||
| Fig. 7 A model of the process of chalcopyrite bioleaching upon introducing different oxidizers. SAG: strain adaptive-growing phase; RIS: rapidly increasing phase; STA: stationary phase. (A) indigenous community; (B) indigenous + L. ferriphilum & F. thermophilum; (C) indigenous + A. caldus & A. thiooxidans; and (D) indigenous + A. ferrooxidans & S. thermosulfidooxidans. Numbers ①–⑩ represent eqn (1)–(10). | ||
Ferrous and ferric ions in bioleaching systems were the main reactants of iron metabolism. It has been suggested that the adsorption behavior of attached cells at the early stage is beneficial to further concentrate ferric ions and attack the chalcopyrite, according to eqn (2).43,44 Subsequently, ferrous ions formed from the former reactions (eqn (2) and (3)) were oxidized by iron oxidizers to promote the dissolution of chalcopyrite.30 Compared with the indigenous leachate, increased ferrous concentration in systems B, C and D indicated stronger iron reactions in exogenous systems (Fig. 2a). This is because the exogenous bacteria greatly increased the cell density and changed the community composition. The mechanism model showed that in the RIC phase, the iron oxidizers in systems A, C and D were mainly A. ferrooxidans and S. thermosulfidooxidans and in system B, the iron oxidizers were A. ferrooxidans and L. ferriphilum. During the STA phase, S. thermosulfidooxidans and L. ferriphilum successively performed an increasingly important role in all the four systems. During this period, the yield of tawny jarosite precipitation was greatly enhanced (Fig. 4) by the excess ferric ions (eqn (5)), which inhibited further copper dissolution.
| 2CuFeS2 + 17/2O2 + 2H+ = 2Cu2+ + 2Fe3+ + 4SO42− + H2O | (1) |
| CuFeS2 + 4Fe3+ = Cu2+ + 5Fe2+ + 2S0 | (2) |
| CuFeS2 + O2 + 4H+ = Cu2+ + Fe2+ + 2S0 + 2H2O | (3) |
| 4Fe2+ + O2 + 4H+ = 4Fe3+ + 2H2O | (4) |
| M+ + 3Fe3+ + 2SO42− + 6H2O = MFe3(SO4)2(OH)6 + 6H+ | (5) |
Sulfur metabolism was closely related to iron metabolism. With the dissolution of chalcopyrite, according to eqn (2) and (3), elemental sulfur was released, which covered the ore surface. Moderate community structure was important to maintain a better balance between iron and sulfur metabolism.34 With the introduction of different oxidizing consortia, the indigenous community could automatically adjust its structure to a new balanced state and continue the oxidizing function. The reduced inorganic sulfur compounds (RISCs) could be effectively consumed by sulfur-oxidizing bacteria, such as A. caldus and A. thiooxidans, according to eqn (6) and (7). The model showed that the sulfur-oxidizing bacteria were more abundant and diverse in system C, and the copper extraction rate of system C was 50.40% higher than that of system A. Thus, the exogenous A. thiooxidans established in the indigenous community successfully enhanced the sulfur oxidizing function. Furthermore, XRD analysis showed an inconspicuous peak of chalcocite in the pattern of system C, which indicated that chalcopyrite was partly dissolved by cupric ions and ferrous ions (eqn (8)). Chalcocite had better leaching features than chalcopyrite; thus, it was oxidized by ferric ions or oxygen to cupric ions (eqn (9) and (10)) more easily.
| 2S0 + 2H2O + 3O2 = 2SO42− + 4H+ | (6) |
| SxOyn− + H2O + O2 → SO42− + H+ | (7) |
| CuFeS2 + 3Cu2+ + 3Fe2+ = 2Cu2S + 4Fe3+ | (8) |
| Cu2S + 4Fe3+ = 2Cu2+ + 4Fe2+ + S0 | (9) |
| Cu2S + O2 + 4H+ = 2Cu2+ + S0 + 2H2O | (10) |
With the synergistic effect of iron and sulfur metabolisms, the copper ions were gradually dissolved, as according to eqn (2), (3) and (9). By introducing different oxidizing consortia, the final copper concentrations in the four systems reached 2308 ± 55, 2383 ± 46, 3276 ± 59 and 2040 ± 50 mg L−1, respectively. Compared with the indigenous system (32.90%), exogenous sulfur oxidizers increased the copper extraction rate to 49.48%, which indicated that exogenous sulfur oxidizers could establish in the new environment and hold stronger leaching ability for chalcopyrite than other exogenous cells.
