Open Access Article
Open Access ArticleTargeting HMGA protein inhibits retinoblastoma cell proliferation†
Akilandeswari Balachandran‡
a,
Ajit Zambre‡b,
Jagjot Singh Kainth be,
Lakshmi Dhevi Nagarajha Selvanc,
Sowmya Parameswarand,
Zahra Afrasiabie,
Subramanian Krishnakumar*acd,
Raghuraman Kannan
be,
Lakshmi Dhevi Nagarajha Selvanc,
Sowmya Parameswarand,
Zahra Afrasiabie,
Subramanian Krishnakumar*acd,
Raghuraman Kannan *bf and
Anandhi Upendran*gh
*bf and
Anandhi Upendran*gh
aDepartment of Nanobiotechnology, Vision Research Foundation, Kamalnayan Bajaj Institute for Research in Vision and Ophthalmology, Chennai, India. E-mail: drkk@snmail.org; drkrishnakumar_2000@yahoo.com
bDepartment of Radiology, University of Missouri, Columbia, MO, USA. E-mail: kannanr@health.missouri.edu
cL&T Ocular Pathology Department, Vision Research Foundation, Kamalnayan Bajaj Institute for Research in Vision and Ophthalmology, Chennai, India
dRadheshyam Kanoi Stem Cell Laboratory, Vision Research Foundation, Kamalnayan Bajaj Institute for Research in Vision and Ophthalmology, Chennai, India
eDepartment of Life Sciences, Lincoln University, Jefferson City, MO, USA
fDepartment of Biological Engineering, University of Missouri, Columbia, MO, USA
gDepartment of Medical Pharmacology and Physiology, University of Missouri, Columbia, MO, USA
hInstitute of Clinical and Translational Sciences (MU-iCATS), School of Medicine, University of Missouri, Columbia, MO, USA. E-mail: upendrana@health.missouri.edu
First published on 7th September 2018
Abstract
We describe a novel synthetic strategy for conjugating HMGA2 siRNA and the HMGA aptamer to the nucleolin aptamer and nucleolin antibody, respectively. Our studies demonstrate that these conjugates inhibit cell proliferation in retinoblastoma cells.
Retinoblastoma (RB) is a childhood eye cancer and approximately 9000 children are diagnosed every year.1 Current treatments include systemic chemotherapy using a combination of drugs such as vincristine, etoposide, and carboplatin with local chemotherapy of intravitreal melphalan and intra-arterial chemotherapy using carboplatin to save the vision of these children. These therapies result in severe toxicity to the retina and retinal pigment epithelial cells and in advanced stages, removal of the eye becomes inevitable.2–5 Therefore, it is important to identify targets that could complement the local chemotherapy to improve the efficiency, reduce the toxicity of these drugs and importantly prevent the vision loss. Studies have shown that targeted molecular therapy has great potential to improve the therapeutic outcome.6 However, the limitation is a lack of a suitable vehicle for selective delivery of molecular therapeutics such as siRNA or aptamers that possess the ability to alter the functionality at genetic levels.
In retinoblastoma, the high mobility group A (HMGA) protein has been identified as an important therapeutic target, and is associated with invasiveness and metastasis of the disease.7 HMGA family proteins comprise two non-histone nuclear protein subsets, HMGA1 and HMGA2, and binds to AT-rich minor groove of the DNA. Upon binding, it alters the DNA chromatin structure and facilitates attachment of transcription factors leading to differential regulation of the transcription of genes. Previous studies indicated that RB tumors over express HMGA proteins and correlated with high risk histopathology features such as diffused invasion of the choroid and optic nerve.8,9 HMGA proteins are important target for molecular cancer therapy as these proteins are overexpressed only in tumors and not expressed in healthy adult tissues.10,11
HMGA proteins and mRNA transcripts can be selectively targeted using siRNA, aptamers, or DNA minor groove binders such as netropsin.12 Previous studies have shown that siRNA targeting HMGA2, promoted cell death through apoptosis and cell cycle arrest in cancer cells.13–15 Similarly, HMGA2 aptamer increased the sensitivity to gemcitabine treatment in pancreatic cancer cell lines.16 In RB tumours, HMGA aptamer reduced cell proliferation by activation of TGFβ-SMAD4-mediated apoptotic pathway and a synergistic effect was also observed with etoposide.17 Indeed, our previous studies revealed that targeting HMGA at the transcript as well as protein levels compromised RB survival in in vitro.13,17 The selective delivery of either HMGA2 siRNA or HMGA aptamer to retinoblastoma can be achieved by targeting cell surface proteins those are overexpressed in RB cells. Previous studies have showed that nucleolin (NCL) is overexpressed on the surface of RB cells, and could be used for selective delivery of siRNA or aptamer.18 NCL shuttles from nucleolus to nucleoplasm, cytoplasm and cell surface. Targeting NCL needs a delivery vehicle and entry into the tumor cells.
