Open Access Article
Open Access ArticleMechanistic insights into geniposide regulation of bile salt export pump (BSEP) expression
Guixin Wu†
a,
Min Wen†a,
Lin Suna,
Huitao Lia,
Yubei Liua,
Rui Lib,
Feihua Wua,
Rong Yanga and
Yining Lin *a
*a
aSchool of Traditional Chinese Pharmacy, China Pharmaceutical University, 639 Longmian Avenue, Nanjing 211198, P. R. China. E-mail: cpuzyzj246@163.com
bSchool of Pharmacy, Nanjing Medical University, Nanjing 211166, P. R. China
First published on 5th November 2018
Abstract
Geniposide (GE) is a major component isolated from Gardenia jasminoides Ellis, which has been used to treat cholestasis liver diseases. Our previous study has shown that GE could notably increase mRNA and protein expressions of BSEP in cholestatic rats. BSEP plays a critical role in maintenance of the enterohepatic circulation of bile acids. BSEP could be regulated by the transactivation pathway of farnesoid X receptor (FXR) and nuclear factor erythroid 2-related factor (Nrf2). Here the mechanisms for BSEP regulation by GE were investigated. GE induced the mRNA levels of BSEP in HepG2 cells and cholestatic mice, and knockdown of FXR and Nrf2 reduced the mRNA expression of BSEP at varying degrees after treatment of GE. FXR acts as the major regulator of BSEP transcription. The involvement of FXR regulated BSEP expression by GE was further investigated. An enhancement was observed in FXR-dependent BSEP promoter activation using luciferase assay. ChIP assay further confirmed the interaction between FXR and BSEP after GE treatment. Using siRNA and ChIP assays, we demonstrated that peroxisome-proliferator-activated receptor γ co-activator-1α (PGC-1α) and co-activator-associated arginine methyltransferase 1 (CARM1) were predominantly recruited to the BSEP promoter upon FXR activation by GE. In conclusion, GE regulated the expression of BSEP through FXR and Nrf2 signaling pathway. The FXR transactivation pathway was enhanced by increasing recruitment of coactivators PGC-1α and CARM1 upon GE treatment, coupled with an increased binding of FXR to the BSEP promoter.
1 Introduction
Bile acids are synthesized in the liver and play important roles in transport, absorption, and elimination of cholesterol and lipids. However, excessive intracellular accumulation of bile acids is toxic to hepatocytes and causes cholestasis liver diseases.1,2 Bile acids homeostasis is mainly maintained through keeping bile acids continuously cycled in the enterohepatic circulation and enterohepatic bile flow is essential for the elimination of toxic compounds, which is conducted by the efflux transporters residing in the canalicular membrane of hepatocytes.3,4 Bile salt export pump (BSEP) is the major bile acids transporter at the mammalian canalicular membrane, which is the rate-limiting step in bile secretion and protects from accumulation of toxic bile acids during cholestasis by facilitating their efflux into the canaliculus.5,6 The impairment of BSEP expression or function has been directly linked to several cholestatic liver diseases such as progressive familial intrahepatic cholestasis type 2, intrahepatic cholestasis of pregnancy, and cholesterol cholelithiasis.7–9 Considering the crucial role of BSEP plays in cholestasis liver disease, regulation of BSEP expression has been a subject of intense research.The expression of BSEP gene is positively regulated by FXR, Nrf2.10,11 LRH-1 transcriptionally regulates BSEP expression through LRHRE1 in the BSEP promoter. Species differences exhibited in LRH-1-mediated transactivation.12 The LRHRE1 site was not completely conserved between human and mouse or rat, LRHRE1-like site in mouse is functional in responding to LRH-1. The transactivation was not observed with the rat BSEP promoter due to the absence of the LRHRE.13 Previous studies have reported that Nrf2 acts as a positive transcriptional regulator of the human BSEP promoter. BSEP expression is decreased after lowered Nrf2 expression in HepG2 cells.14 In addition, many pieces of evidence support that the nuclear receptor FXR acts as the major stimulator of BSEP transcription.15,16 It has been reported that excretion of bile acids from hepatocytes was markedly impaired in FXR knockout mice, leading to excessive intracellular accumulation of bile acids.17 The decreased hepatic bile acids secretion correlated with down-regulated expression of the predominant canalicular bile acids efflux transporter, known as BSEP. Therefore, the induction of BSEP expression by activation of nuclear receptor FXR becomes an important target for curing cholestatic liver disorders.
FXR transcriptional activity is modulated by coactivators, which act as important regulators mediated BSEP transcription. Several FXR coactivators have been reported, including peroxisome-proliferator-activated receptor γ co-activator-1α (PGC-1α), co-activator-associated arginine methyltransferase 1 (CARM1), protein arginine methyltransferase type 1 (PRMT1), and steroid receptor co-activator 1 (SRC-1).18–20 The critical regulator of energy homeostasis PGC-1α has been reported to interact with FXR to increase FXR-dependent BSEP expression in a ligand-dependent manner.21 There are growing evidence for the activation of FXR through their modeling of chromatin by PRMT1, CARM1, as well as SRC-1 through histone modification involving methylation and acetylation.22,23 These results indicated that facilitated ligand-induced FXR transcriptional activity by increasing its interaction with transcriptional coactivators was indispensable for full potentiation of the BSEP promoter.
GE is a major active component of the fruit of Gardenia jasminoides Ellis, which is widely used as a traditional Chinese medicine for the treatment of inflammation and hepatic disorders.24,25 GE exhibits many important activities, such as anti-oxidative, anti-inflammatory, and anti-cholestasis effect.26–28 GE could alleviate cholestasis via regulating uptake and efflux transporters and enzymes involved in bile acids homeostasis in rats and mice.29,30 Our previous study found that GE (100 mg kg−1) could increase gene and protein expression of FXR and notably modulate mRNA and protein levels of BSEP in rats. BSEP played a critical role in regulation of hepatic bile acids homeostasis by mediating the biliary excretion of bile acids. BSEP regulation is complex and several transcriptional factors target BSEP gene expression. To our knowledge, the potential mechanisms of GE effect on BSEP upregulation remain unclear. The aim of this study was to investigate the transcription factors involved in BSEP expression regulated by GE. Considering the highly conserved transactivation pathways between human and mouse, the effects of FXR and Nrf2 pathway on BSEP expression were evaluated in mice and HepG2 cells. The results will help to elucidate the anti-cholestasis effect of GE.
