Open Access Article
Open Access ArticleSensitive and accurate detection of ALP activity using a fluorescence on–off–on switch and mass barcode signal amplification†
Chang Shu *ab,
Duo Liab,
Tengfei Liab,
Shunli Jiab and
Li Ding
*ab,
Duo Liab,
Tengfei Liab,
Shunli Jiab and
Li Ding *ab
*ab
aKey Laboratory of Drug Quality Control and Pharmacovigilance (China Pharmaceutical University), Ministry of Education, Nanjing 210009, China
bDepartment of Pharmaceutical Analysis, School of Pharmacy, China Pharmaceutical University, 24 Tongjiaxiang, Nanjing, 210009, P. R. China. E-mail: shuchang@cpu.edu.cn; dinglidlcpu@sina.com
First published on 29th October 2018
Abstract
Alkaline phosphatase (ALP) is an important biomarker for many diseases. Therefore, the sensitive and accurate detection of ALP activity is essential for fundamental biochemical processes and clinic diagnosis. Herein, we design a fluorescent on–off–on switch for sensitive and visual detection of ALP activity. Meanwhile, mass barcode-modified quantum dots (QDs) amplified the LC-MS/MS detection signal in complex biological samples. Firstly, the QDs were modified with phosphorylated Gly-Gly-Phe-Phe-Tyr (OPO3H2) peptide (GGFFYp) and the mass barcode. The fluorescence of QDs-SS-Yp was quenched by fluorescence resonance energy transfer (FRET) between QDs-SS-Yp and dansyl chloride (DNS). ALP can hydrolyze the phosphorylated peptide to form peptide self-assemblies on the QDs-SS-Yp surfaces. The effective separation distance between the QDs-SS-Yp donor and DNS acceptor becomes larger, restricting FRET between the QDs-SS-Yp and DNS. At this point, the obvious QDs-SS-Yp fluorescence signal can be restored. However, the absence of ALP results in no peptide self-assembly on the QDs-SS-Yp surface and no obvious QDs-SS-Yp fluorescence signal was detected. Therefore, the ALP activity can be analyzed according to the degree of fluorescence restoration by the fluorescence on–off–on switch. Finally, the small tag molecules obtained by cleaving the disulfide bond of the QDs-SS-Yp as a mass barcode were used to amplify the LC-MS/MS detection signal. The proposed approach shows a good linear relationship (from 0.01 to 2.4 U L−1) and has the significant advantage of a low detection limit of 0.001 U L−1.
1. Introduction
As a membrane-bound enzyme, alkaline phosphatase1 (ALP) is widely used as an important biomarker in clinical diagnostics.2 An abnormal level of ALP in human serum is closely related to many diseases, such as cancer, bone disease, liver dysfunction, and diabetes.3 Therefore, sensitive and accurate ALP determination is important. To date, a variety of methods for ALP detection have been reported, including spectroscopy (SERS), colorimetry, fluorometry, surface-enhanced Raman surface plasmon resonance (SPR), electrochemistry, isotopic labeling and chromatography.4–7 Although these methods use different techniques, they usually have some drawbacks. For example, traditional colorimetric and fluorescent methods based on organic dyes have poor photostability and water solubility, and are expensive,8 while, electrochemistry and isotopic labeling methods based on conjugated polyelectrolytes have complicated synthetic, purification, and modification procedures, which affect ALP activity.9 Furthermore, in most cases, ALP is expressed at a low abundance level naturally in biological systems, so the detector signal for ALP levels has been affected by interference in human serum.10 Many of these reported methods have not been sensitive or selective enough to accurately detect ALP levels in biological samples, which greatly limits their practical application in bioassays, quantitative assessment, and medical diagnosis.11 Therefore, it is necessary to develop an ALP activity monitoring method with highly amplified sensitivity, improved selectivity, and reduced sample consumption for limited sample quantities.12Recently, nanomaterial-based signal amplification methods have attracted increased interest for highly sensitive and selective ALP detection. Semiconductor quantum dots (QDs)13 possess excellent properties, such as good photostability, long fluorescence lifetime, broad excitation band, large Stokes shifts, narrow and size-tunable emission spectra, high quantum yield (QY), and large single-QD surface area for modification.14 Therefore, QDs have been used for multicolor imaging, optical barcoding, and multiplex analysis through different sensing mechanisms. Many QDs (CdTe, CdS, CdSe, and CdSe–ZnS) have been reported to detect ALP activity