Antibodies, peptides, or nanoparticles are commonly used as agents for delivery of siRNA or aptamer to cells.19–23 Antibody–aptamer pincers, antibody–siRNA conjugates, chemically programmable antibodies conjugated to RNA aptamers improve the affinity to target molecules.24–28 Aptamer conjugated with drugs are known;29–33 however, antibody–aptamer conjugates are sparse in the literature.25 Although, the above listed strategies improved the delivery,34 the bottle-necks are: poor conjugation yield, and weak stability resulting in poor efficacy and increased toxicity. Therefore, it is important to overcome the above listed problems for successful delivery of siRNA or aptamer to RB tumor.
In this study, we synthesized and investigated two different delivery systems: NCL aptamer mediated delivery of HMGA2 siRNA (NCLap–HMGA2si) and NCL antibody mediated delivery of HMGA aptamer (NCLAb–HMGAap) for targeting HMGA in RB cells.
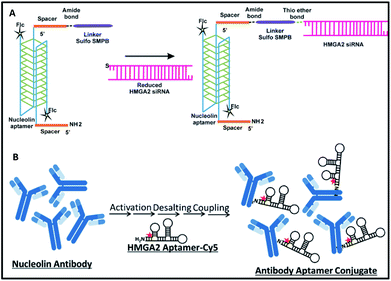
Synthesis of NCLap–HMGA2si and NCLAb–HMGAap are shown in Fig. 1. The newly synthesized conjugates were characterized; subsequently, stability, internalization, and in vitro toxicity were investigated. Internalization studies were performed in WERI-Rb-1 cells to validate the selective delivery of HMGA2si as well as HMGAap through NCLap and NCLAb respectively. Functional activity was demonstrated and compared with transfected HMGA2si or HMGAap in WERI-Rb-1 cells (described in ESI†).
For synthesizing NCLap–HMGA2si conjugate, the 5′ end of NCL aptamer was modified with amine group using a spacer, and the 5′ end of HMGA2 siRNA was modified with thiol.35,36 The 3′ end of NCL aptamer was modified with fluorescein, while the cytosine and uracil bases of siRNA modified with 2′ fluoro to improve its stability. For synthesizing NCLAb–HMGAap, we used amine modified HMGA2-cy5 aptamer and nucleolin antibody.37 The sequence of amine–NCL aptamer, thiol–HMGA2 siRNA, and amine modified HMGA aptamer is shown in ESI-Table 1.†
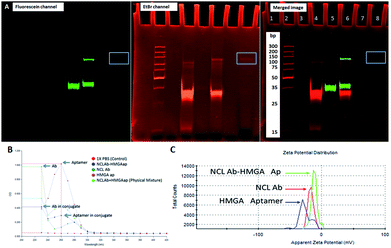
As shown in Fig. 1, we synthesized NCLap–HMGA2si by treating equimolar concentrations of NCL aptamer and HMGA2 siRNA, in the presence of sulfo-SMPB bispecific linker.36 We analyzed the reaction mixture by gel electrophoresis. The conjugate showed gel retention at a higher molecular weight compared to that of free aptamer or siRNA. The presence of both NCL aptamer and HMGA2 siRNA in the conjugate were confirmed by respective bands in EtBr and fluorescein channels (Fig. 2A). The presence of bands corresponding to reactants suggests that the conjugation yield is only modest (40–50%) and required further purification. Previous literature showed that similar reactions, using sulfo-SMPB for conjugation of oligonucleotides with streptavidin, contain unreacted starting materials in the reaction mixture.38 To remove free aptamer and siRNA, NCLap–HMGA2si conjugate was excised from gel and purified by modified crush and soak method.39