2 Materials and methods
2.1 Chemicals and reagents
Geniposide and tert-butyl hydroquinone (t-BHQ) were purchased from Aladdin (Shanghai, China). GW4064 was obtained from Selleck Chemicals (Houston, TX) and prepared as a 5 mM stock solution in DMSO. Dulbecco's modified Eagle's medium (DMEM) and OPTI-MEM were from Gibco (Grand Island, NY). Fetal bovine sera (FBS) was from Sciencell (Carlsbad, CA). Lipofectamine 2000 was from Invitrogen (Carlsbad, CA). A Dual Luciferase assay kit was purchased from Promega (Madison, WI). Antibodies against FXR and BSEP were obtained from Abcam (Cambridge, MA). The antibodies against Nrf2 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were purchased from Bioworld Technology (St. Paul, MN). Antibodies to CARM1 and PGC-1α were purchased from proteintech (Chicago, IL). Antibodies to Lamin B, β-actin, PRMT1 and SRC-1 were from Abways (Danvers, MA). Other supplies and reagents are described below.2.2 Animals and treatments
Animals were maintained in accordance with the Guidelines for Care and Use of Laboratory Animals of China Pharmaceutical University. All the experimental procedures were approved by the Animal Ethics Committee of our university. Male ICR mice (20–25 g) were purchased from Nanjing Qinglong Experimental Animal Co. Ltd., China. Mice were housed at 22–23 °C with a 12 h light/dark cycle. Water and food were available for mice ad libitum. Animals were randomly divided into four groups (n = 8 per group): vehicle, GE (150 mg kg−1, dissolved in normal saline), ANIT (75 mg kg−1, dissolved in olive oil), GE (150 mg kg−1) + ANIT. GE or vehicle was treated to mice by oral gavage once daily for 7 days. On the 5th day, 4 h after GE or vehicle treatment, mice were received oral gavage a single dose of ANIT. On the 7th day, 4 h after GE or vehicle treatment, blood samples were obtained to determine serum biochemical parameters. Liver tissues were isolated and stored at −80 °C until use. The blood samples were centrifuged at 3500 rpm for 10 min to obtain serum, and then stored at −80 °C for further use.2.3 Liver function assay and serum inflammatory cytokines determination
Serum levels of alanine transaminase (ALT), aspartate transaminase (AST), alkaline phosphatase (ALP), total bilirubin (TBIL), direct bilirubin (DBIL), and total bile acid (TBA) were measured to evaluate hepatic injury. The serum IL-1β and IL-6 concentrations were assayed using commercially available kits (Jiancheng Bioengineering Institute, Nanjing, China).2.4 Measurement of bile flow
Mice were fasted for 12 hours with free access to water. A laparotomy was performed under anesthesia, the cystic duct was ligated and the common bile duct was cannulated with a 30-gauge needle. After a 5 min equilibration period, bile was collected for 30 min to measure the flow rate, and the volume was determined gravimetrically assuming a density of 1.0 g mL−1 and normalized to liver weight.2.5 HepG2 cells culture and treatments
HepG2 cell line was purchased from the Institute of Biochemistry Cell (Shanghai, China), and were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS at 37 °C in a humidified atmosphere (5% CO2). Cells were plated at a density of 5 × 105 cells per well. The following day, the cells treated with GE at final concentrations of 25, 50, and 100 μM for 24 h. The positive controls GW4064 and t-BHQ were added to a final concentration of 5 and 25 μM, respectively. The cultures were harvested after 24 h incubation.2.6 Cell viability assay
Cell viability was assessed by the CCK-8 assay. HepG2 cells (5 × 103 per well) were cultured in 96-well plates overnight and then were treated in culture medium with different concentrations of GE (1, 10, 25, 50, 75, and 100 μM). After 24 h incubation, the CCK-8 solution was added to each well at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution followed by further incubation at 37 °C for 2 h. The absorbance of each well was measured at 450 nm using a microplate reader (Varioskan Flash, Thermo Electron).
10 dilution followed by further incubation at 37 °C for 2 h. The absorbance of each well was measured at 450 nm using a microplate reader (Varioskan Flash, Thermo Electron).
2.7 RNA extraction and quantitative real-time PCR analysis
Total RNA were extracted from frozen liver tissues or HepG2 cells using Trizol reagent (Ambion, Austin, TX) and reverse-transcribed into cDNA using a 5× All-In-One RT Master Mix first strand cDNA synthesis kit (Applied Biological Materials Inc., CA) according to the manufacturer's instructions. qRT-PCR using a EvaGreen 2× qPCR Master Mix kit (Applied Biological Materials Inc.) was performed in a Bio-Rad CFX96 real-time system machine to determine the mRNA expression of target gene, whose primers were designed by Sunshine Biotechnology (Shanghai, China) and sequences are listed in Table 1. The TaqMan GAPDH probe was used as reference for normalizing data. All the experiments were repeated at least five times independently to ensure the reproducibility of the results.| Gene name | Direction | Primer sequence 5′ → 3′ |
|---|---|---|
| Primers for qRT-PCR | ||
| hFXR | Forward | TCTCCTGGGTCGCCTGACT |
| Reverse | ACTGCACGTCCCAGATTTCAC | |
| hNrf2 | Forward | GCACATCCAGTCAGAAACCA |
| Reverse | GCCGAAGAAACCTCATTGTC | |
| hBSEP | Forward | ACATGCTTGCGAGGACCTTTA |
| Reverse | GGAGGTTCGTGCACCAGGTA | |
| hGAPDH | Forward | CTTTGGTATCGTGGAAGGACTC |
| Reverse | GTAGAGGCAGGGATGATGTTCT | |
| mFXR | Forward | CTTGATGTGCTACAAAAGCTGTG |
| Reverse | ACTCTCCAAGACATCAGCATCTC | |
| mNrf2 | Forward | GCAACTCCAGAAGGAACAGG |
| Reverse | GGAATGTCTCTGCCAAAAGC | |
| mBSEP | Forward | AGCAGGCTCAGCTGCATGAC |
| Reverse | AATGGCCCGAGCAATAGCAA | |
| mGAPDH | Forward | CTCCCACTCTTCCACCTTCG |
| Reverse | CCACCACCCTGTTGCTGTAG | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Primer for ChIP assay | ||
| BSEP | Forward | TCTCGTATGTCACTGAACTGT |
| Reverse | ATTTGAGGAAGCCAGAGGAA | |
2.8 Western blotting
Protein was extracted using a Nuclear or Membrane and Cytoplasmic Protein Extraction kit (Beyotime Biotechnology, Shanghai, China), respectively. Protein concentrations were determined with a BCA Protein Quantification kit (Beyotime Biotechnology, Shanghai, China). Protein extracted from cultured cells and liver tissues were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA). The membranes were incubated with 5% fat-free dry milk in Tris-buffered saline containing 0.1% Tween-20 (TBST) at room temperature for 2 h, and then probed with specific primary rabbit polyclonal IgG antibodies for overnight at 4 °C. Lamin B, β-actin and GAPDH were used as a loading control to calibrate the target protein signals for data analysis. The dilution ratios for the different antibodies used: BSEP, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; FXR, 1
1000; FXR, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Nrf2, 1
1000; Nrf2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; CARM1, 1
1000; CARM1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800; PRMT1, 1
800; PRMT1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800; SRC-1, 1
800; SRC-1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; PGC-1α, 1
1000; PGC-1α, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; GAPDH, 1
1000; GAPDH, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000; Lamin B, 1
5000; Lamin B, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000; β-actin, 1
3000; β-actin, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000. After incubation with HRP-conjugated secondary antibody (Bioworld Technology, Minnesota, USA, 1
3000. After incubation with HRP-conjugated secondary antibody (Bioworld Technology, Minnesota, USA, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000) for 1 h, visualization was performed using enhanced chemiluminescence imaging system (Bio-Rad, Hercules, CA) and quantification was achieved using ChemidocXRS+ System with Image Lab Software.
5000) for 1 h, visualization was performed using enhanced chemiluminescence imaging system (Bio-Rad, Hercules, CA) and quantification was achieved using ChemidocXRS+ System with Image Lab Software.
