Open Access Article
Open Access ArticleAdsorption of cadmium by live and dead biomass of plant growth-promoting rhizobacteria
Xingjie Li ,
Dongbo Li,
Zhenning Yan and
Yansong Ao*
,
Dongbo Li,
Zhenning Yan and
Yansong Ao*
School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai 200240, China. E-mail: aoys@sjtu.edu.cn; Tel: +86-13916002737
First published on 1st October 2018
Abstract
Plant growth-promoting rhizobacteria (PGPR) have been extensively investigated in combination remediation with plants in heavy metal contaminated soil. However, being biosorbent, few studies of live and dead cells of PGPR have been undertaken. Meanwhile, the application of live or dead biomass for the removal of heavy metals continues to be debated. Therefore, this study uses living and non-living biosorbents of Cupriavidus necator GX_5, Sphingomonas sp. GX_15, and Curtobacterium sp. GX_31 to compare their Cd(II) adsorption capacities by SEM-EDX, FTIR, and adsorption experiments. In the present study, whether the cells were living or dead and whatever the initial Cd(II) concentration was, removal efficiency and adsorption capacity can be arranged as GX_31 > GX_15 > GX_5 (p < 0.05). However, removal efficiency in live and dead biosorbents was quite different and it greatly affected by the initial Cd(II) concentrations. The dead cells exhibited a higher adsorption capacity than the live cells of GX_31. Nevertheless, for GX_5 and GX_15, the loading capacity of the non-living biomass was stronger than that of the living biomass at 20 mg L−1 of Cd(II), but the capacity was similar at 100 mg L−1 of Cd(II). Minor changes of spectra were found after autoclaving and it seemed that more functional groups of the dead biosorbent were involved in Cd(II) binding by FTIR analysis, which also illustrated that the hydroxyl, amino, amide, and carboxyl groups played an important role in complexation with Cd(II). Based on these findings, we concluded that the dead cells were more potent for Cd(II) remediation, especially for GX_31.
1. Introduction
Heavy metal contamination is a serious environmental issue, which primarily results from human activities, such as mining, the application of fertilizer, waste disposal, and sewage irrigation.1 Among these metals, cadmium is a ubiquitous element that is hazardous in the environment. Cd(II) accumulation in soil is of great concern because of its adverse effects on plants, animals, and microorganisms and its characteristics of high toxicity, bioaccumulation, and non-biodegradation.2 More importantly, Cd(II) accumulation may also cause health risks in humans through the food chain.3 For example, an excessive intake of Cd(II) can lead to hepatic and renal damage as well as skeletal dysfunctions, such as osteoporosis, cartilage and bone deformities. Therefore, research on the remediation of heavy metal contaminated soil is an urgent and challenging task.4–6Physicochemical methods have been widely used for the removal of heavy metals from contaminated sites. However, these applications are mostly ineffective and very expensive and nonspecific, especially when the concentrations of heavy metals are low.7 In contrast, biosorption has appeared to be an alternative to overcome these shortcomings of conventional methods for heavy metal removal,7–9 which utilizes bacteria, fungi, yeast, algae, industrial wastes, agricultural wastes, other polysaccharide materials,10 and exogenous carbonaceous materials.11 The advantages of biosorption consist of a flexible operation, free availability, high adsorption capacity, low cost, and possible reuse.12
Combination remediation refers to both plants and plant growth-promoting rhizobacteria (PGPR) or fungi involved in the process of remediation, which has been accepted as a promising approach for Cd(II) removal.13 PGPR are beneficial bacteria that colonize the rhizosphere of plants and facilitate plant growth and development by direct or indirect mechanisms.14 Developing a fundamental knowledge of the interaction between PGPR and metal ions allows for a better understanding of the bioavailability of heavy metals in the soil, which is also imperative for the development of combination remediation technologies. PGPR not only promote growth and heavy metal uptake by plants, but they are also effective biosorbents for the adsorption of heavy metals, like other bacteria.
The adsorption of heavy metals using the biomass of live or dead bacteria, algae, and yeast has emerged as a potential strategy.15–17 For example, Tangaromsuk et al. investigated Cd(II) uptake by Sphingomonas paucimobilis biomass and reported that the removal capacity of live cells was markedly higher than that of dead cells.18 Xu et al. reported that both dead and live cells of Enterobacter cloacae TU had the ability to remove Cd(II), and that live cells were superior to dead cells.19 Conversely, a study conducted by Huang et al. demonstrated that dead cells could adsorb cadmium to a greater degree than living cells.20 Guo et al. also observed that the live and dead biomass of Pseudomonas plecoglossicida demonstrated almost the same uptake capacities for Cd(II).21 Despite previous research, however, there continues to be a lack of information about the comparative study of live and dead cells for Cd(II) removal. There are also debates about removal capacities for heavy metals between live and dead cells based on the studies mentioned above. Moreover, little work has been done to compare the adsorption of Cd(II) using the live and dead biomass of various PGPR under different concentrations of Cd(II). As a result, the debated question about the use of live or dead biomass for the removal of heavy metals remains.
The objectives of this study are: (1) to investigate the capacities of PGPR (i.e. Cupriavidus necator GX_5 (CP002878), Sphingomonas sp. GX_15 (MF959440), and Curtobacterium sp. (MF959445) GX_31) for Cd(II) adsorption under the same experimental conditions; (2) to analyse morphology and functional group changes of live and dead cells after interaction with Cd(II) using SEM-EDX and FTIR; (3) to compare the removal capacities of the live and dead biomass of these strains; and (4) to evaluate the advantages of live and dead biosorbents for the remediation of contaminated sites.