The NCLAb–HMGAap conjugate was prepared using EDC/sulfo-NHS procedure40 and the excess aptamer was removed by passing through centrifugal filter (30 kD Amicon). The NCLAb–HMGAap conjugate was characterized by gel electrophoresis, UV absorption, fluorescence and zeta potential measurements. The successful conjugation and isolation of pure NCLAb–HMGAap conjugate was confirmed by gel electrophoresis through fluorescent imaging (ESI-Fig. 1†). The presence of individual components of the conjugate, NCLAb and HMGAap was confirmed by coomassie blue and gel red staining respectively (ESI-Fig. 2 and 3†). The UV absorption spectrum of the conjugate showed characteristic peaks corresponding to antibody at 230 nm and aptamer at 260 nm (Fig. 2B). The zeta potential of the conjugate shifted towards less negative value (−13 ± 3 mV) compared to antibody (−17 ± 3 mV) and the aptamer (−40 ± 5 mV) confirming the formation of the conjugate. The zeta potential of NCLAb–HMGAap conjugate compared with free antibody and aptamer is shown in Fig. 2C. To confirm that conjugate contains both protein and aptamer, we quantified the protein using Bradford and Nanodrop, and aptamer by both fluorescent and Nanodrop measurements. The studies confirmed conjugation efficiency of ∼65% and ∼25% composition of the conjugate. There are fewer literature precedence on detailed characterization of the aptamer–oligo body conjugates25 and therefore, the analytical measurements used to confirm NCLAb–HMGAap could serve as a predicate to analyze these types of conjugates.
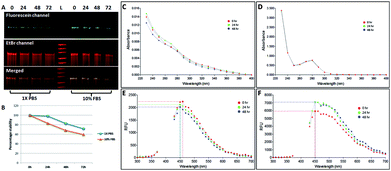
We investigated the stability of both the conjugates, NCLap–HMGA2si and NCLAb–HMGAap, in 1× PBS and 10% FBS at 24 h, 48 h, and 72 h and monitored using electrophoresis and optical spectroscopy (Fig. 3). The presence of single band in PAGE and stable peak in UV and fluorescence spectra confirmed the conjugates are stable.
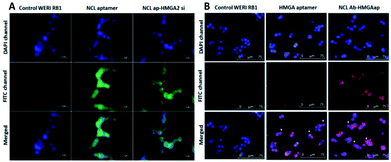
The targeting ability of the newly synthesized conjugates were evaluated in WERI-Rb-1 cells by analyzing the receptor binding affinity. The cells were treated with the fluorescent labeled conjugates for two hours. Fluorescent microscopy was used to analyze the uptake of the conjugates in the cells. Co-localization of DAPI and FITC channels showed NCLap–HMGA2si internalize in to the nuclear region (Fig. 4A). The surface and nuclear localization confirmed the binding ability of NCL aptamer to nucleolin protein present on the surface and subsequent internalization. Similar studies were performed with NCLAb–HMGAap conjugate to visualize aptamer internalization through NCL antibody. Co-localization of DAPI and CY-5 channels confirmed surface localization of the NCLAb–HMGA2ap conjugate. Fluorescent images showed selective uptake of NCLAb–HMGAap conjugate on the surface of the WERI-Rb-1 cells confirming receptor mediated uptake of HMGA aptamer (Fig. 4B). The fluorescent image shows higher degree of cellular internalization of NCLAb–HMGAap conjugate compared to that of transfected HMGA aptamer. These results confirm that NCL antibody is effective in transporting the HMGA aptamer into the cells through the nucleolin receptors and retention of structural integrity.