due to their simplicity, sensitivity, and convenience.15 However, the high biotoxicity of Cd2+ restricts its applications in biosensing and bioanalysis. Ag or Au nanoclusters16 are used to detect ALP activity with the assistance of Cu2+, which has poor optical stability in biosystems. Carbon dots17, a new kind of fluorescent nanomaterial, have been widely used in drug delivery, bioimaging, biosensing, and photocatalysis owing to their biocompatibility and aqueous solubility.18 However, an extremely low fluorescence quantum yield (lower than 2.2%) and lengthy preparation process have limited their further biomedical applications.19 Heteroatom doping20 (Cu-doped CdS, Mn-doped ZnS, and nitrogen doping) could have potential in bioanalysis studies owing to photostability and tunable fluorescent properties. However, doping poses several challenges, including sophisticated synthetic processes, organic reaction facilities, and discrimination against interference.21
Mass spectrometry (MS)22 is another promising analytical technique for bioanalysis owing to its sensitivity, high throughput, and easy automation. Matrix-assisted laser desorption ionization-time-of-flight23 (MALDI-TOF) MS coupled with soft ionization can detect molecular ion peaks of biomolecules, such as proteins, peptides, and polymers, with minimum fragmentation. However, MALDI-TOF MS still has some problems, such as heterogeneous cocrystallization with analytes, interference in the low mass region (m/z < 500), insufficient sensitivity for detecting low abundance analytes, and difficulty in finding appropriate matrix compositions. Therefore, MS-based signal amplifying strategies have been developed. MS signal amplification based on mass barcodes24 is widely used in kinase activity analysis, DNA assays, immunoassays, protein quantitative assessment, and tissue imaging. Mass barcodes are mass-sensitive enhancement reagents or small tag molecules that are easily detected by MS. Mass barcodes usually conjugate with the target biomolecules to amplify their MS signal.25
Based on the above analysis, we proposed a fluorescent on–off–on switch22 for sensitive and accurate detection of ALP activity and amplified signal detection of mass barcode-modified QDs by LC-MS/MS and tested this approach in complex biological samples. Firstly, the mass barcode and phosphorylated Gly-Gly-Phe-Phe-Tyr (OPO3H2) peptide (GGFFYp) were sequentially conjugated with QDs. In the absence of ALP, the fluorescence intensity of QDs-SS-Yp was low (OFF state) owing to FRET between the QD-SS-Yp and DNS. After ALP addition, the phosphate ester of phosphorylated peptide was hydrolyzed by ALP and then formed a self-assembly on the QD surfaces. The distance between the QDs-SS-Yp donor and DNS acceptor became larger and restricted FRET occurring between the QDs-SS-Yp and DNS. The highly fluorescent signal of QDs-SS-Yp can then be measured (ON state). Finally, the mass barcode was collected by cleaving the disulfide bond of QDs-SS-Yp and used to amplify LC-MS/MS detection signal (Fig. 1).
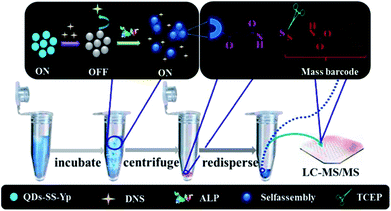 | ||
| Fig. 1 Schematic diagram of sensitive and accurate detection of ALP activity using a fluorescence on–off–on switch and mass barcode signal amplification. | ||
2. Experimental
2.1 Preparation of ZnSe QDs
ZnSe QDs were prepared according to our previous report with modifications. In brief, NaHSe solution was prepared by reacting Se powder (7.8 mg, 0.1 mmol) with NaBH4 (14 mg, 0.4 mmol) in ultrapure water (1 mL) in an ice-water bath under N2 for 40 min, during which the black color changed to colorless. ZnSO4·7H2O (87.7 mg, 0.4 mmol) and L-GSH (151.7 mg, 0.4 mmol) were dissolved in ultrapure water (100 mL) and the mixture was adjusted to pH 10.5 using NaOH (1 M) and stirred at 30 °C under N2 for 30 min. Next, the NaHSe solution was rapidly injected into the above nitrogen-saturated aqueous solution and reacted for 30 min at 30 °C. The solution was then refluxed at 100 °C for 90 min. The obtained transparent solution was purified by adding three times the volume of anhydrous ethanol and centrifuging at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. The precipitate was collected and dried overnight at 40 °C under vacuum. White or off-white QD powder was obtained, redispersed in ultrapure water, and stored at 4 °C until use.
000 rpm for 10 min. The precipitate was collected and dried overnight at 40 °C under vacuum. White or off-white QD powder was obtained, redispersed in ultrapure water, and stored at 4 °C until use.