2.9 Immunofluorescence and confocal microscopy
HepG2 cells were cultured on 4 chamber glass bottom dishes (In vitro Scientific, Sunnyvale, CA) overnight prior to addition of compounds, and then treated either with 0.1% DMSO or GE (100 μM) for 24 h. Cells were washed thrice with PBS (pH 7.4), fixed with 4% paraformaldehyde and then incubated with polyclonal antibody FXR and BSEP for overnight at 4 °C. Alexa488 goat anti-rabbit IgG (Abcam, Cambridge, MA) was used as the secondary antibody for 1 h at room temperature in the dark, and nucleic DNA was stained with 1 μg mL−1 DAPI (4′-6-diamidino-2-phenylindole, Bioworld, St. Paul, MN). The fluorescent image was visualized using a 63× oil immersion objective with the Zeiss LSM 700 scanning laser confocal microscope and image software (Zen 2010, Carl Zeiss Micro Imaging GHBH, Jena, Germany).2.10 Gene knockdown with small interfering RNA
Small interfering RNA (siRNAs) for FXR, Nrf2, CARM1, PRMT1, SRC-1, and PGC-1α were constructed by TranSheep Bio Co. Ltd (Shanghai, China). The sequences for siRNAs are given in Table 2. HepG2 cells were seeded in 6-well plates approximately 20 h prior to transfection. Transfection reagent and negative control siRNA or siRNA targeting were pre-incubated at room temperature for 20 min to facilitate formation of lipid-siRNA complexes. Cells were then incubated in 100 nM siRNA or control (non-targeting) siRNA with lipofectamine 2000 (Invitrogen). Media was replaced after 6 h and following a further 48 h before treatment with either DMSO or GE (100 μM) for 24 h. Then cells were collected for qRT-PCR.| Gene name | Direction | Primer sequence 5′ → 3′ |
|---|---|---|
| siRNA-FXR | Sence | GAACCAUACUCGCAAUACATT |
| Antisence | UGUAUUGCGAGUAUGGUUCTT | |
| siRNA-Nrf2 | Sence | GGUUGAGACUACCAUGGUUTT |
| Antisence | AACCAUGGUAGUCUCAACCTT | |
| siRNA-CARM1 | Sence | GGUUCACCUCACACUUGAATT |
| Antisence | UUCAAGUGUGAGGUGAACCTT | |
| siRNA-PRMT1 | Sence | GGUUGCAUUUACGCCAAUATT |
| Antisence | UAUUGGCGUAAAUGCAACCTT | |
| siRNA-SRC-1 | Sence | GGAGCGAUAGGAAACAACATT |
| Antisence | UGUUGUUUCCUAUCGCUCCTT | |
| siRNA-PGC-1α | Sence | GACUCUAGACAACUAGAAATT |
| Antisence | UUUCUAGUUGUCUAGAGUCTT |
2.11 Plasmids construction, transient transfection and luciferase reporter assay
A 2.0 kb DNA fragment containing the human BSEP proximal promoter region was generated by PCR and cloned into pGL3-basic luciferase promoter vector as described previously.31 FXR expression plasmid was constructed by cloning genes encoding FXR into pcDNA 3.1 mammalian expression vector. HepG2 cells were cotransfected with 100 ng for BSEP promoter construct, 100 ng for nuclear receptor FXR expression plasmids, and 10 ng for the null-Renilla luciferase plasmid as the internal control, and transient transfection was conducted with Lipofectamine 2000. After transfection for 24 h, cells were treated with vehicle DMSO (0.1%), GW4064 (5 μM) and GE with increasing concentrations of 25, 50, and 100 μM for 24 h, and then harvested for the determination of luciferase activity. The firefly and renilla luciferase activities were measured using a Dual Luciferase Reporter assay (Promega). Cell lysates were transferred to a 96-well reader plate, and luciferase activities were detected by Luminoskan Ascent (Thermo Scientific, MA). The pRL-TK Renilla luciferase plasmid was used to normalize firefly luciferase activities and activity was expressed as fold induction after treatment with compounds compared with the control. Data are presented as mean ± S.E.M of at least five separate experiments.2.12 ChIP assay
ChIP analysis was performed using the pierce agarose ChIP kit (Thermo Scientific, Fair Lawn, NJ) according to the manufacturer's instructions. HepG2 cells were seeded in 100 mm dishes and treated with either 0.1% DMSO or GE (100 μM) for 24 h. Chromatin samples were digested with nuclease into length averaging between 150 to 900 bp. The sheared chromatin was incubated with FXR, CARM1, PGC-1α rabbit polyclonal antibody, and rabbit IgG (as a negative control) at 4 °C overnight, respectively. Antibody–chromatin complexes were captured by incubation with protein A/G plus agarose. Following washing of the agarose beads, reversal of cross-linking and purification, the precipitated DNA was used in a PCR (standard and quantitative) using specific primers flanking the FXR element (FXRE) site of human BSEP promoters. PCR products were run in a 2% agarose gel and stained with ethidium bromide to confirm the amplicon size. qRT-PCR was performed to quantify the precipitated DNA and expressed as fold relative to the IgG control. ChIP primers used in the studies are provided in Table 1.2.13 Statistical analysis
All values were presented as the mean ± S.E.M. The software program GraphPad Prism version 5 (San Diego, CA) was used for statistical analysis. The statistical significance of differences between two groups were determined by Student's t-test, and One-way ANOVA was employed to compare the differences of multiple groups. Differences with P < 0.05 were considered statistically significant.3 Results
3.1 GE ameliorated ANIT-induced hepatic injury and serum levels of IL-1β and IL-6
The serum levels of ALT, AST, ALP, TBIL, DBIL, and TBA were measured as markers of liver injury with cholestasis. As shown in Table 3, all of them in ANIT group were markedly increased compared with the vehicle group. However, mice in GE + ANIT cotreatment group had significant lower levels of the above parameters when compared to ANIT group. These data demonstrated that GE could attenuate ANIT-induced hepatic injury.| Group | ALT (IU L−1) | AST (IU L−1) | ALP (IU L−1) | TBIL (μmol L−1) | DBIL (μmol L−1) | TBA (μmol L−1) | IL-1β (pg mL−1) | IL-6 (pg mL−1) | Bile flow (μg min−1 g−1 LW) |
|---|---|---|---|---|---|---|---|---|---|
| a Data are the mean ± S.E.M. (n = 8).b P < 0.01.c P < 0.001 vs. vehicle.d P < 0.05.e P < 0.01 vs. ANIT group. | |||||||||
| Vehicle | 6.3 ± 0.9 | 6.7 ± 2.0 | 8.1 ± 0.4 | 4.3 ± 0.5 | 9.4 ± 1.6 | 7.3 ± 1.5 | 52.9 ± 10.4 | 13.2 ± 5.1 | 0.94 ± 0.13 |
| GE | 7.8 ± 2.1 | 3.4 ± 0.7 | 11.1 ± 0.1 | 6.3 ± 2.7 | 6.1 ± 1.0 | 8.2 ± 3.6 | 48.4 ± 8.1 | 11.0 ± 4.0 | 1.07 ± 0.10 |
| ANIT | 87.5 ± 12.8b | 31.6 ± 3.6b | 154.6 ± 10.4b | 205.9 ± 17.1b | 31.6 ± 2.4c | 52.8 ± 6.0b | 133.1 ± 16.6b | 52.9 ± 8.2b | 0.52 ± 0.04c |
| GE + ANIT | 53.8 ± 4.0d | 21.6 ± 3.4d | 73.2 ± 1.7e | 146.5 ± 2.7d | 24.5 ± 1.5d | 38.2 ± 4.8d | 79.5 ± 7.7d | 26.4 ± 3.8d | 0.72 ± 0.05e |
The result showed that significantly elevated serum levels of IL-1β and IL-6 in ANIT group were significantly attenuated by GE treatment.