2. Materials and methods
2.1. Bacterial strains and growth conditions
In this study, three PGPR, Gram-positive strain Curtobacterium sp. GX_31 (MF959445) and Gram-negative strains Cupriavidus necator GX_5 (CP002878) and Sphingomonas sp. GX_15 (MF959440) were used, and isolated from a 60 year-old lead–zinc core from rhizosphere soil of local (Guangxi, China) dominant plants, with an average Cd(II) concentration of 59.43 mg kg−1. The strains were cultured in a Luria-Bertani (LB) broth medium and maintained at 28 °C.2.2. Preparation of the live and dead biosorbents
The strains were incubated in a LB culture medium at pH 7.0 ± 0.2 on a rotary shaker at 28 °C, with a rotating speed of 180 rpm. After 24 hours of incubation, the cultured cells were harvested and washed three times with sterile deionized distilled water (ddH2O), and then separated by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min at 4 °C, collected, pre-cooled at −80 °C, and lyophilized overnight in a Labconco freeze dryer.22 The dried cells were ground into a fine powder and used as the live biosorbent. For the dead biomass, the cultured bacterial suspensions were firstly autoclaved at 121 °C for 20 min. Next, the non-living cells were collected, lyophilized, and ground into a powder just as for the live cells mentioned above, and were used as the dead biosorbent.
000 × g for 10 min at 4 °C, collected, pre-cooled at −80 °C, and lyophilized overnight in a Labconco freeze dryer.22 The dried cells were ground into a fine powder and used as the live biosorbent. For the dead biomass, the cultured bacterial suspensions were firstly autoclaved at 121 °C for 20 min. Next, the non-living cells were collected, lyophilized, and ground into a powder just as for the live cells mentioned above, and were used as the dead biosorbent.
2.3. Preparation of Cd(II) solutions
A Cd(II) stock solution of 1000 mg L−1 was prepared by dissolving CdCl2·2.5H2O in Milli-Q water. Working Cd(II) solutions were prepared by diluting the stock solution to the desired concentrations, which were used for Cd(II) adsorption experiments of the live and dead biomass. The initial pH of the working solutions was adjusted to 6.0 by the addition of 0.1 mol L−1 HCl or NaOH solution. All the chemicals used in this study were of analytical grade and the solutions were prepared using Milli-Q water.2.4. Adsorption experiments of Cd(II) by live and dead biosorbents
The adsorption behaviours of the live and dead biomass were conducted at 28 °C by mixing 0.02 g of each biosorbent in 50 mL plastic tubes with 20 mL of 20, 50, and 100 mg L−1 working solution. In other words, the concentration of the biomass was 1 g L−1 of dry cells. The mixture was shaken at 180 rpm for 6 hours on a shaker and then centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min. Next, the concentration of Cd(II) in the supernatant was determined by ICP (inductively coupled plasma). The difference between the initial Cd(II) concentration and the equilibrium ion concentration was assumed to have been adsorbed by the live or dead cells. The removal efficiency adsorption (%) for Cd(II) was calculated using the following eqn (1):
000 × g for 10 min. Next, the concentration of Cd(II) in the supernatant was determined by ICP (inductively coupled plasma). The difference between the initial Cd(II) concentration and the equilibrium ion concentration was assumed to have been adsorbed by the live or dead cells. The removal efficiency adsorption (%) for Cd(II) was calculated using the following eqn (1):
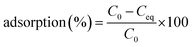 | (1) |
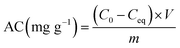 | (2) |
2.5. SEM-EDX observation