2.2 Preparation of QDs-SS-Yp
The ALP-hydrolyzed phosphate ester reaction involved three steps. Firstly, Fmoc-SS (9.5 mg, 0.02 mmol) was activated with EDC (7.7 mg, 0.04 mmol) and NHS (9.2 mg, 0.08 mmol) in methanol (500 μL) for 2 h at room temperature. QD solution (10 mL, 0.01 mmol) was then mixed with the above activated solution and stirred gently for 12 h in the dark. The obtained QDs-SS was purified by adding three times the volume of anhydrous ethanol and centrifuging at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min to remove unreacted impurities. Secondly, QDs-SS (0.01 mmol) was redispersed in ultrapure water (10 mL) and activated with EDC (3.8 mg, 0.02 mmol) and NHS (4.6 mg, 0.04 mmol) at room temperature for 2 h. GGFFYp peptide (13.4 mg, 0.02 mmol) was then added and stirred for 12 h at room temperature while avoiding light. The QDs-SS-Yp solution was filtered through a dialysis tube (MWCO = 1000) to remove the free peptide and other impurities. As-prepared QDs-SS-Yp was redispersed in Tris–HCl buffer (10 mL, 10 mM, pH 7.4) and stored at 4 °C for use.
000 rpm for 10 min to remove unreacted impurities. Secondly, QDs-SS (0.01 mmol) was redispersed in ultrapure water (10 mL) and activated with EDC (3.8 mg, 0.02 mmol) and NHS (4.6 mg, 0.04 mmol) at room temperature for 2 h. GGFFYp peptide (13.4 mg, 0.02 mmol) was then added and stirred for 12 h at room temperature while avoiding light. The QDs-SS-Yp solution was filtered through a dialysis tube (MWCO = 1000) to remove the free peptide and other impurities. As-prepared QDs-SS-Yp was redispersed in Tris–HCl buffer (10 mL, 10 mM, pH 7.4) and stored at 4 °C for use.
2.3 ALP activity detection procedure
The detection procedure for evaluating our fluorescent approach to ALP activity was as follows: A series of different concentrations (0–2.4 U L−1) of ALP in Tris–HCl buffer solution (200 μL, pH 7.4) containing QDs-SS-Yp (100 μL, 0.4 μmol mL−1 final concentration), DNS (5 μL, 0.0167 μg mL−1 final concentration). The fluorescence signal was measured after incubation at 37 °C for 30 min. The excitation wavelength was 310 nm and the emission spectra scan range was 320–600 nm. The maximum emission intensity of QDs at 385 nm and the maximum emission intensity of DNS at 500 nm were used for data analysis.2.4 LC-MS/MS signal amplification by mass barcode
A series of different concentrations (0–2.4 U L−1) of ALP in Tris–HCl buffer solution (200 μL) contained QDs (100 μL, 0.4 μmol mL−1 final concentration), DNS (5 μL, 0.0167 μg mL−1 final concentration). After incubation at 37 °C for 30 min, the self-assembly complex was centrifuged for 3 min at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and carefully rinsed three times with washing buffer to remove unassembled reactants. The precipitate was redispersed in water (200 μL) and incubated with TCEP (1 mM final concentration) for 30 min at room temperature, which dissociated the disulfide bonds of QDs and released the mass barcode. Mifepristone (10 μL, 1 μmol mL−1 final concentration) as internal standard (IS) and methanol (790 μL) were added to samples. After vigorous shaking for 3 min, the mixture was centrifuged for 10 min at 13
000 rpm and carefully rinsed three times with washing buffer to remove unassembled reactants. The precipitate was redispersed in water (200 μL) and incubated with TCEP (1 mM final concentration) for 30 min at room temperature, which dissociated the disulfide bonds of QDs and released the mass barcode. Mifepristone (10 μL, 1 μmol mL−1 final concentration) as internal standard (IS) and methanol (790 μL) were added to samples. After vigorous shaking for 3 min, the mixture was centrifuged for 10 min at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and the supernatant was used for LC-MS/MS detection.
000 rpm and the supernatant was used for LC-MS/MS detection.
Chromatographic separation and mass detection were performed on an Agilent Technologies Series system (Agilent, USA) using an ODS-2 column (2.1 mm × 150 mm, 5 μm, Hanbon, China) with a mobile phase consisting of methanol and ultrapure water (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, v/v) at a flow rate of 0.4 mL min−1. The autosampler temperature was kept at 8 °C and a 5 μL volume was injected for LC-MS/MS analysis. The selected multiple reaction monitoring (MRM) transitions were m/z 322.1 → 322.1 for the mass barcode and m/z 430.1 → 372.2 for the IS. The fragmentor voltage values for the mass barcode and IS were 120 and 100 V, respectively. The collision energy sets for the mass barcode and IS were 30 and 0 eV, respectively.
20, v/v) at a flow rate of 0.4 mL min−1. The autosampler temperature was kept at 8 °C and a 5 μL volume was injected for LC-MS/MS analysis. The selected multiple reaction monitoring (MRM) transitions were m/z 322.1 → 322.1 for the mass barcode and m/z 430.1 → 372.2 for the IS. The fragmentor voltage values for the mass barcode and IS were 120 and 100 V, respectively. The collision energy sets for the mass barcode and IS were 30 and 0 eV, respectively.