To investigate functional effects of the conjugates, in vitro cytotoxicity assays were performed in WERI-Rb-1 cells. MTT cell proliferation assay showed increased cytotoxicity in both the conjugates compared to HMGA2siRNA and HMGA aptamer (Table 1). NCLap–HMGA2si showed IC50 of 72 nM (0.99 μg ml−1), whereas, NCLAb–HMGAap showed IC50 of 90 nM (9.75 μg ml−1). Both NCLap–HMGA2si and NCLAb–HMGAap conjugates showed significant decrease in cell viability compared to their respective controls (Fig. 5A). The percentage cell viability for NCLap–HMGA2si conjugate was found to be 59% and 47% with 50 nM and 100 nM conjugate respectively. Likewise, the cell viability for NCLAb–HMGAap conjugate was 67% and 41% at 65 nM and 130 nM respectively (Fig. 5B). The enhanced cytotoxicity of the NCLap–HMGA2si conjugate is probably due to the synergistic effect of NCL aptamer and HMGA2 siRNA as both aptamer and siRNA are functional.13,18
| Conjugate | IC50 (nM) | IC50 (μg mL−1) |
|---|---|---|
| NCLap | 200 | 2.4 |
| HMGA2 siRNA | >200 | >2.7 |
| NCLap–HMGAsi | 72 | 1.0 |
| NCLAb | >200 | >20 |
| HMGAAp | 1400 | >10 |
| NCLab–HMGAap | 90 | 9.8 |
| PM (Ab + Ap) | >200 | >20 |
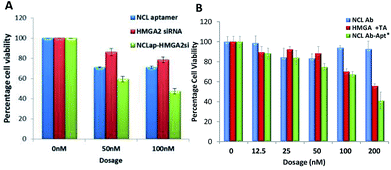 | ||
| Fig. 5 Cytotoxicity of conjugates. (A) WERI-RB1 cells treated with NCLap–HMGAsi, NCL aptamer and HMGA2 siRNA. (B) WERI-RB1 cells treated with NCLAb–HMGAap, NCLAb, HMGA aptamer and physical mixture of NCLAb and HMGA aptamer. *Dosage represented here are based on pristine antibody concentration (ESI-Table 2†). | ||
We compared the cytotoxicity of NCLAb–HMGAap with free HMGA aptamer, NCL antibody and its physical mixture (Table 1). Based on the quantification data, we found that 90 nM of NCLAb–HMGAap conjugate contains 26 nM of HMGAap. The IC50 of NCLAb–HMGAap conjugate was 90 nM suggesting that it is ∼50 fold (p < 0.001; ESI-Fig. 4†) more cytotoxic compared to the free aptamer (1400 nM). The NCLAb showed no signs of cytotoxicity (IC50 > 200 nM); therefore, we believe that the enhanced cytotoxicity of the NCLAb–HMGAap conjugate could be attributed to targeted delivery of HMGA aptamer to RB cells. In addition to its role as a targeting vector, NCLAb camouflages the HMGA aptamer thereby facilitating the retention of structural integrity and functional property.
In summary, we synthesized, characterized, and investigated the in vitro HMGA targeting ability of both NCL aptamer conjugated siRNA and NCL antibody conjugated aptamer in human RB cells. Our data demonstrate that the NCLAb–HMGAap conjugate has unique and translatable characteristics compared to that of NCLAp–HMGA2si conjugate and listed as follows: (i) easier synthesis; (ii) scalable and better conjugation efficiency; (iii) higher degree of cellular internalization in WERI-RB1 cells through receptor mediated internalization; and (iv) enhanced cytotoxicity, ∼50 fold higher, in WERI-RB1 compared to free HMGA aptamer. Taken together, our data suggests that NCLAb-HMGAap is a potential candidate for future pre-clinical applications.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is funded by research grant from Childhood Eye Cancer Trust (CHECT foundation), London, UK and Michael J and Sharon R Bukstein Cancer Research Endowment Funds.Notes and references
- https://wechope.org/retinoblastoma/retinoblastoma-overview/global-incidence/.
- J. H. Francis, S. E. Brodie, B. Marr, E. C. Zabor, I. Mondesire-Crump and D. H. Abramson, Ophthalmology, 2017, 124, 488–495 CrossRef PubMed.