The surface structure and morphology of live and dead biosorbents before and after interaction with Cd(II) was studied using a SEM (Sirion 200, USA). For SEM analysis, 0.02 of g lyophilized samples of live and dead cells were mixed with 20 mL of the Cd(II) working solution (100 mg L−1). After adsorption, the centrifugally collected cells were fixed with 2.5% glutaraldehyde at 4 °C overnight. The fixed samples were then smeared on the coverslip, air dried, dehydrated using a gradient series with ethanol (30%, 50%, 70%, 90% and 100%) for 15 min each, and sputter-coated with gold.23 Energy dispersive X-rays (EDX) of both the live and dead biosorbents before and after the interaction with Cd(II) were simultaneously analysed.2.6. Fourier transformed infrared (FTIR) spectroscopy analysis
Infrared spectra of the live and dead biomass loaded with and without Cd(II) were obtained using a FTIR spectrometer (Nicolet 6700, USA) at room temperature. Cd(II)-loaded (100 mg L−1) samples of live and dead biosorbents were freeze-dried overnight. Then, 1 mg of dried cells was mixed and ground with 100 mg of KBr in an agate mortar to obtain the samples. Finally, the samples were immediately recorded with the spectrometer in the range of 4000–400 cm−1 with a resolution of 4 cm−1 in order to investigate the functional groups and possible Cd(II) binding sites.242.7. Statistical analysis
Data were subjected to analysis of variance and are represented as the mean ± standard deviation. Pairwise differences among treatments were tested using least significant difference at p = 0.05. All the statistical analyses were performed using SAS8.1 software.3. Results and discussion
3.1. Removal efficiency and adsorption capacity of live and dead biosorbents
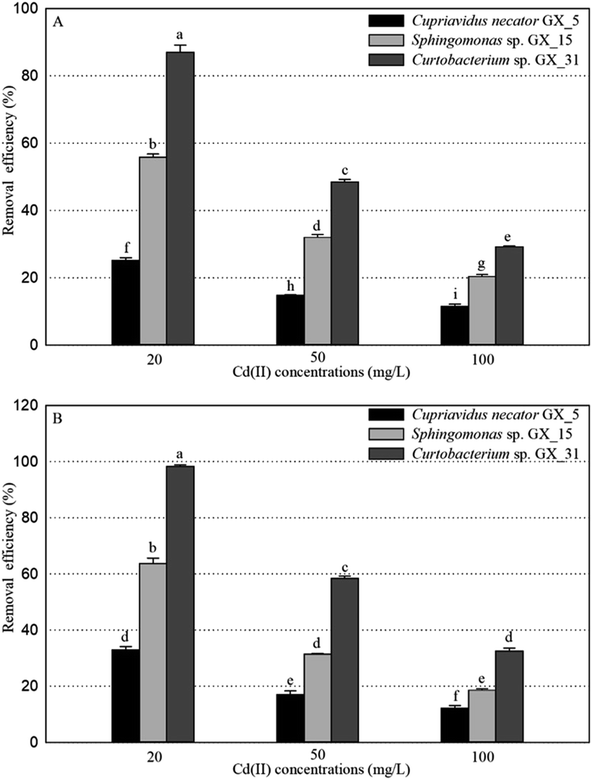
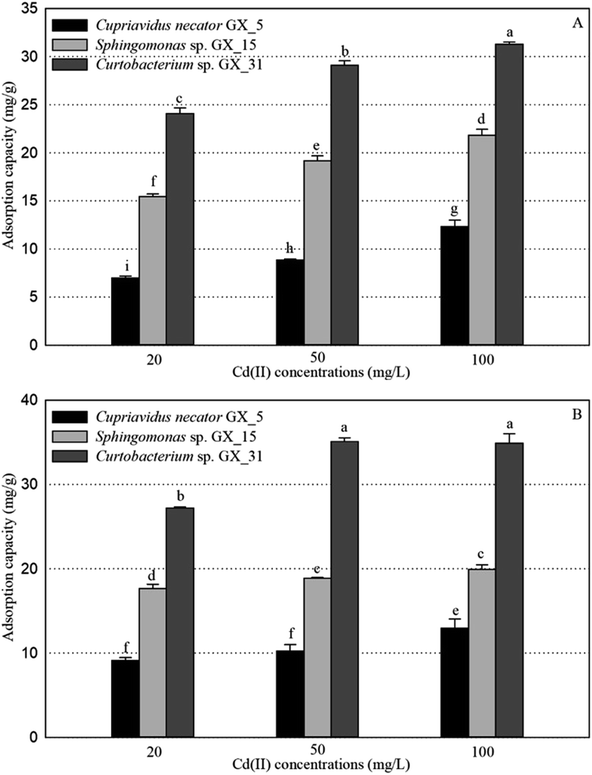
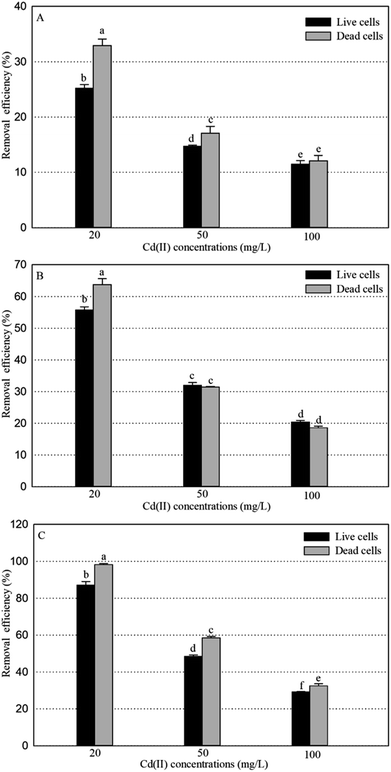
Under optimal conditions (pH, 6.0; reaction time, 6 h; biomass dosage, 1.0 g L−1) based on the preliminary data,25 the adsorption experiments for Cd(II) of live and dead biosorbents of the three PGPR (Cupriavidus necator GX_5, Sphingomonas sp. GX_15, and Curtobacterium sp. GX_31) were conducted with initial Cd(II) concentrations of 20, 50, and 100 mg L−1. There were significant differences in the removal efficiency and adsorption capacity of live and dead biosorbents between the three strains, and the ranking can be ordered as GX_31 > GX_15 > GX_5 (p < 0.05), whatever the initial Cd(II) concentration is (20, 50 or 100 mg L−1) (Fig. 1 and 2). Other biosorbents have also been used for Cd(II) adsorption. For example, the maximum sorption capacity of NTAA-LCM for Cd(II) reached 143.4 mg g−1 with an initial Cd(II) concentration of 200 mg L−1,26 higher than that in this study (31.28 and 35.09 mg g−1 for the live and dead biomass of GX_31 with 100 mg L−1 of Cd(II)) (Fig. 2). On the contrary, the Cd(II) adsorption capacity of Penicillium simplicissimum was 21.5 mg g−1,27 lower than that of GX_31. In another investigation, the maximum adsorption capacity of the dry waste biofilms for Cd(II) (42 mg g−1) was also higher. However, the removal efficiency of Cd(II) was 89.3%,28 which was lower than that of the dead biosorbent of GX_31 (98.27%) (Fig. 1B). It seems that the results from different studies may not be directly comparable on account of differences in experimental conditions,29 but it is reasonable to make a comparison in the present study under the same conditions. The adsorption capacity and removal efficiency varied among the live and dead biomass of these strains due to their own adsorption mechanisms.30Regarding the live biosorbent of the same strain, whatever the strain was, the removal efficiency and adsorption capacity also had significant variations under different Cd(II) concentrations (Fig. 1A and 2A). Fig. 1A shows that the removal efficiency was clearly higher