3.2 Effect of GE on bile excretion
As shown in Table 3, ANIT decreased bile flow at 48 h after administration. The bile flow in ANIT group was about half that in the vehicle group (P < 0.001). The bile flow of GE + ANIT cotreatment group was increased 1.4-fold (P < 0.01) compared with ANIT group, indicating that GE could ameliorate the ANIT-suppressed bile flow.3.3 GE displayed no influence on HepG2 cell viability
The effects of administered concentrations of GE on HepG2 cell viability were examined by CCK-8 assay. The result indicated that incubation with GE (1–100 μM) for 24 h had no effect on the viability of HepG2 cells (Table 4), which was consistent with those of previous studies.32,33| GE (μM) | Cell viability (% of control) |
|---|---|
| a Date are expressed as mean ± S.D. of three independent experiments. | |
| 0 | 100.00 ± 2.09 |
| 1 | 101.47 ± 1.16 |
| 10 | 102.81 ± 2.82 |
| 25 | 100.50 ± 2.27 |
| 50 | 99.41 ± 2.40 |
| 75 | 98.70 ± 2.81 |
| 100 | 99.54 ± 2.30 |
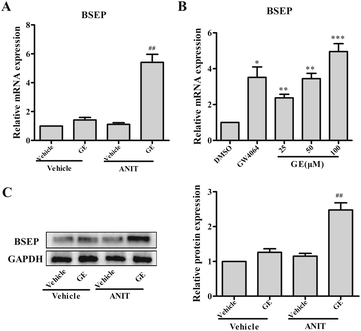
3.4 GE up-regulated BSEP expression in vivo and in vitro
GE exhibited anti-cholestasis effect. BSEP is the main bile acids efflux transporter, which is important to maintain bile acids homeostasis in the cholestatic disease. Fig. 1A illustrated that ANIT group showed no significant change in the expression of BSEP. Notably, GE cotreatment significantly promoted BSEP mRNA and protein expression (4.9- and 2.5-fold, respectively) in vivo (Fig. 1A and C). These results indicated that the significant elevation of BSEP may contribute to GE anti-cholestasis effect against ANIT. To further corroborate experimental results in vivo, HepG2 cells were treated with 25, 50, and 100 μM GE and measured the mRNA levels of BSEP. GW4064 (5 μM) and DMSO (0.1%) were included in the treatments as positive and negative control, respectively. As shown in Fig. 1B, the induction of BSEP expression by GE showed concentration-dependent manner with doses of 25–100 μM and a marked increase at the concentration of 100 μM compared with vehicle control. These results confirmed that GE could induce BSEP expression in vivo and in vitro.3.5 GE induced FXR and Nrf2 expression in vivo and in vitro
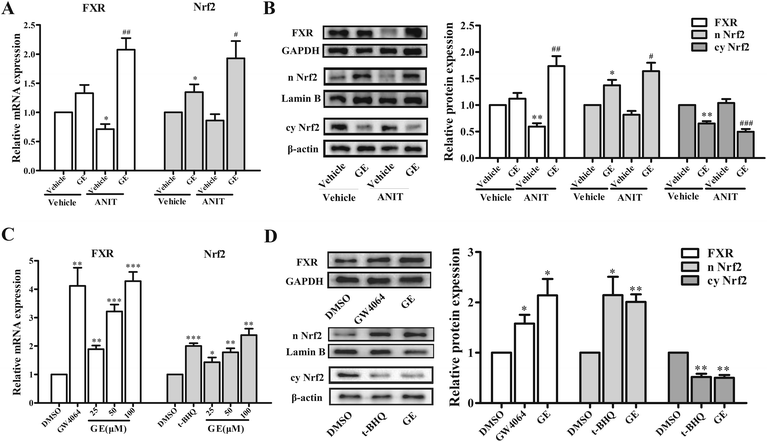
The expression of hepatic BSEP gene was regulated by a network of transactivation pathways, including FXR and Nrf2. To further investigate which transcriptional factors modulate the expression of BSEP, the effects of GE on mRNA expression of FXR and Nrf2 had been measured in ANIT-induced cholestatic mice and HepG2 cells. Fig. 2A showed that GE treatment alone had little impact on the mRNA levels of FXR, while the mRNA expression of Nrf2 was increased in vivo. GE co-treatment resulted in a significant up-regulation of FXR and Nrf2 mRNA expression by 2.9- and 2.3-fold over ANIT group, respectively. To determine whether changes in mRNA expressions of FXR and Nrf2 after GE treatment corresponded with protein levels, western blotting analysis was performed. FXR and Nrf2 nuclear protein expressions were significantly increased in GE co-treatment group compared with ANIT group (Fig. 2B). The results confirmed that FXR and Nrf2 expression were up-regulated by GE in ANIT-induced cholestatic mice.The effects of GE on mRNA expressions of FXR and Nrf2 were also measured in HepG2 cells. As shown in Fig. 2C, GE dose-dependently increased mRNA expression of FXR and Nrf2, which increased approximately 4.3- and 2.4-fold at the concentration of 100 μM in HepG2 cells. To confirm the qRT-PCR results regarding induction of FXR and Nrf2 by GE, the protein levels were further detected using western blotting analysis, and the results were similar to mRNA results (Fig. 2D). The marked induction of FXR and Nrf2 nuclear protein levels were detected in the presence of GE.