- H. A. Aziz, J. W. Kim, F. L. Munier and J. L. Berry, Ocul. Oncol. Pathol., 2017, 3, 34–40 CrossRef PubMed.
- S. T. Michaels, T. A. Abruzzo, J. J. Augsburger, Z. M. Correa, A. Lane and J. I. Geller, J. Pediatr. Hematol. Oncol., 2016, 38, 65–69 CrossRef PubMed.
- D. Susskind, U. Hagemann, M. Schrader, K. Januschowski, S. Schnichels and S. Aisenbrey, Acta. Ophthalmol., 2016, 94, 471–478 CrossRef PubMed.
- A. Ferrario, M. Luna, N. Rucker, S. Wong, A. Lederman, J. Kim and C. Gomer, PLoS One, 2016, 11, e0153011 CrossRef PubMed.
- P. Pallante, R. Sepe, F. Puca and A. Fusco, Front. Med. (Lausanne), 2015, 2, 15 Search PubMed.
- G. Mu, H. Liu, F. Zhou, X. Xu, H. Jiang, Y. Wang and Y. Qu, Hum. Pathol., 2010, 41, 493–502 CrossRef PubMed.
- N. Venkatesan, M. Kandalam, G. Pasricha, V. Sumantran, G. Manfioletti, S. J. Ono, M. A. Reddy and S. Krishnakumar, J. Pediatr. Hematol. Oncol., 2009, 31, 209–214 CrossRef PubMed.
- A. Fusco and M. Fedele, Nat. Rev. Cancer, 2007, 7, 899–910 CrossRef PubMed.
- R. Sgarra, A. Rustighi, M. A. Tessari, J. Di Bernardo, S. Altamura, A. Fusco, G. Manfioletti and V. Giancotti, FEBS Lett., 2004, 574, 1–8 CrossRef PubMed.
- Y. Miao, T. Cui, F. Leng and W. D. Wilson, Anal. Biochem., 2008, 374, 7–15 CrossRef PubMed.
- N. Venkatesan, S. Krishnakumar, P. Deepa, M. Deepa, V. Khetan and A. M. Reddy, Mol. Vision, 2012, 18, 2420–2437 Search PubMed.
- S. Esmailzadeh, B. Mansoori, A. Mohammadi, D. Shanehbandi and B. Baradaran, J. Gastrointest. Cancer, 2017, 48, 156–163 CrossRef PubMed.
- Z. Shi, D. Wu, R. Tang, X. Li, R. Chen, S. Xue, C. Zhang and X. Sun, J. Biosci., 2016, 41, 229–236 CrossRef PubMed.
- M. Watanabe, S. Sheriff, K. B. Lewis, S. L. Tinch, J. Cho, A. Balasubramaniam and M. A. Kennedy, Cancer Lett., 2012, 315, 18–27 CrossRef PubMed.
- V. Nalini, P. R. Deepa, R. Raguraman, V. Khetan, M. A. Reddy and S. Krishnakumar, Ocul. Oncol. Pathol., 2016, 2, 262–269 CrossRef PubMed.
- N. Subramanian, A. Srimany, J. R. Kanwar, R. K. Kanwar, B. Akilandeswari, P. Rishi, V. Khetan, M. Vasudevan, T. Pradeep and S. Krishnakumar, Mol. Ther.–Nucleic Acids, 2016, 5, e358 CrossRef PubMed.
- J. Zhou and J. Rossi, Nat. Rev. Drug Discovery, 2017, 16, 181–202 CrossRef PubMed.
- K. Chen, B. Liu, B. Yu, W. Zhong, Y. Lu, J. Zhang, J. Liao, J. Liu, Y. Pu, L. Qiu, L. Zhang, H. Liu and W. Tan, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2017, 9, e1438 Search PubMed.
- L. Yang, X. Zhang, M. Ye, J. Jiang, R. Yang, T. Fu, Y. Chen, K. Wang, C. Liu and W. Tan, Adv. Drug Delivery Rev., 2011, 63, 1361–1370 CrossRef PubMed.