2.5 Human serum sample detection
Healthy adult volunteers gave informed consent and their blood samples were collected for ALP activity detection. All experiments were performed in accordance with the guidelines of the Declaration of Helsinki, and experiments were approved by the ethics committee at China Pharmaceutical University. ALP solutions of different concentrations (0.05, 0.1, 0.6, 1.0 and 1.8 U L−1) were added into the 1% diluted human serum. The detection procedures outlined in Sections 2.3 and 2.4 were used.3. Results and discussion
3.1 Characterization of QDs-SS-Yp
The optical properties and composition of QDs-SS-Yp were further investigated by UV-vis absorption, fluorescence spectroscopy (FL), X-ray diffraction (XRD), and Fourier transform infrared (FT-IR), respectively. The UV-vis spectra (Fig. 2A) showed a maximum absorption peak at 355 nm, which was attributed to the n–π* transition of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond. Meanwhile, the peak emission of QDs-SS-Yp occurred at 385 nm with maximum excitation at 310 nm. The narrow half peak width of the emission spectrum was probably attributed to a more uniform size and less surface defects. Quantum yields of QDs-SS-Yp were calculated to be 57.2% using quinine sulfate as reference. The XRD spectrum (Fig. 2B) showed a broad peak at 27.1°, which was consistent with the crystal form of ZnSe QDs. The emission peak of ZnSe QDs was at 380 nm (Fig. 2C). After QDs-SS-Yp formation, the emission peak was redshifted to 385 nm. This result indicated that Fmoc-SS and GGFFYp peptide were conjugated onto the QDs. Furthermore, FT-IR analysis (Fig. 2D) was performed to verify the conclusion. In the ZnSe QDs, abundant amino and carboxyl groups were present on the surface, as demonstrated by the O–H band at 3273 cm−1, N–H band at 1568 cm−1, and C–O band at 1400 cm−1. In QDs-SS, Fmoc-SS was conjugated with the surface of the ZnSe QDs, as demonstrated by the N–H band at 3324 cm−1, C–H band at 3068 cm−1, N–H band at 1550 cm−1, C–N band at 1402 cm−1, and C–S band at 708 cm−1. In QDs-SS-Yp, the P–O band at 1186 cm−1 indicated that the GGFFYp peptide was bonded to the QD surface.
O bond. Meanwhile, the peak emission of QDs-SS-Yp occurred at 385 nm with maximum excitation at 310 nm. The narrow half peak width of the emission spectrum was probably attributed to a more uniform size and less surface defects. Quantum yields of QDs-SS-Yp were calculated to be 57.2% using quinine sulfate as reference. The XRD spectrum (Fig. 2B) showed a broad peak at 27.1°, which was consistent with the crystal form of ZnSe QDs. The emission peak of ZnSe QDs was at 380 nm (Fig. 2C). After QDs-SS-Yp formation, the emission peak was redshifted to 385 nm. This result indicated that Fmoc-SS and GGFFYp peptide were conjugated onto the QDs. Furthermore, FT-IR analysis (Fig. 2D) was performed to verify the conclusion. In the ZnSe QDs, abundant amino and carboxyl groups were present on the surface, as demonstrated by the O–H band at 3273 cm−1, N–H band at 1568 cm−1, and C–O band at 1400 cm−1. In QDs-SS, Fmoc-SS was conjugated with the surface of the ZnSe QDs, as demonstrated by the N–H band at 3324 cm−1, C–H band at 3068 cm−1, N–H band at 1550 cm−1, C–N band at 1402 cm−1, and C–S band at 708 cm−1. In QDs-SS-Yp, the P–O band at 1186 cm−1 indicated that the GGFFYp peptide was bonded to the QD surface.
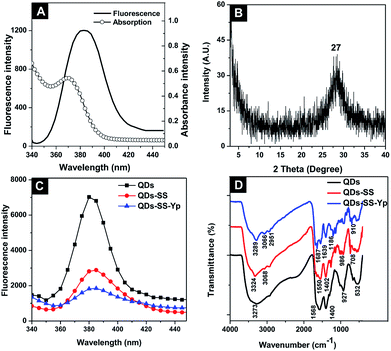 | ||
| Fig. 2 (A) Absorption and emission spectra and (B) XRD spectrum of QDs-SS-Yp solution. (C) Fluorescence emission spectra and (D) IR spectra of as-prepared QDs, QDs-SS, and QDs-SS-Yp. | ||
The fluorescence signal remained almost constant under 365 nm light illumination for 1 h (Fig. S10A, ESI†) or after 3 months at room temperature while avoiding light (Fig. S10B, ESI†). The excellent optical properties of QDs-SS-Yp facilitated their further applications. The interference of HCl, NaOH, and NaCl with the fluorescence intensity of QDs-SS-Yp was also investigated (Fig. S10C, ESI†). QDs-SS-Yp was not stable at higher NaOH concentrations and was sensitive to HCl. QDs-SS-Yp was stable in NaCl up to a NaCl concentration of 0.7 M. Therefore, the QDs-SS-Yp exhibited good stability under different pH and salt concentrations, which ensured its application to biological samples analysis.