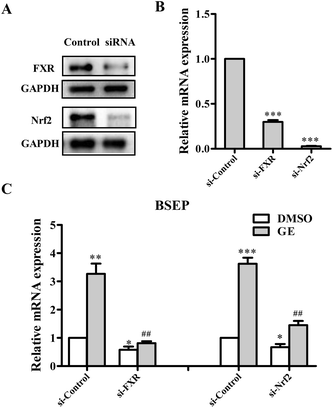
3.6 Downregulation of FXR and Nrf2 diminished the effect of GE on BSEP expression in HepG2 cells
To further demonstrate whether FXR and Nrf2 activation are required for GE-mediated up-regulation of BSEP, gene silencing experiments were performed using HepG2 cells. The knockdown efficiencies were assessed by qRT-PCR and western blotting. As anticipated, the protein expressions of FXR and Nrf2 were all reduced over 70% compared with that cells transfected with the negative control-siRNA (Fig. 3A), and decreased mRNA expressions were also observed (Fig. 3B). Following FXR and Nrf2 knockdown and cotreatment with 100 μM GE for 24 h, the mRNA expressions of BSEP were reduced to different degrees. FXR and Nrf2 silence reduced the BSEP mRNA expressions by 59% and 35% compared with si-Control, respectively (Fig. 3C). BSEP expression was efficiently blocked in FXR knocked-down cells after treatment of GE. The results suggested that FXR activation mediated BSEP up-regulation by GE, while Nrf2 activation is mildly involved in the BSEP expression by GE.3.7 Expressions and localizations of FXR and BSEP after GE treatment in HepG2 cells
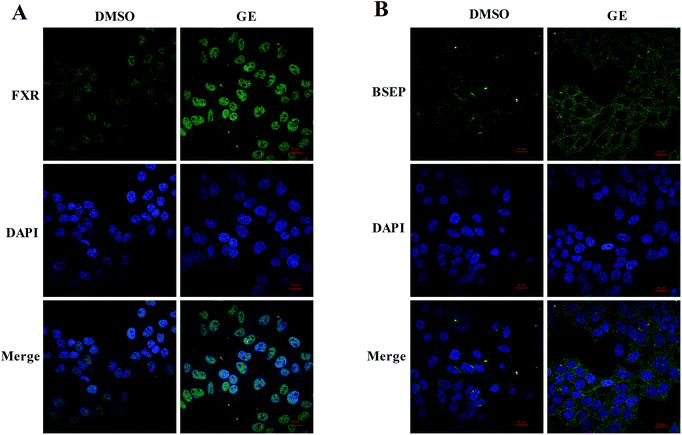
Confocal microscopy was carried out to investigate the expressions and localizations of FXR and BSEP after GE treatment. After immunostaining, labeled cells were scanned under 488 nm wavelength excitation and images were obtained as single transcellular optical sections. As shown in Fig. 4A and B, relatively weaker signals were measured with DMSO-treated cells and signals were increased after treatment of GE. Endogenous FXR was observed to accumulate predominantly in the nucleus and BSEP was observed to locate in the membrane of HepG2 cells after treatment of GE. Immunofluorescence studies in HepG2 cells further confirmed that GE activated the nuclear receptor FXR and promoted the key efflux transporter BSEP expression.3.8 GE activated FXR to modulate BSEP transactivation in HepG2 cells
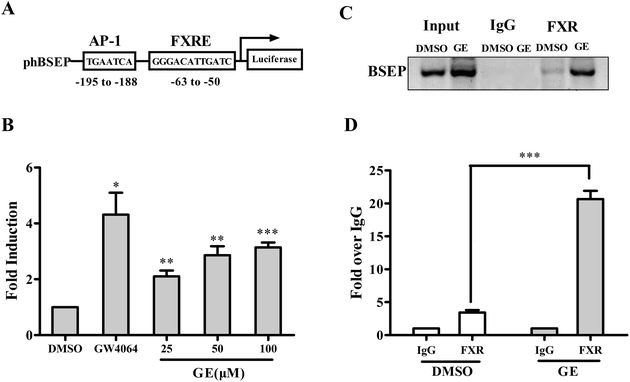
To investigate whether GE-mediated modulation of BSEP mRNA expression through FXR signaling pathway, we subsequently transiently cotransfected with pBSEP-Luc and FXR expression plasmid in HepG2 cells, followed by treating transfected cells with DMSO (0.1%), GW4064 (5 μM) and increasing concentrations of GE (25, 50, and 100 μM), and examined using luciferase reporter assay. As shown in Fig. 5B, in the presence of FXR, the level of basal transactivation of BSEP promoter was increased by GE in a dose-dependent manner, and an additional more than 3.2-fold induction was measured after treatment with GE (100 μM). The results indicated that GE regulated BSEP expression at the transcriptional level through FXR signaling pathway.To further determine whether GE increased the binding of FXR to BSEP promoter, a ChIP assay was performed in HepG2 cells. As shown in Fig. 5C, a band was observed in all inputs. Upon GE (100 μM) treatment, the recruitment of FXR to the BSEP promoter was robustly enhanced when chromatins immunoprecipitated with FXR antibody. In contrast, no enrichment was detected after chromatins immune-precipitated with IgG, indicating the association of BSEP with FXR is specific. Results of qRT-PCR of the BSEP promoter locus showed a 6.1-fold increase in the signal relative to IgG control when antibody to FXR was used for ChIP (Fig. 5D). These data supported the notion that FXR was specifically recruited by GE and bound to the BSEP promoter.
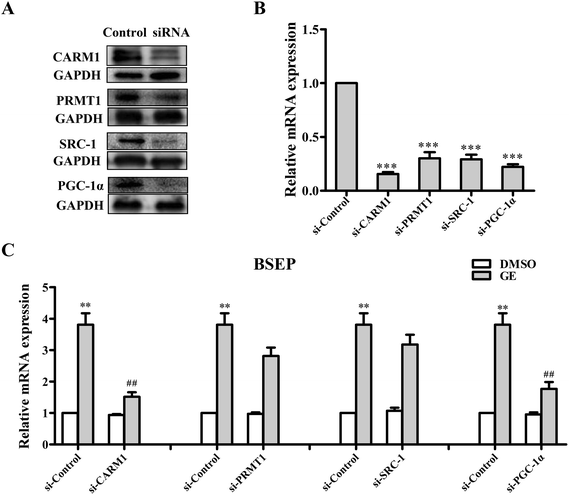
3.9 siRNA knockdown of CARM1 or PGC-1α abolished BSEP mRNA induction by GE
Several coactivators CARM1, PRMT1, SRC-1 and PGC-1α are recruited to BSEP promoter upon FXR activation. To address the question of whether these coactivators are involved in regulation of FXR transcriptional activity in the presence of GE, siRNA silencing experiments were performed in HepG2 cells. As a result of CARM1 or PGC-1α depletion, the BSEP mRNA expression declined significantly (Fig. 6C), whereas PRMT1 or SRC-1 knockdown slightly modulated BSEP expression in response to GE. It is speculated that GE enhanced endogenous CARM1 and PGC-1α interaction with FXR in HepG2 cells.3.10 Coactivators CARM1 and PGC-1α were predominantly recruited to the BSEP promoter upon FXR activated by GE
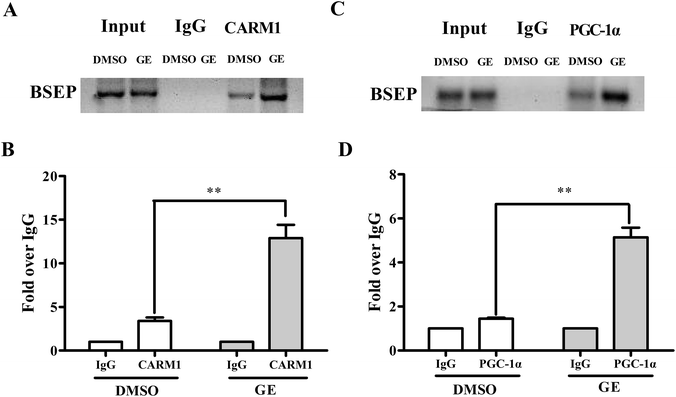
Having demonstrated that CARM1 and PGC-1α are involved in BSEP transcriptional activation, ChIP assays were carried out to confirm that GE increased CARM1 and PGC-1α associated with the FXRE in the BSEP promoter in HepG2 cells. Fig. 7A and C showed a representative gel of PCR reaction from ChIP experiment, indicating recruitment of CARM1 and PGC-1α to BSEP promoter in the presence of GE in HepG2 cells. Similar results were observed in qRT-PCR detection (Fig. 7B and D), which showed a 3.8-fold increase in the signal relative to IgG control when antibody to CARM1 was used for ChIP, while a 3.5-fold increase when antibody to PGC-1α was used. These findings provided evidence that GE increased recruitment of coactivators CARM1 and PGC-1α to the FXRE in the BSEP promoter.4 Discussion
The present study demonstrated that GE acted as an FXR activator to regulate the expression of BSEP. The conclusion is supported by the following results. (1) GE induced BSEP, FXR, and Nrf2 expression in vivo and in vitro; (2) depletion of FXR and Nrf2 by siRNA diminished the increase in BSEP mRNA expression in the presence of GE; (3) reporter assays demonstrated that GE dose-dependently activated FXR to induce BSEP transactivation, and ChIP assays further verified that FXR bound to the genomic BSEP promoter locus after GE treatment; (4) siRNA and ChIP assays demonstrated that PGC-1α and CARM1 were the predominant coactivators recruited to the BSEP promoter upon FXR activation by GE.The recommended clinical dose for G. jasminoides is 6–10 g.34 The average content of GE was 60 μg mg−1.35 Thus, the highest daily dose of GE for 60 kg human is 10 mg kg−1. According to the dose conversion method between animal and human,36 dose of GE 150 mg kg−1 used in mice in this study could be converted to estimate human equivalent dose 10.6 mg kg−1. According to our previous study at dose of GE 100 mg kg−1 in rats29 and reported studies at the dose of GE 100–200 mg kg−1 in mice,37,38 dose of GE 150 mg kg−1 was used in mice in this study.