under a lower Cd(II) concentration than under a higher Cd(II) concentration, which adheres to findings in other investigations.31,32 At a lower Cd(II) concentration, there were easily enough free binding sites of live biosorbent for Cd(II) to interact, which resulted in a high removal efficiency. Therefore, the maximum Cd(II) removal efficiency of the live biomass of GX_5, GX_15 and GX_31 displayed with an initial Cd(II) concentration of 20 mg L−1 was 25.21%, 55.79% and 87.03%, respectively, and the minimum removal efficiency was 11.48%, 29.16% and 31.45%, respectively (100 mg L−1 of Cd(II)) (Fig. 1A). On the other hand, the adsorption capacity of live biosorbent increased significantly with an increasing initial Cd(II) concentration for an identical strain (Fig. 2A), which was consistent with the study by Cheng et al.33 The strongest and weakest adsorption capacities were 12.31, 21.83, and 31.28 mg g−1 and 6.98, 15.44, and 24.08 mg g−1 for GX_5, GX_15, and GX_31 at 100 mg L−1 and 20 mg L−1 of Cd(II), respectively (Fig. 2A). Considering the same amount of biomass dosage, a high initial concentration could supply a driving force to interact with limited Cd(II) binding sites and facilitate adsorption by the live biomass.34
There was also a significant difference in the removal efficiency for Cd(II) of the dead biosorbent of the same strain under different Cd(II) concentrations (Fig. 1B). This displayed the same tendency as that of the live biosorbent, namely, that the removal efficiency decreased along with the increasing initial Cd(II) concentration, because at a high Cd(II) concentration, a lack of adequate binding sites restricted the removal efficiency.35 Fig. 1B shows that the highest and lowest removal efficiencies of the dead biosorbents of GX_5, GX_15, and GX_31 were 32.95%, 63.77%, and 98.27% and 12.09%, 18.58%, and 32.51% under 20 and 100 mg L−1 of Cd(II), respectively. However, the adsorption capacity of the dead biosorbent did not show the same tendency as the live biosorbent did, except GX_5, which had the largest and smallest adsorption capacities of 12.97 and 9.12 mg g−1 at 100 and 20 mg L−1 of Cd(II), respectively (Fig. 2B). For the dead biomass of GX_15 and GX_31, the adsorption capacity increased with a higher metal concentration and it reached a saturation value due to finite binding sites. When the concentration of Cd(II) changed from 20 to 50 mg L−1, the adsorption capacities varied from 17.65 to 18.87 mg L−1 for GX_15 and 27.19 to 35.09 mg L−1 for GX_31 (Fig. 2B). A higher Cd(II) concentration did not lead to a higher adsorption capacity. This phenomenon agrees with the study by Khadivinia et al., who point out that at a higher Cd(II) concentration, the binding sites become fewer and the biosorbent sites adsorb metal ions more quickly at lower concentrations, which leads to the decrease of adsorption yield.36
3.2. SEM-EDX analysis
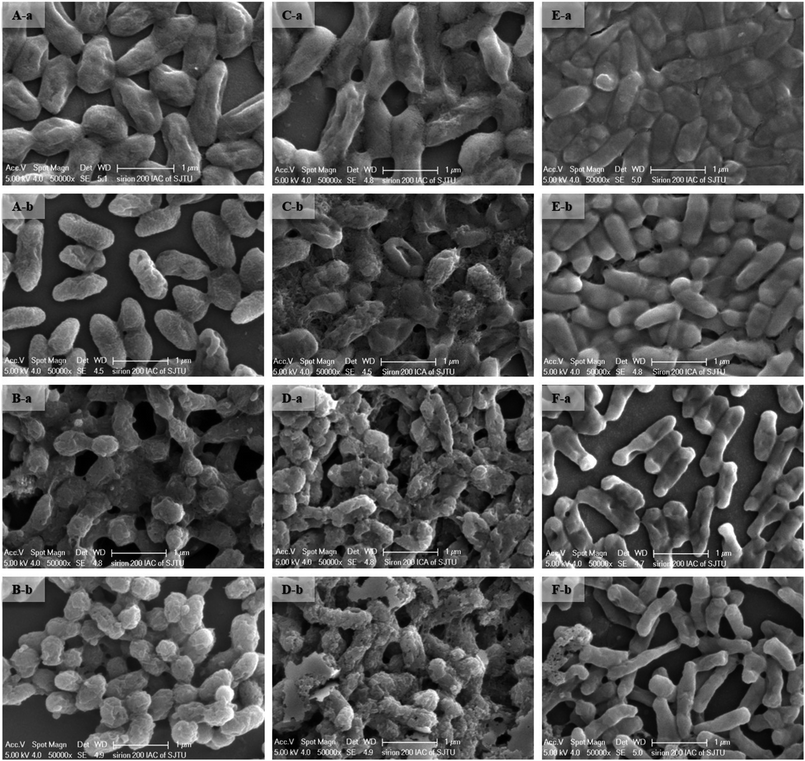
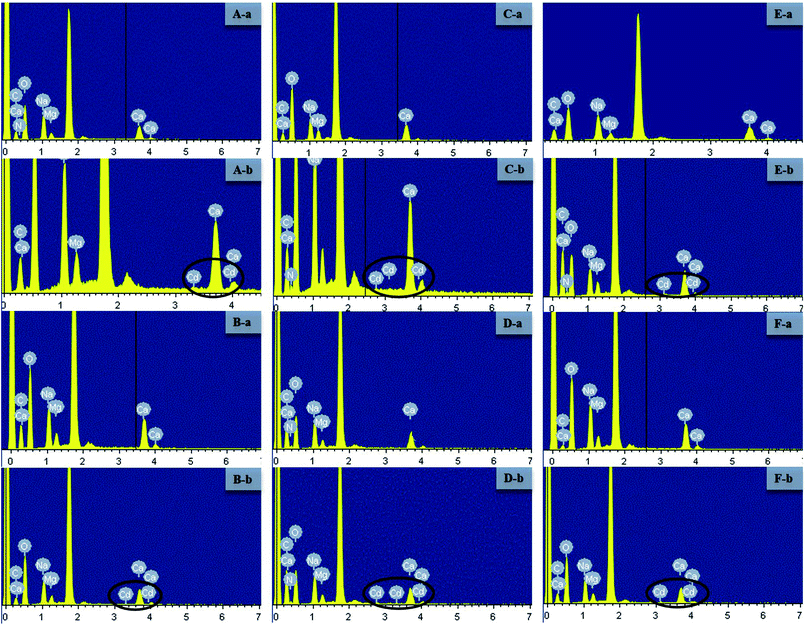
The surface structure and cell morphology changes of the live and dead biosorbent were determined by SEM coupled with EDX before and after interaction with 100 mg L−1 of Cd(II), as depicted in Fig. 3 and 4. Before adsorption, all the live biosorbents were observed to be rod shapes with clear boundaries (Fig. 3A-a–C-a) and the dead biosorbents evidently seemed to join together, especially for GX_5 and GX_15 (Fig. 3B-a and D-a). However, after binding, the surfaces of both live and dead cells became rough, irregular, and noticeably covered with silvery white sediments (Fig. 3). The SEM micrographs also indicated that Cd(II) exposure caused anomalous aggregation of the L. plantarum CCFM8610.37 Similarly, the surface of the live and dead biomass of Spirulina sp. became rough after Cd(II) uptake,38 which was in accordance with the SEM observations. For GX_15, some floccus precipitation was found on their surfaces (Fig. 3C and D), which was also observed by Lin et al.39 As pointed out in another study, after Cd(II) adsorption, more flocculated sediments appeared near the cell surface of E. cloacae TU.19 Likewise, under heavy metal stress, many flocculated particles could be discovered, which suggested the presence of heavy metals on the cell wall.40 EDX spectra recorded the signals of carbon, nitrogen, oxygen, sodium, magnesium, and calcium, which were likely in the polysaccharides and proteins of the biomass (Fig. 4). No Cd(II) signals were detected in unloaded biosorbent, but clear peaks for Cd(II) were observable after Cd(II) exposure, which indicates the presence of Cd(II) in the biosorbents after adsorption. SEM-EDX confirmed that both the live and dead biomass of these strains had potential for Cd(II) remediation in cadmium polluted environments. SEM-EDX is a useful tool for visual confirmation of surface morphology changes of cells after absorbing metal ions and it has been extensively applied in research.24,41,423.3. FTIR analysis
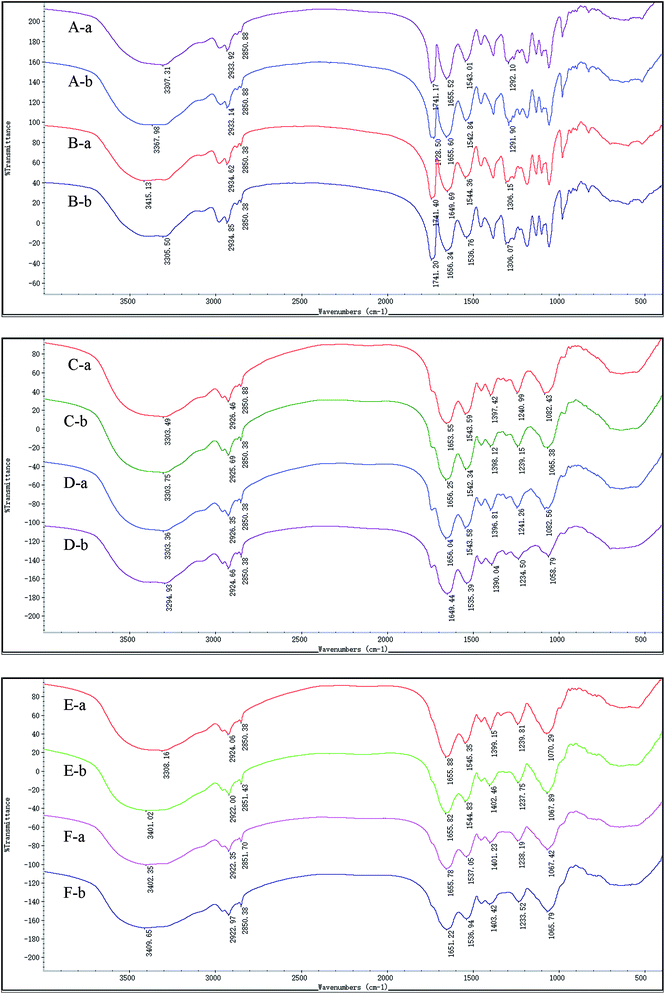
The surface functional group of the biosorbent is an important factor for metal adsorption. FTIR spectra of the live and dead biosorbents of GX_5, GX_15, and GX_31 were recorded before and after Cd(II) uptake. Fig. 5 shows various functional groups present on the cell surface, and reveals the complex nature of these strains. The spectra of GX_15 and GX_31 are similar to each other but they differ from GX_5, especially in the region of 1400–700 cm−1.In this study, live and dead cells displayed similar spectra, and all the characteristic peaks were present on both biosorbents, which was in consistent with other studies (Fig. 5).16,43,44 Broad bands in the range of 3200–3500 cm−1 correspond to the hydroxyl group as well as the –NH bond of the amino group.45 Two peaks at approximately 2930 and 2850 cm−1 were ascribed to symmetric and asymmetric –CH vibration in lipids.36 The band at 1741 cm−1 represented the carbonyl group in the polyhydroxyalkanoic acid, identified by nuclear magnetic resonance analysis of a chloroform extract.46 The peak positions around 1655 and 1543 reflected the presence of amide I (–CO– stretching vibration) and amide II (–NH bending and –CN stretching) in proteins, respectively.47 Peaks at 1240 and 1071 cm−1 were assigned to the alcoholic –CN and –CO– stretching vibration, revealing the presence of amino and hydroxyl groups,48 and a band at 1400 cm−1 also represented a hydroxyl group.27 The –CO–, –CN, P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, and S
O, and S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations all existed at the band at about 1291.09 cm−1.49 An adsorption band was positioned at 1080 cm−1, corresponding to the –CO– group vibration in the cyclic structure of carbohydrates.36
O stretching vibrations all existed at the band at about 1291.09 cm−1.49 An adsorption band was positioned at 1080 cm−1, corresponding to the –CO– group vibration in the cyclic structure of carbohydrates.36
Although they had similar spectra, there were minor changes when we compared the spectra between live and dead biosorbents before adsorption. The bands were shifted from 3307.31, 1655.52, 1543.01, and 1292.01 cm−1 to 3415.13, 1649.69, 1544.36, and 1306.15 cm−1 for GX_5 (Fig. 5A-a and B-a); from 1653.55, and 1397.42 cm−1 to 1656.04, and 1396.81 cm−1 for GX_15 (Fig. 5C-a and D-a); and from 3308.16, 1545.35, and 1070.29 cm−1 to 3402.35, 1537.05, and 1067.42 cm−1 for GX_31 (Fig. 5E-a and F-a), which indicated that autoclave had some effect on the functional groups, though not significant.
After adsorption, for GX_5, shifts were observed for the live biomass from 3307.31, and 1741.17 cm−1 to 3367.98, and 1728.50 cm−1, and from 3415.13, 1649.69, and 1544.36 cm−1 to 3305.5, 1656.34, and 1536.76 cm−1 for dead biomass, revealing that hydroxyl, amino, carbonyl, or amide groups were involved for binding Cd(II) (Fig. 5A and B). Fig. 5C and D show that peaks at 1653.55, 1397.42, and 1082.43 cm−1 of live cells of GX_15 shifted to 1656.25, 1398.12, and 1065.38 cm−1, while bands down shifted from 3303.36, 1656.04, 1543.38, 1396.81, 1241.26, and 1082.56 cm−1 to 3294.93, 1649.44, 1535.39, 1390.04, 1234.50, and 1058.79 cm−1, indicating that hydroxyl, amino, amide, or carboxyl groups were responsible for the Cd(II) removal. However, it seemed that more bands were involved in the adsorption of Cd(II) in the dead biosorbent, which corresponds to the findings of a previous study by Huang et al.20 In the case of GX_15, the peaks of live cells at 3308.16, 1399.15 and 1070.29 cm−1 were shifted to 3401.02, 1402.46, and 1067.89 cm−1 and those of dead cells, 3402.35, 1655.78, 1401.23, 1238.19, and 1067.42 cm−1 to 3409.65, 1651.22, 1403.42, 1233.52, and 1065.79 cm−1 (Fig. 5E and F), demonstrating that hydroxyl, amino, carboxyl, or amide groups played an important role for Cd(II) adsorption. It also appeared that more groups participated in Cd(II) binding.
The present FTIR spectra analysis provides evidence that functional groups like hydroxyl, amino, amide, carbonyl, and carboxyl groups are involved in the binding of Cd(II) on surface of both live and dead biosorbents. Such findings have also been reported in other research.50–53
3.4. Live and dead biosorbent comparison
The removal efficiencies for Cd(II) of live and dead biosorbents of GX_5, GX_15, and GX_31 were compared, with different Cd(II) concentrations. It was clear that the performance of the removal efficiency was different between the live and dead biomass of these three strains (Fig. 6). The Cd(II) removal capacity of dead cells was markedly higher than that of live cells with an initial Cd(II) concentration of 20 mg L−1 for all the strains (Fig. 6). The increase of the metal adsorption capacity of the dead biomass can be explained in the following ways. A loss of cell membrane integrity during autoclaving allows the exposure of binding sites inside the cell.16 According to the hypothesis, an increase of Cu(II) adsorption by Penicillium spinulosum and of U uptake by S. cerevisiae cells permeabilized by the action of detergents53 or by the action of HCHO or HgCl2,54 respectively, was also described. Machado assessed the membrane integrity of heat-treated S. cerevisiae cells with propidium iodide by fluorescence microscopy and observed membrane integrity was lost during the thermal treatment.44 In addition, more functional groups are involved in Cd(II) uptake as depicted in the former section. Li et al. also pointed out that in their study more functional groups participated in the adsorption processes of the dead biomass, compared with those linked to the live biomass.55 The third reason is the efflux mechanism of live cells, which could reduce Cd(II) adsorption. The most frequent mechanism of toxic divalent cation resistance is energy-dependent pumping out, that is membrane efflux pumps.56 Many studies have focused on the Cd(II) efflux mechanism of live cells for coping with high Cd(II) stress.57,58 When the Cd(II) concentrations were 50 and 100 mg L−1, the dead cells of GX_31 still outperformed the live cells (Fig. 6C). However, for GX_15, the adsorption efficiency of the live biomass was observed to be slightly higher, but there were no differences in the dead biomass (Fig. 6B). Similarly, the adsorption capacity of GX_5 was significantly higher in the dead biosorbent than in the live biosorbent with a Cd(II) concentration of 50 mg L−1. On the contrary, under a Cd(II) concentration of 100 mg L−1, the metal loading capacity of the dead cells of GX_5 was equivalent to that of the living cells (Fig. 6A).With an increase of the Cd(II) concentration, the adsorption efficiency of the live biomass got close to that of the dead biomass, which may be because of the additional intercellular accumulation of living cells.2 The initial Cd(II) concentration had a significant effect on the adsorption. However, in this study, the live biosorbents did not exhibit a significantly higher adsorption capacity than the dead biosorbents, which is reported by other researchers.18,38 The adsorption capacity of the live cells may be greater to, equal to, or less than that of the dead cells and it may depend on the bacteria under consideration, experimental methods and type of metal ions being used.59 As shown by Machado et al., the inactivated biomass of Saccharomyces cerevisiae displayed a greater Zn2+ and Ni2+ accumulation, but a similar accumulation of Cu2+ than in the live biomass.16
Cd(II) adsorption results of dead and live cells may suggest that the dead biomass is a preferred alternative for remediating Cd(II) in a contaminated environment. Furthermore, the dead biosorbent possessed some advantages: no requirements for nutrients, no toxicity limitations, easy recovery of metals, easy regeneration and reuse of biomass, and less affected by pH and temperature.60–62 Despite the obvious advantages of using dead biomass over live biomass, many attributes of living microbes should be emphasized as well. Live strains could degrade organic pollutants and can adsorb, transport, complex and transform metals, metalloids and radionuclides,63 especially the use of live PGPR for remediating heavy metal contaminated soils.64
4. Conclusions
The removal efficiencies and adsorption capacities of live and dead biosorbents of Cupriavidus necator GX_5, Sphingomonas sp. GX_15, and Curtobacterium sp. GX_31 are quite different and strongly affected by the initial Cd(II) concentrations. Whether the cells are living or dead, the adsorption capacity among strains can be listed as GX_31 > GX_15 > GX_5. SEM-EDX, FTIR analysis and adsorption studies indicate that both live and dead cells have the ability to adsorb Cd(II), but the dead biomass is superior to the live biomass within the experimental conditions tested, especially when the concentration of Cd(II) is low. However, to obtain more accurate results and apply them in practice, more strains should be employed to compare the remediation capacity between live and dead biosorbents for different metals, under a series of metal concentrations.Conflicts of interest
There is no conflict of interest and all the authors are interested to publish the manuscript.Acknowledgements
This work was supported by Agriculture Committee of Shanghai (2014/5-2). We are thankful to MS Yao Han for SEM and EDX analysis, and Prof. Bangshang Zhu for FTIR spectra analysis (Instrumental Analysis Center of Shanghai Jiao Tong University, Shanghai, China).References
- R. A. Wuana and F. E. Okieimen, ISRN Ecol., 2011, 2011, 1–20 CrossRef.
- W. Zhou, D. Liu, H. Zhang, W. Kong and Y. Zhang, Bioresour. Technol., 2014, 165, 145–151 CrossRef CAS PubMed.
- M. Beldowska, A. Zgrundo and J. Kobos, Water, Air, Soil Pollut., 2018, 229, 168 CrossRef PubMed.
- G. C. R. Casagrande, C. dos Reis, R. Arruda, R. L. T. de Andrade and L. D. Battirola, Water, Air, Soil Pollut., 2018, 229, 116 CrossRef.
- J. Marrugo-Negrete, J. Pinedo-Hernández and S. Díez, Environ. Res., 2017, 154, 380–388 CrossRef CAS PubMed.
- R. Strachel, J. Wyszkowska and M. Baćmaga, Water, Air, Soil Pollut., 2018, 229, 235 CrossRef PubMed.
- J. Wang and C. Chen, Biotechnol. Adv., 2009, 27, 195–226 CrossRef CAS PubMed.
- J. Wang and C. Chen, Biotechnol. Adv., 2006, 24, 427–451 CrossRef CAS PubMed.
- Y. Cheng, C. Yang, H. He, G. Zeng, K. Zhao and Z. Yan, J. Environ. Eng. Div. (Am. Soc. Civ. Eng.), 2016, 142, C4015001 CrossRef.
- K. Vijayaraghavan and Y. S. Yun, Biotechnol. Adv., 2008, 26, 266–291 CrossRef CAS PubMed.
- X. Ren, G. Zeng, L. Tang, J. Wang, J. Wan, H. Feng, B. Song, C. Huang and X. Tang, Soil Biol. Biochem., 2018, 116, 70–81 CrossRef CAS.
- C. Yang, J. Wang, M. Lei, G. Xie, G. Zeng and S. Luo, J. Environ. Sci., 2010, 22, 675–680 CrossRef CAS.
- B. Prapagdee, M. Chanprasert and S. Mongkolsuk, Chemosphere, 2013, 92, 659–666 CrossRef CAS PubMed.
- B. R. Glick, Biotechnol. Adv., 2010, 28, 367–374 CrossRef CAS PubMed.
- M. M. Areco, S. Hanela, J. Duran and S. Afonso Mdos, J. Hazard. Mater., 2012, 213–214, 123–132 CrossRef CAS PubMed.
- M. D. Machado, S. Janssens, H. M. Soares and E. V. Soares, J. Appl. Microbiol., 2009, 106, 1792–1804 CrossRef CAS PubMed.
- L. Velasquez and J. Dussan, J. Hazard. Mater., 2009, 167, 713–716 CrossRef CAS PubMed.
- J. Tangaromsuk, P. Pokethitiyook, M. Kruatrachue and E. Upatham, Bioresour. Technol., 2002, 85, 103–105 CrossRef CAS PubMed.
- C. Xu, S. He, Y. Liu, W. Zhang and D. Lu, Chemosphere, 2017, 173, 622–629 CrossRef CAS PubMed.
- F. Huang, Z. Dang, C. L. Guo, G. N. Lu, R. R. Gu, H. J. Liu and H. Zhang, Colloids Surf., B, 2013, 107, 11–18 CrossRef CAS PubMed.
- J. Guo, X. D. Zheng, Q. B. Chen, L. Zhang and X. P. Xu, Curr. Microbiol., 2012, 65, 350–355 CrossRef CAS PubMed.
- L. Fang, P. Cai, W. Chen, W. Liang, Z. Hong and Q. Huang, Colloids Surf., A, 2009, 347, 50–55 CrossRef CAS.
- M. Oves, M. S. Khan and H. A. Qari, J. Taiwan Inst. Chem. Eng., 2017, 80, 540–552 CrossRef CAS.
- O. A. Mohamad, X. Hao, P. Xie, S. Hatab, Y. Lin and G. Wei, Microbes Environ., 2012, 27, 234–241 CrossRef PubMed.
- X. J. Li, D. B. Li, Z. N. Yan and Y. S. Ao, RSC Adv., 2018, 8, 30902–30911 RSC.
- Y. Huang, C. Yang, Z. Sun, G. Zeng and H. He, RSC Adv., 2015, 5, 11475–11484 RSC.
- T. Fan, Y. Liu, B. Feng, G. Zeng, C. Yang, M. Zhou, H. Zhou, Z. Tan and X. Wang, J. Hazard. Mater., 2008, 160, 655–661 CrossRef CAS PubMed.
- H. J. He, Z. H. Xiang, X. J. Chen, H. Chen, H. Huang, M. Wen and C. P. Yang, Int. J. Environ. Sci. Technol., 2017, 15, 1491–1500 CrossRef.
- J. Bai, X. Yang, R. Du, Y. Chen, S. Wang and R. Qiu, J. Environ. Sci., 2014, 26, 2056–2064 CrossRef PubMed.
- T. Limcharoensuk, N. Sooksawat, A. Sumarnrote, T. Awutpet, M. Kruatrachue, P. Pokethitiyook and C. Auesukaree, Ecotoxicol. Environ. Saf., 2015, 122, 322–330 CrossRef CAS PubMed.
- P. Arivalagan, D. Singaraj, V. Haridass and T. Kaliannan, Ecol. Eng., 2014, 71, 728–735 CrossRef.
- M. Oves, M. S. Khan and A. Zaidi, Saudi J. Biol. Sci., 2013, 20, 121–129 CrossRef CAS PubMed.
- J. Cheng, W. Yin, Z. Chang, N. Lundholm and Z. Jiang, J. Appl. Phycol., 2016, 29, 211–221 CrossRef.
- N. Masoudzadeh, F. Zakeri, T. bagheri Lotfabad, H. Sharafi, F. Masoomi, H. S. Zahiri, G. Ahmadian and K. A. Noghabi, J. Hazard. Mater., 2011, 197, 190–198 CrossRef CAS PubMed.
- P. Chakravarty, N. S. Sarma and H. Sarma, Chem. Eng. J., 2010, 162, 949–955 CrossRef CAS.
- E. Khadivinia, H. Sharafi, F. Hadi, H. S. Zahiri, S. Modiri, A. Tohidi, A. Mousavi, A. H. Salmanian and K. A. Noghabi, J. Ind. Eng. Chem., 2014, 20, 4304–4310 CrossRef CAS.
- Q. Zhai, F. Tian, G. Wang, J. Zhao, X. Liu, K. Cross, H. Zhang, A. Narbad and W. Chen, RSC Adv., 2016, 6, 5990–5998 RSC.
- H. Doshi, A. Ray and I. L. Kothari, Curr. Microbiol., 2007, 54, 213–218 CrossRef CAS PubMed.
- Y. Lin, X. Wang, B. Wang, O. Mohamad and G. Wei, Ecotoxicol. Environ. Saf., 2012, 77, 7–17 CrossRef CAS PubMed.
- F. Huang, Z. H. Wang, Y. X. Cai, S. H. Chen, J. H. Tian and K. Z. Cai, Ecotoxicol. Environ. Saf., 2018, 157, 216–226 CrossRef CAS PubMed.
- M. Polak-Berecka, P. Boguta, J. Ciesla, A. Bieganowski, T. Skrzypek, T. Czernecki and A. Wasko, Appl. Microbiol. Biotechnol., 2017, 101, 3415–3425 CrossRef CAS PubMed.
- Y. Yang, M. Hu, D. Zhou, W. Fan, X. Wang and M. Huo, RSC Adv., 2017, 7, 18793–18802 RSC.
- N. Chubar, T. Behrends and P. Van Cappellen, Colloids Surf., B, 2008, 65, 126–133 CrossRef CAS PubMed.
- X. Lu, X. J. Zhou and T. S. Wang, J. Hazard. Mater., 2013, 262, 297–303 CrossRef CAS PubMed.
- W. Huang and Z. M. Liu, Colloids Surf., B, 2013, 105, 113–119 CrossRef CAS PubMed.
- S. S. Zamil, M. H. Choi, J. H. Song, H. Park, J. Xu, K. W. Chi and S. C. Yoon, Appl. Microbiol. Biotechnol., 2008, 80, 531–544 CrossRef CAS PubMed.
- H. Wu, Q. Wu, G. Wu, Q. Gu and L. Wei, PLoS One, 2016, 11, e0151479 CrossRef PubMed.
- J. Yu, M. Tong, X. Sun and B. Li, J. Hazard. Mater., 2007, 143, 277–284 CrossRef CAS PubMed.
- J. Pan, R. Liu and H. Tang, J. Environ. Sci., 2007, 19, 403–408 CrossRef CAS.
- N. N. N. A. Rahman, M. Shahadat, C. A. Won and F. M. Omar, RSC Adv., 2014, 4, 58156–58163 RSC.
- G. Sathiyanarayanan, S. K. Bhatia, H. J. Kim, J.-H. Kim, J.-M. Jeon, Y.-G. Kim, S.-H. Park, S. H. Lee, Y. K. Lee and Y.-H. Yang, RSC Adv., 2016, 6, 96870–96881 RSC.
- T. Wang, X. Zheng, X. Wang, X. Lu and Y. Shen, J. Environ. Radioact., 2017, 167, 92–99 CrossRef CAS PubMed.
- G. M. Gadd, Fungi and yeasts for metal accumulation, New York, McGraw-Hill, 1990, pp. 249–276 Search PubMed.
- G. W. Strandberg, S. E. II Shumate and J. R. Jr Parrot, Appl. Environ. Microbiol., 1981, 41, 237–245 CAS.
- H. Li, Y. Lin, W. Guan, J. Chang, L. Xu, J. Guo and G. Wei, J. Hazard. Mater., 2010, 179, 151–159 CrossRef CAS PubMed.
- S. Silver and T. Phung le, J. Ind. Microbiol. Biotechnol., 2005, 32, 587–605 CrossRef CAS PubMed.
- D. H. Nies, FEMS Microbiol. Rev., 2003, 27, 313–339 CrossRef CAS PubMed.
- F. Huang, C. Guo, G. Lu, X. Yi, L. Zhu and Z. Dang, Chemosphere, 2014, 109, 134–142 CrossRef CAS PubMed.
- T. Akar and S. Tunali, Bioresour. Technol., 2006, 97, 1780–1787 CrossRef CAS PubMed.
- N. R. Bishnoi and Garima, J. Sci. Ind. Res., 2005, 64, 93–100 CAS.
- A. Hansda, V. Kumar and Anshumali, World J. Microbiol. Biotechnol., 2016, 32, 170 CrossRef PubMed.
- E. V. Soares and H. M. Soares, Environ. Sci. Pollut. Res., 2012, 19, 1066–1083 CrossRef PubMed.
- M. Fomina and G. M. Gadd, Bioresour. Technol., 2014, 160, 3–14 CrossRef CAS PubMed.
- H. Chauhan, D. J. Bagyaraj, G. Selvakumar and S. P. Sundaram, Appl. Soil Ecol., 2015, 95, 38–53 CrossRef.
| This journal is © The Royal Society of Chemistry 2018 |