Conclusions
The introduction of acidophilic consortia with different oxidizing functions was performed prior to the rapid growth phase of an indigenous community. Exogenous ferrous oxidizers and sulfur oxidizers greatly changed the community structure and microbial succession and promoted cell growth rate during the bioleaching process, while ferrous/sulfur oxidizers showed no significant effects on the indigenous community. Exogenous ferrous oxidizers, particularly L. ferriphilum, and sulfur oxidizers, particularly A. thiooxidans, successfully established and colonized in the indigenous community. However, only colonized A. thiooxidans, rather than L. ferriphilum, showed the advantage of enhancing the dissolution of chalcopyrite. At the end of bioleaching, the copper extraction rate of the system invaded by sulfur oxidizers was 50.40% higher than that of the indigenous leachate. These results indicated that exogenous sulfur oxidizer A. thiooxidans, which was scarce in the indigenous community, could colonize easily, execute its function sufficiently, and greatly enhance copper dissolution.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 31570113 and 41573072) and the Fundamental Research Funds for the Central Universities of Central South University, China (No. 2018dcyj054).Notes and references
- S. Yin, L. Wang, X. Chen, R. Yan, K. An, L. Zhang and A. Wu, Minerals, 2018, 8, 32 CrossRef.
- D. E. Rawlings and D. B. Johnson, Microbiology, 2007, 153, 315 CrossRef CAS PubMed.
- D. B. Johnson, Curr. Opin. Biotechnol., 2014, 30, 24–31 CrossRef CAS PubMed.
- S. Panda, A. Akcil, N. Pradhan and H. Deveci, Bioresour. Technol., 2015, 196, 694–706 CrossRef CAS PubMed.
- C. Méndezgarcía, A. I. Peláez, V. Mesa, J. Sánchez, O. V. Golyshina and M. Ferrer, Front. Microbiol., 2015, 6, 475 Search PubMed.
- T. Rohwerder, T. Gehrke, K. Kinzler and W. Sand, Appl. Microbiol. Biotechnol., 2003, 63, 239 CrossRef CAS PubMed.
- P. Spolaore, C. Joulian, J. Gouin, D. Morin and P. D'Hugues, Appl. Microbiol. Biotechnol., 2011, 89, 441–448 CrossRef CAS PubMed.
- H. R. Watlinga, E. L. Watkinb and D. E. Ralphe, Environ. Technol., 2010, 31, 915–933 CrossRef CAS PubMed.
- S. Li, Z. Hui, Y. Hu, J. Zhao, Z. He and G. Gu, Bioresour. Technol., 2014, 153, 300–306 CrossRef CAS PubMed.
- L. Ma, X. Wang, X. Feng, Y. Liang, Y. Xiao, X. Hao, H. Yin, H. Liu and X. Liu, Bioresour. Technol., 2017, 223, 121 CrossRef CAS PubMed.
- Z. G. He, F. L. Gao, J. C. Zhao, Y. H. Hu and G. Z. Qiu, FEMS Microbiol. Ecol., 2010, 74, 155–164 CrossRef CAS PubMed.
- Y. Xiao, X. Liu, L. Ma, Y. Liang, J. Niu, Y. Gu, X. Zhang, X. Hao, W. Dong and S. She, Appl. Microbiol. Biotechnol., 2016, 100, 6871–6880 CrossRef CAS PubMed.
- Y. Wang, W. Zeng, G. Qiu, X. Chen and H. Zhou, Appl. Environ. Microbiol., 2014, 80, 741 CrossRef PubMed.
- E. S. Jules, M. J. Kauffman, W. D. Ritts and A. L. Carroll, Ecology, 2002, 83, 3167–3181 CrossRef.
- Y. Liu, H. Yin, W. Zeng, Y. Liang, Y. Liu, N. Baba, G. Qiu, L. Shen, X. Fu and X. Liu, Bioresour. Technol., 2011, 102, 8092–8098 CrossRef CAS PubMed.
- L. Zhang, W. Zhou, K. Li, F. Mao, L. Wan, X. Chen, H. Zhou and G. Qiu, Biochem. Eng. J., 2015, 93, 142–150 CrossRef CAS.
- C. Richter, H. Kalka and H. Märten, Solid State Phenom., 2017, 262, 456–460 Search PubMed.
- L. Xia, P. Uribe, X. Liu, C. Yu, L. Chai, J. Liu, W. Qiu and G. Qiu, World J. Microbiol. Biotechnol., 2013, 29, 275–280 CrossRef CAS PubMed.
- N. Chimpalee, D. Chimpalee, S. Srithawepoon, T. Patjarut and D. T. Burns, Anal. Chim. Acta, 1995, 304, 97–100 CrossRef CAS.
- K. B. Hallberg, E. González-Toril and D. B. Johnson, Extremophiles, 2010, 14, 9 CrossRef CAS PubMed.
- J. G. Caporaso, C. L. Lauber, W. A. Walters, D. Berg-Lyons, J. Huntley, N. Fierer, S. M. Owens, J. Betley, L. Fraser and M. Bauer, ISME J., 2012, 6, 1621 CrossRef CAS PubMed.
- J. Tao, X. Liu, Y. Liang, J. Niu, Y. Xiao, Y. Gu, L. Ma, D. Meng, Y. Zhang and W. Huang, Appl. Microbiol. Biotechnol., 2016, 101, 1289–1299 CrossRef PubMed.
- R. C. Edgar, Bioinformatics, 2010, 26, 2460 CrossRef CAS PubMed.
- Q. Wang, G. M. Garrity, J. M. Tiedje and J. R. Cole, Appl. Environ. Microbiol., 2007, 73, 5261 CrossRef CAS PubMed.
- R Development Core Team, Vienna, Austria: R Foundation for Statistical Computing, 2014, 1, pp. 12–21 Search PubMed.
- P. Dixon, J. Veg. Sci., 2003, 14, 927–930 CrossRef.
- D. H. Parks and R. G. Beiko, STAMP: Statistical Analysis of Metagenomic Profiles, Springer US, 2015 Search PubMed.
- Z. He, F. Gao, J. Zhao, Y. Hu and G. Qiu, FEMS Microbiol. Ecol., 2010, 74, 155–164 CrossRef CAS PubMed.
- C. L. Brierley and J. A. Brierley, Appl. Microbiol. Biotechnol., 2013, 97, 7543–7552 CrossRef CAS PubMed.
- Y. Wang, L. Su, W. Zeng, L. Wan, Z. Chen, L. Zhang, G. Qiu, X. Chen and H. Zhou, Miner. Eng., 2014, 61, 66–72 CrossRef CAS.
- J. J. Plumb, R. Muddle and P. D. Franzmann, Miner. Eng., 2008, 21, 76–82 CrossRef CAS.
- Y. Huang, Int. J. Electrochem. Sci., 2017, 10493–10510 CrossRef CAS.
- Å. Sandström, A. Shchukarev and J. Paul, Miner. Eng., 2007, 18, 505–515 CrossRef.
- N. Pradhan, K. C. Nathsarma, K. S. Rao, L. B. Sukla and B. K. Mishra, Miner. Eng., 2008, 21, 355–365 CrossRef CAS.
- N. Hiroyoshi, H. Miki, T. Hirajima and M. Tsunekawa, Hydrometallurgy, 2000, 57, 31–38 CrossRef CAS.
- S. Deng, G. Gu, J. Ji and B. Xu, Anal. Bioanal. Chem., 2018, 410, 1725 CrossRef CAS PubMed.
- S. Feng, H. Yang and W. Wang, Bioresour. Technol., 2015, 192, 75–82 CrossRef CAS PubMed.
- W. Zeng, G. Qiu, H. Zhou and M. Chen, Hydrometallurgy, 2011, 105, 259–263 CrossRef CAS.
- L. Xia, S. Dai, C. Yin, Y. Hu, J. Liu and G. Qiu, J. Ind. Microbiol. Biotechnol., 2009, 36, 845–851 CrossRef CAS PubMed.
- H. C. Liu, Z. Y. Nie, J. L. Xia, H. R. Zhu, Y. Yang, C. H. Zhao, L. Zheng and Y. D. Zhao, Int. J. Miner. Process., 2015, 137, 1–8 CrossRef.
- D. E. Rawlings, Microb. Cell Fact., 2005, 4, 1–15 CrossRef PubMed.
- D. B. Johnson, Trans. Nonferrous Met. Soc. China, 2008, 18, 1367–1373 CrossRef CAS.
- W. Zeng, G. Qiu, H. Zhou, J. Peng, M. Chen, S. N. Tan, W. Chao, X. Liu and Y. Zhang, Bioresour. Technol., 2010, 101, 7079–7086 Search PubMed.
- S. Feng, H. Yang and W. Wang, Bioresour. Technol., 2015, 191, 37–44 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2018 |