Conventional and high-dose GE exhibited opposite effect on the mRNA expression of FXR and BSEP. High-dose GE (≥280 mg kg−1) could cause distinct liver injury in rats. Tian et al.39 investigated GE-induced hepatic injury after intragastric administration of 100 mg kg−1 or 300 mg kg−1 GE to healthy rats. The 300 mg kg−1 GE caused liver injury and increased serum biochemical parameters. Author assumed that 300 mg kg−1 GE-induced rat liver injury was likely cholestatic. High dose of GE inhibited the mRNA expression of FXR, BSEP, and multiple transporters involved in bile acids transportation. In comparison, administration of 100 mg kg−1 GE showed no signs of liver damage in rats. The mRNA expression of FXR and BSEP showed weak difference between the control group and 100 mg kg−1 GE.
In this study, FXR and BSEP expression were not induced by GE in the vehicle control group in mice. The result is consistent with the reported study.39 However, BSEP and FXR expression were significantly elevated in mice treated with GE + ANIT. Bile acids synthesis, metabolism, and transport are regulated via nuclear receptors and transcription factors to maintain bile acids homeostasis. Endogenous bile acids have differing abilities to bind to nuclear receptors to facilitate the positive feedforward and negative feedback regulation of hepatic transport. Bile acids-mediated BSEP expression is likely to be influenced by intracellular bile acids flux rather than steady state concentration.40 Endogenous bile acids exhibit different ability to activate FXR.19 ANIT can cause intrahepatic cholestasis which is characterized by bile acids accumulation in hepatocytes. Under cholestatic conditions, adaptive mechanisms are activated to counteract cholestatic liver injury. However, these adaptive response mechanisms are not always sufficient to prevent bile acids toxicity. Xenobiotics can augment the intrinsic adaptive pathways in animal models of cholestasis.41 In this study, both luciferase and ChIP assay confirmed the interaction between FXR and BSEP after GE treatment. Therefore, GE can induce hepatic export through activating FXR under pathologic condition. In contrast, GE (150 mg kg−1) showed no hepatic injury after intragastric administration to healthy mice. The total bile acids in general remained unchanged compared to vehicle control group (Table 3). Thus, mRNA and protein expressions of FXR and BSEP resulted no changes.
The current work demonstrated that GE increased the FXR mRNA and protein expressions in vivo and in vitro. FXR is a key regulator of bile, fat, and glucose homeostasis and suppresses the inflammatory response. FXR activation can inhibit inflammation in the intestine and liver in vivo.42 Thus, FXR activation by GE may be beneficial in attenuation of inflammation and treating cholestasis disorders.
It is already known that BSEP was regulated by transcription factors FXR and Nrf2. BSEP expression was induced significantly through Nrf2 binding to the antioxidant responsive element (ARE) in the BSEP promoter. Nrf2 is a major regulator of antioxidant and cytoprotective genes, which can reduce the oxidative burden by decreasing detoxification of chemicals via efflux transportation.43,44 ANIT administration led to intrahepatic cholestasis. Cholestasis results in increased production of oxidative stress and toxic bile acids, which are capable of activating Nrf2 through induction of cytoprotective genes that help protect cells against bile acids toxicity.45 GE antagonized oxidative stress through Nrf2 pathway.46 In this study, it was demonstrated that GE increased the expression of Nrf2 at both mRNA and protein in cholestatic rats and HepG2 cells. The induction of GE on BSEP expression was mildly attenuated by transfection with Nrf2 siRNA, which suggested that Nrf2 is mildly involved in BSEP up-regulation effect of GE in HepG2 cells.
To confirm FXR mediating the up-regulation of the BSEP expression by GE, gene silencing experiment was performed in HepG2 cells. In vitro results demonstrated that the significant induction of BSEP by GE was abolished after FXR silencing. To further ascertain whether transcriptional activation of the BSEP gene was caused after FXR activation by GE, the dual luciferase reporter assays and ChIP assays were performed in HepG2 cells. Luciferase assay confirmed that GE dose-dependently induced transactivation of BSEP through activating FXR in HepG2 cells. ChIP assays further demonstrated that FXR can directly interact with the promoter region in the BSEP gene and is important for BSEP gene regulation. Taken together, these results strongly indicate a specific role of GE in activating FXR and mediating BSEP expression.
It has been reported that bile acids increased coactivators including CARM1, PRMT1, SRC-1, and PGC-1α recruitment in the BSEP promoter through FXR. CARM1 and PRMT1 are important histone methyltransferases in mammalian cells, and SRC-1 is a member of p160 family of acetyltransferases. CARM1, PRMT1, and SRC-1 activate FXR through the remodeling of chromatin by histone modification.18,47 PGC-1α, a key regulator of energy homeostasis, is the primary coactivator recruited to the BSEP promoter upon FXR activation.21,48 The present study demonstrated that GE significantly decreased the mRNA levels of BSEP after knockdown of CARM1 and PGC-1α by siRNA. In addition, the association of coactivators CARM1 and PGC-1α with the FXRE in the BSEP promoter were prominently increased after treatment with GE. To sum up, GE induced BSEP expression through rising occupation of coactivators CARM1 and PGC-1α to FXR-regulated BSEP promoter.
5 Conclusion
In summary, GE induced BSEP expression in vivo and in vitro. GE induced BSEP transactivation through increasing FXR interacting with the BSEP promoter. Furthermore, coactivators PGC-1α and CARM1 interact with FXR to enhance the transcription of BSEP in the presence of GE. Given that G. jasminoides is one of the most extensively-used herbal medicines for the treatment of cholestatic hepatic disorders, these findings may represent a mechanism for the clinical use of Fructus Gardeniae to alleviate cholestasis. GE may be a candidate compound for maintaining bile acids homeostasis.Conflicts of interest
The authors have declared no conflict of interest.Abbreviations
| GE | Geniposide |
| ANIT | α-Naphthylisothiocyanate |
| BSEP | Bile salt export pump |
| FXR | Farnesoid X receptor |
| Nrf2 | Nuclear factor erythroid 2-related factor |
| ChIP | Chromatin immunoprecipitation |
| siRNA | Small interfering RNA |
| CARM1 | Co-activator-associated arginine methyltransferase 1 |
| PGC-1α | Peroxisome-proliferator-activated receptor γ co-activator-1α |
| PRMT1 | Protein arginine methyl-transferase type 1 |
| SRC-1 | Steroid receptor coactivator-1 |
| t-BHQ | tert-Butyl hydroquinone |
| CDCA | Chenodeoxycholic acid |
| DMSO | Dimethyl sulfoxide |
| FXRE | FXR-responsive element |
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81573836); Natural Science Foundation of Jiangsu Province (No. BK20151440); and China Pharmaceutical University for College Students Innovation Project for the R&D of Novel Drugs (No. 201710316127).References
- C. D. Klaassen and L. M. Aleksunes, Xenobiotic, bile acid, and cholesterol transporters: function and regulation, Pharmacol. Rev., 2010, 62(1), 1–96 CrossRef CAS PubMed.