- J. G. Bruno, Pharmaceuticals, 2013, 6, 340–357 CrossRef PubMed.
- M. P. Melancon, M. Zhou, R. Zhang, C. Xiong, P. Allen, X. Wen, Q. Huang, M. Wallace, J. N. Myers, R. J. Stafford, D. Liang, A. D. Ellington and C. Li, ACS Nano, 2014, 8, 4530–4538 CrossRef PubMed.
- S. Kang and S. S. Hah, Bioconjugate Chem., 2014, 25, 1421–1427 CrossRef PubMed.
- K. Heo, S. W. Min, H. J. Sung, H. G. Kim, H. J. Kim, Y. H. Kim, B. K. Choi, S. Han, S. Chung, E. S. Lee, J. Chung and I. H. Kim, J. Controlled Release, 2016, 229, 1–9 CrossRef PubMed.
- U. Wuellner, J. I. Gavrilyuk and C. F. Barbas 3rd, Angew. Chem. Int. Ed. Engl., 2010, 49, 5934–5937 CrossRef PubMed.
- Y. Wan, L. Wang, C. Zhu, Q. Zheng, G. Wang, J. Tong, Y. Fang, Y. Xia, G. Cheng, X. He and S. Y. Zheng, Cancer Res., 2018, 78, 798–808 CrossRef PubMed.
- S. Dickgiesser, N. Rasche, D. Nasu, S. Middel, S. Horner, O. Avrutina, U. Diederichsen and H. Kolmar, ACS Chem. Biol., 2015, 10, 2158–2165 CrossRef PubMed.
- K. Chen, B. Liu, B. Yu, W. Zhong, Y. Lu, J. Zhang, J. Liao, J. Liu, Y. Pu, L. Qiu, L. Zhang, H. Liu and W. Tan, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2017, 9(3) DOI:10.1002/wnan.1438.
- R. Wang, G. Zhu, L. Mei, Y. Xie, H. Ma, M. Ye, F. L. Qing and W. Tan, J. Am. Chem. Soc., 2014, 136, 2731–2734 CrossRef PubMed.
- S. Yoon, K. W. Huang, V. Reebye, D. Spalding, T. M. Przytycka, Y. Wang, P. Swiderski, L. Li, B. Armstrong, I. Reccia, D. Zacharoulis, K. Dimas, T. Kusano, J. Shively, N. Habib and J. J. Rossi, Mol. Ther.–Nucleic Acids, 2017, 6, 80–88 CrossRef PubMed.
- N. Zhao, S. N. Pei, J. Qi, Z. Zeng, S. P. Iyer, P. Lin, C. H. Tung and Y. Zu, Biomaterials, 2015, 67, 42–51 CrossRef PubMed.
- G. Zhu, G. Niu and X. Chen, Bioconjugate Chem., 2015, 26, 2186–2197 CrossRef PubMed.
- J. H. Jeong, H. Mok, Y. K. Oh and T. G. Park, Bioconjugate Chem., 2009, 20, 5–14 CrossRef PubMed.
- C. L. Esposito, S. Catuogno and V. de Franciscis, Journal of RNAi and Gene Silencing: An International Journal of RNA and Gene Targeting Research, 2014, 10, 500–506 Search PubMed.
- W. Y. Lai, W. Y. Wang, Y. C. Chang, C. J. Chang, P. C. Yang and K. Peck, Biomaterials, 2014, 35, 2905–2914 CrossRef PubMed.
- M. Brinkley, Bioconjugate Chem., 1992, 3, 2–13 CrossRef PubMed.
- C. M. Niemeyer, T. Sano, C. L. Smith and C. R. Cantor, Nucleic Acids Res., 1994, 22, 5530–5539 CrossRef PubMed.
- Z. Chen and D. E. Ruffner, Biotechniques, 1996, 21, 820–822 CrossRef PubMed.
- G. T. Hermanson, in Bioconjugate Techniques, Academic Press, Boston, 3rd edn, 2013, pp. 549–587 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra06026f |
| ‡ Equal contribution to work. |
| This journal is © The Royal Society of Chemistry 2018 |