3.2 Principle of ALP activity assay and fluorescence detection
The principle of ALP activity detection is based on a fluorescence on–off–on switch, as shown in Fig. 1. The fluorescence of QDs-SS-Yp can be quenched sufficiently by DNS owing to FRET between QDs-SS-Yp and DNS. When ALP is added, the fluorescence of QDs-SS-Yp was restored immediately. This was due to ALP being able to hydrolyze phosphorylated peptide (GGFFYp) and subsequently cause peptide self-assembly on the QD surface to restrain FRET occurring between the QDs and DNS. Therefore, the fluorescence of QDs-SS-Yp was recovered again owing to integration of the released free QDs. Based on this concept, real-time visual detection of ALP activity could be spatially and temporally achieved using a single fluorescence on–off–on switch.In this work, we selected the 385 nm emitting QDs-SS-Yp as donor and 500 nm emitting DNS dye as acceptor. This pair overlapped between the excitation spectrum of DNS and the emission spectrum of QDs-SS-Yp (Fig. 3A). Generally, QDs can accommodate multiple acceptors to improve the FRET efficiency in the QD-based FRET assay. To achieve the best performance in the FRET assay, we mixed different amounts of DNS with a fixed amount of QDs-SS-Yp and measured the fluorescence of both the QDs-SS-Yp and DNS at an excitation wavelength of 310 nm. As shown in Fig. 3B, the fluorescence intensity of QDs-SS-Yp at 385 nm decreased with increasing DNS fluorescence intensity at 500 nm. The maximum fluorescence intensity of DNS and the minimum fluorescence intensity of QDs-SS-Yp were obtained at a DNS concentration of 0.0167 μg mL−1.
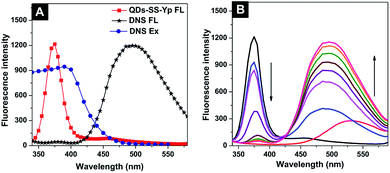 | ||
| Fig. 3 (A) Excitation and emission spectra of QDs-SS-Yp and DNS solution. (B) Fluorescence emission spectra of QDs-SS-Yp with different concentrations of DNS solution. | ||
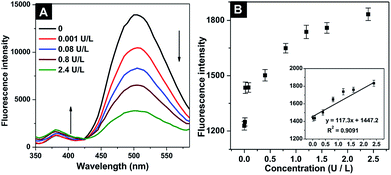
The emission spectra of QDs-SS-Yp (Fig. 4A) were investigated to visually detect the ALP activity after incubating with different concentrations of ALP (0–2.4 U L−1). With the addition of ALP, the QDs-SS-Yp emission intensity at 385 nm increased. While the DNS fluorescence intensity at 500 nm decreased. This phenomenon demonstrated that ALP hydrolyzed the phosphorylated GGFFYp peptide and subsequently formed a self-assembly on the QD surface. This caused the effective separation distance between the QDs-SS-Yp donor and DNS acceptor to become larger, and restricted FRET between QDs-SS-Yp and DNS. The fluorescence of QDs-SS-Yp was recovered by the re-release of QDs. The fluorescence intensity and ALP activity showed a good linear relationship ranging from 0.02 to 2.4 U L−1 (Fig. 4B inset). The linear regression equation was y = 117.3x + 1447.2 (R2 = 0.9091). Variable y was the fluorescence intensity of QDs-SS-Yp and variable x was the ALP concentration. In this work, the detection limit was 0.001 U L−1, which was sensitive enough to detect ALP activity in complex biological samples assays. The real-time visual detection of ALP activity could be achieved, although the accurate quantitative evaluation of ALP did not meet our analysis standards.
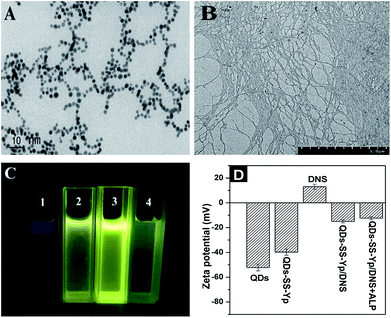
This observation was further verified and characterized by TEM from a microcosmic perspective. Fig. 5A shows that QDs-SS-Yp without ALP was nearly spherical and monodisperse, with a diameter of 2–3 nm. However, reticular structures of nanofibers were present after adding ALP because ALP hydrolyzed the phosphate ester of the phosphorylated peptide and formed peptide self-assemblies on the QD surface (Fig. 5B). The corresponding response photograph of the fluorescence on–off–on switch at different stages is shown in Fig. 5C, as obtained under UV light (365 nm). Fig. 5C(1 and 2) show blue and yellow fluorescence in water for QDs-SS-Yp and DNS, respectively. In the presence of DNS, QDs-SS-Yp with blue fluorescence was efficiently quenched and strong yellow fluorescence (Fig. 5C(3)) was observed, suggesting that the energy transfer process between QDs and DNS had occurred. When ALP was added into the FRET system, the strong yellow fluorescence became weak and light blue fluorescence appeared (Fig. 5C(4)). Zeta potential measurements were used to investigate the interaction between QDs-SS-Yp and ALP. As shown in Fig. 5D, the zeta potentials of ZnSe QDs and QDs-SS-Yp solution were −52.29 ± 2.66 mV and −39.77 ± 2.49 mV, respectively. These results suggested that the Fmoc-SS and phosphorylated GGFFYp peptide were successfully conjugated on the QD surface. The zeta potential of the DNS solution was 13.02 ± 2.08 mV. After adding DNS into the QDs-SS-Yp solution, the zeta potential became −15.02 ± 1.17 mV, suggesting that FRET occurred between the QDs and DNS. The zeta potential of the ALP solution was 0 ± 0.14 mV. After the ATP and QDs-SS-Yp solution were incubated for 30 min at 37 °C, the zeta potential became −12.41 ± 1.25 mV, suggesting that the phosphate ester group of GGFFYp peptide was hydrolyzed to an OH group by ALP. These OH groups on surface of the QDs then joined together through hydrogen bonds to form the self-assembly.
3.3 Optimization of ALP activity assay conditions
Before the ALP activity was monitored by QDs-SS-Yp containing DNS solution, we first optimized the assay conditions for ALP activity detection, such as buffer solution, QDs-SS-Yp concentration, and incubation time. As shown in Fig. S11A (ESI),† PBS buffer solutions (pH 7.4) had a serious impact on the ALP activity and fluorescence intensity of QDs-SS-Yp, whereas the water had little effect. In Tris–HCl solution (pH 7.4), the fluorescence intensity gradually increased with increasing ALP concentration (0–0.8 U L−1). Considering the quenching efficiency and ALP activity, Tris–HCl solution (pH 7.4) was the optimal reaction system solution in this work. As shown in Fig. S11B (ESI),† lower concentrations of QDs-SS-Yp (0.02 M and 0.01 M) did not cause an obvious change in fluorescence intensity with increasing ALP concentration. Therefore, 0.1 M was chosen as the QDs-SS-Yp concentration for subsequent experiments. As shown in Fig. S11C (ESI),† the fluorescence intensity of QDs-SS-Yp containing ALP gradually increased with reaction time, with only a small difference observed with time. Considering the hydrolyzation and self-assembly process, 30 min was selected as the appropriate reaction time for subsequent experiments. We also examined appropriate TCEP dosages and incubation times for dissociating the QD disulfide bonds. As shown in Fig. S11D (ESI),† the response intensity of the mass with ALP concentration gradually increased with increasing reaction time and TCEP concentration. Therefore, we chose 30 min and 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (molar ratio of TCEP and QDs-SS-Yp) as the optimal conditions.
1 (molar ratio of TCEP and QDs-SS-Yp) as the optimal conditions.
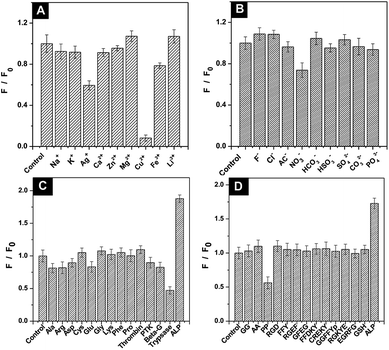
Interference with the fluorescence intensity of QDs-SS-Yp containing DNS solution was investigated to explore the specificity in serum containing several cations (such as Na+, K+, Ag+, Ca2+, Zn2+, Mg2+, Cu2+, Fe3+, Li3+), anions (such as F−, Cl−, Ac−, NO3−, HCO3−, HSO3−, SO42−, CO32−, PO43−), amino acids (such as Pro, Ala, Arg, Glu, Gly, Phe, Lys, Asp, Cys), different enzymes (such as thrombin, protein tyrosine kinase, trypsase, and β-glucuronidase), GSH, and peptides (dipeptide, tripeptide, tetrapeptide, pentapeptide) under the same conditions. Besides the decrease in fluorescence intensity induced by Ag+, Cu2+, Fe3+, NO3−, all cations and anions showed little effect on the fluorescence intensity of QDs-SS-Yp (Fig. 6A and B). For the amino acids, different enzymes (Fig. 6C) and peptides (Fig. 6D), only ALP led to an obvious increase in fluorescence intensity, while others had a negligible effect. The interference experiments clearly demonstrated that the above elements had little effect on the fluorescent intensity of QDs-SS-Yp, indicating the high selectivity for ALP activity detection.
3.4 LC-MS/MS signal amplification by mass barcode
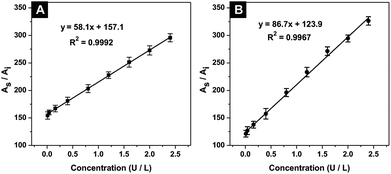
In LC-MS/MS analysis, the retention times achieved for mass barcode and IS were 2.882 and 3.659 min, respectively (Fig. S12, ESI†). As shown in Fig. 7A, the response number increased with increasing ALP concentration and showed a good linear correlation range from 0.01 to 2.4 U L−1. The linear regression equation was y = 58.1x + 157.1 (y is the mass response number and x is the ALP concentration), R2 = 0.9992. Notably, the sensitivity and accuracy of the proposed method were improved owing to the amount of mass barcode dissociated from QDs.The established strategy was investigated for ALP activity detection in real serum samples to demonstrate its feasibility. The detection procedure was the same as described above. Fig. 7B shows good linear correlation ranging from 0.01 to 2.4 U L−1. The linear regression correlation equation was y = 86.7x + 123.9 (R2 = 0.9967). The detection limit was calculated to be 0.001 U L−1. The five serum samples containing different ALP concentrations were measured using the linear regression equation in serum. As shown in Table 1, recoveries between 95.3% and 103.4% with a relative standard deviation (RSD) below 1.9% were achieved. Compared with reported detection methods for ALP activity (Table 2), our work showed good sensitivity, accuracy, and reliability.
| Add ALP (U L−1) | Detected ALP (U L−1) | RSD (%) | Recovery (%) |
|---|---|---|---|
| 0.05 | 0.0486 | 1.4 | 97.2 |
| 0.1 | 0.103 | 0.9 | 103.4 |
| 0.6 | 0.572 | 1.6 | 95.3 |
| 1.0 | 0.976 | 1.9 | 97.6 |
| 1.8 | 1.825 | 1.0 | 101.4 |
| Material | LOD (U L−1) | Linear range (U L−1) | R2 | Assay time (min) | Reference |
|---|---|---|---|---|---|
| N-doped carbon dots | 0.001 | 0.01–25 | 0.996 | 30 | 9 |
| Gold nanoclusters | 0.002 | 0.02–50 | 0.98 | 92 | 10 |
| Upconversion nanoparticles | 19 | 62.5–875 | 0.9985 | 130 | 13 |
| Silver nanocluster | 5 | 30–240 | 0.984 | 195 | 16 |
| Carbon QDs | 1.4 | 4.6–383.3 | 0.996 | 63 | 17 |
| Carbon QDs | 1.1 | 16.7–782.6 | 0.97 | 33 | 18 |
| QDs-mass barcode | 0.001 | 0.01–2.4 | 0.9967 | 80 | This work |
4. Conclusions
In conclusion, we have presented a fluorescent on–off–off switch based on FRET for the sensitive visualization and detection of ALP activity, and that mass barcode-modified QDs showed an amplified detection signal by LC-MS/MS in complex biological samples. The assay is based on the principle that the fluorescence of QDs-SS-Yp can be efficiently quenched by DNS and recovered by adding ALP, which hydrolyzes the phosphorylation peptide of QDs-SS-Yp and subsequently forms a self-assembly on the QD surfaces. The mass barcode is then removed by TCEP and collected for LC-MS/MS analysis. The proposed method is based on three favorable properties: (i) FRET signal detection can increase the accuracy to avoid false positivity; (ii) mass barcode counting techniques by LC-MS/MS have high sensitivity through a signal amplification method; and (iii) the method can detect trace ALP activity with a very low detection limit of 0.001 U L−1. Therefore, this method exhibits significant advantages, including good selectivity, high sensitivity, and low cost, which have great potential in further biological and clinical applications.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors acknowledge financial support from the National Natural Science Foundation of China (No. 81603072 and 81573387). This study was supported by the Open Project Program of MOE Key Laboratory of Drug Quality Control and Pharmacovigilance (No. DQCP2017QN03), and the College Students Innovation Project for the R&D of Novel Drugs (201810316131) of the National Fund for Fostering Talents of Basic Science (NFFTBS).Notes and references
- R. Ahmad, H. Jang, B. S. Batule and H. G. Park, Anal. Chem., 2017, 89, 8966–8973 CrossRef CAS PubMed.
- W. Wang, X. Ji, A. Kapur, C. Zhang and H. Mattoussi, J. Am. Chem. Soc., 2015, 137, 14158–14172 CrossRef CAS PubMed.
- C. Chen, Z. Song, X. Zheng, Z. He, B. Liu, X. Huang, D. Kong, D. Ding and B. Z. Tang, Chem. Sci., 2017, 8, 2191–2198 RSC.
- N. Hildebrandt, C. M. Spillmann, W. R. Algar, T. Pons, M. H. Stewart, E. Oh, K. Susumu, S. A. Díaz, J. B. Delehanty and I. L. Medintz, Chem. Rev., 2017, 117, 536–711 CrossRef CAS PubMed.
- M. Fei, L. Chen and Z. Chun, J. Mater. Chem. B, 2018, 6(39), 6173–6190 RSC.
- F. Ma, W. J. Liu, L. Liang, B. Tang and C. Y. Zhang, Chem. Commun., 2018, 54(19), 2413–2416 RSC.
- J. Zhou, Y. Yang and C. Y. Zhang, Chem. Rev., 2015, 115(21), 11669–11717 CrossRef CAS PubMed.
- X. Jiao, Y. Li, J. Niu, X. Xie, X. Wang and B. Tang, Anal. Chem., 2018, 90, 533–555 CrossRef CAS PubMed.
- G. Li, H. Fu, X. Chen, P. Gong, G. Chen, L. Xia, H. Wang, J. You and Y. Wu, Anal. Chem., 2016, 88, 2720–2726 CrossRef CAS PubMed.
- H. Liu, M. Li, Y. Xia and X. Ren, ACS Appl. Mater. Interfaces, 2017, 9, 120–126 CrossRef CAS PubMed.
- C. Wang, J. Ouyang, Y. Y. Wang, D. K. Ye and X. H. Xia, Anal. Chem., 2014, 86, 3216–3221 CrossRef CAS PubMed.
- D. J. Sobczynski and O. Eniola-Adefeso, Acta Biomater., 2017, 48, 186–194 CrossRef CAS PubMed.
- F. Wang, C. Zhang, Q. Xue, H. Li and Y. Xian, Biosens. Bioelectron., 2017, 95, 21–26 CrossRef CAS PubMed.
- L.-j. Wang, Y. Yang and C.-y. Zhang, Anal. Chem., 2015, 87, 4696–4703 CrossRef CAS PubMed.
- H. Wang, J. Liu, A. Han, N. Xiao, Z. Xue, G. Wang, J. Long, D. Kong, B. Liu, Z. Yang and D. Ding, ACS Nano, 2014, 8, 1475–1484 CrossRef CAS PubMed.
- J.-L. Ma, B.-C. Yin, X. Wu and B.-C. Ye, Anal. Chem., 2016, 88, 9219–9225 CrossRef CAS PubMed.
- Z. Qian, L. Chai, C. Tang, Y. Huang, J. Chen and H. Feng, Anal. Chem., 2015, 87, 2966–2973 CrossRef CAS PubMed.
- Z. S. Qian, L. J. Chai, Y. Y. Huang, C. Tang, J. Jia Shen, J. R. Chen and H. Feng, Biosens. Bioelectron., 2015, 68, 675–680 CrossRef CAS PubMed.
- T.-Y. Wang, C.-Y. Chen, C.-M. Wang, Y. Z. Tan and W.-S. Liao, ACS Sens., 2017, 2, 354–363 CrossRef CAS PubMed.
- J. Zhang, X. Lu, Y. Lei, X. Hou and P. Wu, Nanoscale, 2017, 9, 15606–15611 RSC.
- B. Wu, Y. Lin, B. Li, C. Zhan, F. Zeng and S. Wu, Anal. Chem., 2018, 90, 9359–9365 CrossRef CAS PubMed.
- Z. Wang, C. Wang, S. Liu, W. He, L. Wang, J. Gan, Z. Huang, Z. Wang, H. Wei, J. Zhang and L. Dong, ACS Nano, 2017, 11, 1659–1672 CrossRef CAS PubMed.
- Z. Zhang, C. Wang, Y. Zha, W. Hu, Z. Gao, Y. Zang, J. Chen, J. Zhang and L. Dong, Corona-directed nucleic acid delivery into hepatic stellate cells for liver fibrosis therapy, ACS Nano, 2015, 9, 2405–2419 CrossRef CAS PubMed.
- X. Zhong, L. Qiao, N. Gasilova, B. Liu and H. H. Girault, Anal. Chem., 2016, 88, 6184–6189 CrossRef CAS PubMed.
- J. Wang, J. Li, J. Li, F. Liu, X. Zhou, Y. Yao, C. Wang, L. Qiu and P. Jiang, Anal. Chim. Acta, 2015, 895, 112–117 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra06973e |
| This journal is © The Royal Society of Chemistry 2018 |