- G. Zollner and M. Trauner, Molecular mechanisms of cholestasis, Wien. Med. Wochenschr., 2006, 156(13–14), 380–385 CrossRef PubMed.
- F. J. Cuperus, T. Claudel, J. Gautherot, E. Halilbasic and M. Trauner, The role of canalicular ABC transporters in cholestasis, Drug Metab. Dispos., 2014, 42(4), 546–560 CrossRef PubMed.
- T. Matsubara, F. Li and F. J. Gonzalez, FXR signaling in the enterohepatic system, Mol. Cell. Endocrinol., 2013, 368(1–2), 17–29 CrossRef CAS PubMed.
- Á. Telbisz and L. Homolya, Recent advances in the exploration of the bile salt export pump (BSEP/ABCB11) function, Expert Opin. Ther. Targets, 2015, 20(4), 501–514 CrossRef PubMed.
- M. Trauner and J. L. Boyer, Bile salt transporters: molecular characterization, function, and regulation, Physiol. Rev., 2003, 83(2), 633–671 CrossRef CAS PubMed.
- L. Ping, C. J. Soroka and J. L. Boyer, The bile salt export pump: clinical and experimental aspects of genetic and acquired cholestatic liver disease, Semin. Liver Dis., 2010, 30(2), 125–133 CrossRef PubMed.
- R. Kubitz, C. Dröge, S. Kluge, C. Stross, N. Walter, V. Keitel, D. Häussinger and J. Stindt, Autoimmune BSEP disease: disease recurrence after liver transplantation for progressive familial intrahepatic cholestasis, Clin. Rev. Allergy Immunol., 2015, 48(2–3), 273–284 CrossRef CAS PubMed.
- X. Song, A. Vasilenko, Y. Chen, L. Valanejad, R. Verma, B. Yan and R. Deng, Transcriptional dynamics of bile salt export pump during pregnancy: mechanisms and implications in intrahepatic cholestasis of pregnancy, Hepatology, 2014, 60(6), 1993–2007 CrossRef CAS PubMed.
- G. Zollner and M. Trauner, Nuclear receptors as therapeutic targets in cholestatic liver diseases, Br. J. Pharmacol., 2009, 156(1), 7–27 CrossRef CAS PubMed.
- J. R. M. Plass, O. Mol, J. Heegsma, M. Geuken, K. N. Faber, P. L. M. Jansen and M. Muller, Farnesoid X receptor and bile salts are involved in transcriptional regulation of the gene encoding the human bile salt export pump, Hepatology, 2002, 35(3), 589–596 CrossRef CAS PubMed.
- F. Chen, L. Ma, P. A. Dawson, C. J. Sinal, E. Sehayek, F. J. Gonzalez, J. Breslow, M. Ananthanarayanan and B. L. Shneider, Liver receptor homologue-1 mediates species- and cell line-specific bile acid-dependent negative feedback regulation of the apical sodium-dependent bile acid transporter, J. Biol. Chem., 2003, 278(22), 19909–19916 CrossRef CAS PubMed.
- Z. Tan, A. Liu, M. Luo and B. Guo, Screening of effective ingredients of Yinzhihuang injection against cholestasis and their mechanism, China J. Chin. Mater. Med., 2016, 41(6), 1113–1118 Search PubMed.
- J. Weerachayaphorn, S. Y. Cai, C. J. Soroka and J. L. Boyer, NF-E2-related factor 2 (Nrf2) is a positive regulator of human bile salt export pump (BSEP) expression, Hepatology, 2009, 50(5), 1588–1596 CrossRef CAS PubMed.
- M. Ananthanarayanan, N. Balasubramanian, M. Makishima, D. J. Mangelsdorf and F. J. Suchy, Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor, J. Biol. Chem., 2001, 276(31), 28857–28865 CrossRef CAS PubMed.
- R. Deng, D. Yang, A. Radke, J. Yang and B. Yan, The hypolipidemic agent guggulsterone regulates the expression of human bile salt export pump: dominance of transactivation over farsenoid X receptor-mediated antagonism, J. Pharmacol. Exp. Ther., 2007, 320(3), 1153–1162 CrossRef CAS PubMed.
- C. J. Sinal, M. Tohkin, M. Miyata, J. M. Ward, G. Lambert and F. J. Gonzalez, Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis, Cell, 2000, 102(6), 731–744 CrossRef CAS PubMed.
- F. J. Suchy and M. Ananthanarayanan, Bile salt excretory pump: biology and pathobiology, J. Pediatr. Gastroenterol. Nutr., 2006, 43(1), S10–S16 CrossRef CAS PubMed.
- A. Baghdasaryan, P. Chiba and M. Trauner, Clinical application of transcriptional activators of bile salt transporters, Mol. Aspects Med., 2014, 37(100), 57–76 CrossRef CAS PubMed.
- R. S. Savkur, J. S. Thomas, K. S. Bramlett, Y. Gao, L. F. Michael and T. P. Burris, Ligand-dependent coactivation of the human bile acid receptor FXR by the peroxisome proliferator-activated receptor gamma coactivator-1alpha, J. Pharmacol. Exp. Ther., 2005, 312(1), 170–178 CrossRef CAS PubMed.
- E. Kanaya, T. Shiraki and H. Jingami, The nuclear bile acid receptor FXR is activated by PGC-1alpha in a ligand-dependent manner, Biochem. J., 2004, 382(Pt 3), 913–921 CrossRef CAS PubMed.
- M. Ananthanarayanan, S. Li, N. Balasubramaniyan, F. J. Suchy and M. J. Walsh, Ligand-dependent activation of the farnesoid X-receptor directs arginine methylation of histone H3 by CARM1, J. Biol. Chem., 2004, 279(52), 54348–54357 CrossRef CAS PubMed.
- G. Rizzo, B. Renga, E. Antonelli, D. Passeri, R. Pellicciari and S. Fiorucci, The methyl transferase PRMT1 functions as co-activator of farnesoid X receptor (FXR)/9-cis retinoid X receptor and regulates transcription of FXR responsive genes, Mol. Pharmacol., 2005, 68(2), 551–558 CrossRef CAS PubMed.
- M. Shan, S. Yu, H. Yan, S. Guo, W. Xiao, Z. Wang, L. Zhang, A. Ding, Q. Wu and S. Li, A review on the phytochemistry, pharmacology, pharmacokinetics and toxicology of geniposide, a natural product, Molecules, 2017, 22(10), 1689 CrossRef PubMed.
- Z. Chen, X. Ma, Y. Zhao, J. Wang, Y. Zhang, J. Li, R. Wang, Y. Zhu, L. Wang and X. Xiao, Yinchenhao decoction in the treatment of cholestasis: a systematic review and meta-analysis, J. Ethnopharmacol., 2015, 168, 208–216 CrossRef PubMed.
- Y. Jiang, G. Chang, Y. Wang, D. Zhang, L. Cao and J. Liu, Geniposide prevents hypoxia/reoxygenation-induced apoptosis in H9C2 cells: improvement of mitochondrial dysfunction and activation of GLP-1R and the PI3K/AKT signaling pathway, Cell. Physiol. Biochem., 2016, 39(1), 407–421 CrossRef CAS PubMed.
- Y. Fu, B. Liu, J. Liu, Z. Liu, D. Liang, F. Li, D. Li, Y. Cao, C. Zhang, N. Zhang and Z. Yang, Geniposide, from Gardenia jasminoides Ellis, inhibits the inflammatory response in the primary mouse macrophages and mouse models, Int. Immunopharmacol., 2012, 14(4), 792–798 CrossRef CAS PubMed.
- Z. Tian, A. Liu, M. Luo, X. Yin, D. Song, M. Dai, P. Li, Z. Chu, Z. Zou, M. Ma, B. Guo and B. Chen, Geniposide inhibits alpha-naphthylisothiocyanate-induced intrahepatic cholestasis: the downregulation of STAT3 and NFκB signaling plays an important role, Am. J. Chin. Med., 2016, 44(4), 721–736 CrossRef PubMed.
- L. Wang, G. Wu, F. Wu, N. Jiang and Y. Lin, Geniposide attenuates ANIT-induced cholestasis through regulation of transporters and enzymes involved in bile acids homeostasis in rats, J. Ethnopharmacol., 2017, 196, 178–185 CrossRef CAS PubMed.
- X. Song, R. Kaimal, B. Yan and R. Deng, Liver receptor homolog 1 transcriptionally regulates human bile salt export pump expression, J. Lipid Res., 2008, 49(5), 973–984 CrossRef CAS PubMed.
- R. Deng, D. Yang, J. Yang and B. Yan, Oxysterol 22(R)-hydroxycholesterol induces the expression of the bile salt export pump through nuclear receptor farsenoid X receptor but not liver X receptor, J. Pharmacol. Exp. Ther., 2006, 317(1), 317–325 CrossRef CAS PubMed.
- T. Khanal, H. G. Kim, J. H. Choi, M. T. Do, M. J. Kong, M. J. Kang, K. Noh, H. K. Yeo, Y. T. Ahn, W. Kang, D. H. Kim, T. C. Jeong and H. G. Jeong, Biotransformation of geniposide by human intestinal microflora on cytotoxicity against HepG2 cells, Toxicol. Lett., 2012, 209(3), 246–254 CrossRef CAS PubMed.
- L. Guo, X. Zheng, J. Liu and Z. Yin, Geniposide suppresses hepatic glucose production via AMPK in HepG2 cells, Biol. Pharm. Bull., 2016, 39(4), 484–491 CrossRef CAS PubMed.
- Pharmacopoeia Commission of the People's Republic of China, China Medical Science Press, Beijing, 2015, vol. 1, pp. 231–232 Search PubMed.
- E. J. Lee, J. K. Hong and W. K. Whang, Simultaneous determination of bioactive marker compounds from Gardeniae fructus by high performance liquid chromatography, Arch. Pharmacal Res., 2014, 37(8), 992–1000 CrossRef CAS PubMed.
- A. B. Nair and S. Jacob, A simple practice guide for dose conversion between animals and human, J. Basic Clin. Pharm., 2016, 7(2), 27–31 CrossRef PubMed.
- T. T. Ma, X. F. Li, W. X. Li, Y. Yang, C. Huang, X. M. Meng, L. Zhang and J. Li, Geniposide alleviates inflammation by suppressing MeCP2 in mice with carbon tetrachloride-induced acute liver injury and LPS-treated THP-1 cells, Int. Immunopharmacol., 2015, 29(2), 739–747 CrossRef CAS PubMed.
- Z. Zhang, Y. Li, P. Shen, S. Li, X. Lu, J. Liu, Y. Cao, B. Liu, Y. Fu and N. Zhang, Administration of geniposide ameliorates dextran sulfate sodium-induced colitis in mice via inhibition of inflammation and mucosal damage, Int. Immunopharmacol., 2017, 49, 168–177 CrossRef CAS PubMed.
- J. Tian, J. Zhu, Y. Yi, C. Li, Y. Zhang, Y. Zhao, C. Pan, S. Xiang, X. Li, G. Li, J. W. Newman, X. Feng, J. Liu, J. Han, L. Wang, Y. Gao, M. R. La Frano and A. Liang, Dose-related liver injury of Geniposide associated with the alteration in bile acid synthesis and transportation, Sci. Rep., 2017, 7(1), 8938 CrossRef PubMed.
- H. Wolters, B. M. Elzinga, J. F. Baller, R. Boverhof, M. Schwarz, B. Stieger, H. J. Verkade and F. Kuipers, Effects of bile salt flux variations on the expression of hepatic bile salt transporters in vivo in mice, J. Hepatol., 2002, 37(5), 556–563 CrossRef CAS PubMed.
- G. Zollner, H. U. Marschall, M. Wagner and M. Trauner, Role of Nuclear Receptors in the Adaptive Response to Bile Acids and Cholestasis: Pathogenetic and Therapeutic Considerations, Mol. Pharm., 2006, 3(3), 231–251 CrossRef CAS PubMed.
- D. A. Hollman, A. Milona, K. J. van Erpecum and S. W. van Mil, Anti-inflammatory and metabolic actions of FXR: Insights into molecular mechanisms, Biochim. Biophys. Acta, 2012, 1821(11), 1443–1452 CrossRef CAS PubMed.
- T. Nguyen, P. Nioi and C. B. Pickett, The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress, J. Biol. Chem., 2009, 284(20), 13291–13295 CrossRef CAS PubMed.
- H. Gong, B. Zhang, M. Yan, P. Fang, H. Li, C. Hu, Y. Yang, P. Cao, P. Jiang and X. Fan, A protective mechanism of licorice (Glycyrrhiza uralensis): isoliquiritigenin stimulates detoxification system via Nrf2 activation, J. Ethnopharmacol., 2015, 162, 134–139 CrossRef CAS PubMed.
- J. Weerachayaphorn, A. Mennone, C. J. Soroka, K. Harry, L. R. Hagey, T. W. Kensler and J. L. Boyer, Nuclear factor-E2-related factor 2 is a major determinant of bile acid homeostasis in the liver and intestine, Am. J. Physiol.: Gastrointest. Liver Physiol., 2012, 302(9), G925–G936 CrossRef CAS PubMed.
- Y. Fei, J. H. Liu, X. X. Zheng, L. X. Guo and H. Xiao, Geniposide induces the expression of heme oxygenase-1 via PI3K/Nrf2-signaling to enhance the antioxidant capacity in primary hippocampal neurons, Biol. Pharm. Bull., 2010, 33(11), 1841–1846 CrossRef.
- Y. Chen, A. Vasilenko, X. Song, L. Valanejad, R. Verma, S. You, B. Yan, S. Shiffka, L. Hargreaves, C. Nadolny and R. Deng, Estrogen and estrogen receptor-α-mediated transrepression of bile salt export pump, Mol. Endocrinol., 2015, 29(4), 613–626 CrossRef CAS PubMed.
- Y. Zhang, L. W. Castellani, C. J. Sinal, F. J. Gonzalez and P. A. Edwards, Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha) regulates triglyceride metabolism by activation of the nuclear receptor FXR, Genes Dev., 2004, 18(2), 157–169 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |