Open Access Article
Open Access ArticleAdaptations of Bacillus shacheensis HNA-14 required for long-term survival under osmotic challenge: a multi-omics perspective†
Xiufeng Longa,
Jiewei Tiana,
Xuepin Liao*ab and
Yongqiang Tian *ab
*ab
aDepartment of Biomass and Leather Engineering, Sichuan University, Chengdu 610065, PR China. E-mail: xpliao@scu.edu.cn; yqtian@scu.edu.cn
bNational Engineering Laboratory for Clean Technology of Leather Manufacture, Sichuan University, Chengdu 610065, PR China
First published on 2nd August 2018
Abstract
Genomic sequence, transcriptomic, metabolomic and fatty acid analyses of strain HNA-14 were performed to understand the mechanism of salt tolerance for long-term survival. The results indicated that strain HNA-14 has different osmotic resistance mechanisms for long-term survival and short-term salt stress. The cells mainly synthesized compatible solutes to resist osmotic pressure when cultured under nutrient deficient conditions, while they can slow down the synthesis rate and uptake from the environment when cultured under a nutritionally rich environment. Also, the amounts of branched and unsaturated fatty acids in the cell membrane are maintained to a high degree (>50%) to maintain the fluidity of the cell membrane; when the cells are cultured in a high osmotic environment for long-term survival, they may increase the content of branched fatty acids and phosphoric fatty acids to increase the fluidity of the cell membrane to resist the high osmotic pressure.
1. Introduction
Halophiles (salt-lovers) are microorganisms that require salt (NaCl) to grow and can be found in all three domains of life (Archaea, Bacteria, and Eukarya).1 Halophilic bacteria inhabit a wide range of hypersaline environments such as saline lakes, saltern ponds, deserts, hypersaline soils, and salted foods. According to the salt concentration for optimal growth, halophiles can be further divided into slight halophiles that grow optimally in 3% (w/v) salt, moderate halophiles [optimal growth at 3–15% (w/v) salt], and extreme halophiles that grow optimally at 25% (w/v) salt.2 Due to their unique halophilic properties, halophiles are strong candidates for use in the development of open, unsterile and continuous fermentation processes that use seawater or other mixed substrates. Johnson et al.3 achieved a record 3 year fermentation process without contamination by feeding acetate to a mixed bacterial culture.To thrive in a hypersaline environment, halophiles have two main adaptation mechanisms to prevent NaCl from diffusing into cells: the salt-in-cytoplasm mechanism and the organic-osmolyte mechanism.4 The first mechanism can be found in extremely halophilic archaea and halophilic bacteria,2,5 wherein inorganic ions (mainly K+ and Cl−) accumulate in the cytoplasm until the internal salt concentration is similar to that of the extracellular environment. According to Shabala et al.6 bacteria exposed to hyperosmotic stress have an increased cellular accumulation of K+ creating conditions of hyperosmotic stress that determines the direction of net K+ flux across the membrane. Ultimately, the intracellular K+ content suggested that the process was at least partially explained by the voltage gating of K+ transporters. In many bacteria, primary active P-type ATPases of the Kdp-type are present, in addition to secondary carriers of the Trk-type. The Trk-type systems (trkA, trkB) are related to Trk transporters and are mainly present in many Gram-positive bacteria.7,8 Furthermore, it has been predicted that some related proteins, such as KefA9,10 in Escherichia coli, may regulate mechano-sensitive channels and probable voltage-gated potassium channel subunit beta.
The vast majority of prokaryotes cope with increasing osmolality by an organic-osmolyte mechanism which controls uptake or synthesis of compatible solutes which do not interfere with cell metabolism.11 These molecules are highly soluble and do not carry a net charge at physiological pH. In contrast to inorganic salts, they can reach high intracellular concentrations without disturbing vital cellular functions, such as DNA replication, DNA–protein interactions, or the cellular metabolic machinery.12–15 Compatible solutes, also called osmolytes, are used by microorganisms and include sugars, amino acids and derivatives, polyols and derivatives, glycine betaines, ectoines and occasionally peptides. In addition, disaccharides such as sucrose, maltose, cellobiose, gentiobiose, turanose, and palatinose, can act as osmoprotectants in bacteria.16 Furthermore, bacteria can also autosynthesize and take up suitable compounds from their surroundings.17–20 Ectoine and hydroxyectoine from halophiles are commercially used as protective agents for mammalian cells and skin.21 There is also growing interest in the alkaline enzymes produced by halophilic microorganisms.22 Moreover, many halophilic microorganisms are producers of bio-surfactants and bioemulsifiers.23
Many studies have focused on the resistance of halophilic bacteria to high osmotic pressure, but most of these are focused on the specific mechanisms. Some studies have used metabolomic or transcriptomic approaches,24–26 and have usually focused on the changes inside cells during the early stage (stress period) of increased osmotic pressure. In the natural environment and in industrial applications, halophilic bacteria produce a stress reaction to osmotic pressure, but the reaction time occupies only a small period of the whole growth. Following the stress period, how can cells maintain normal growth? Do they continue to accumulate a high content of K+ or compatible solute, or change the composition of the cell membrane to resist high osmotic pressure? Limited research has focused on this area.
In our previous study, we isolated a moderately halophilic bacteria, Bacillus shacheensis HNA-14, identified as a novel strain from saline-alkaline soil samples collected in Shache County, Xinjiang province, in northwestern China.27 Genomic, transcriptomic, metabolomic and fatty acid analyses of the strain were performed. Based on these three-omic analyses, the salt tolerance mechanism of halophilic bacteria Bacillus shacheensis HNA-14 for long-term survival is discussed.
2. Materials and methods
2.1 Bacterial strain and growth conditions
The strain HNA-14 was isolated from saline-alkali soil samples and identified by morphological, biochemical, chemotaxonomic and phylogenetic methods, as outlined in our previous study.27 The strain was maintained and propagated on TSA agar plates containing tryptone (15 g L−1), soya peptone (5 g L−1), NaCl (100 g L−1) and agar (20 g L−1). For metobolomics and transcriptomics experiments, the strain was also incubated in minimal medium containing: 10 g glucose, 2 g (NH4)2SO4, 1 g K2HPO4, 1 g MgSO4·7H2O, 0.01 g CaCl2 per liter, and 1 mL trace elements mixture (1 g FeSO4·7H2O, 1 g MnCl2·4H2O, 1 g ZnSO4·7H2O per liter). The medium was supplemented with 5%, 10% or 15% NaCl for HNA-14, and the pH was adjusted to 8.0 by HCl/NaOH before sterilization.2.2 Salt tolerance
Different concentrations of NaCl [0, 0.5, 1, 3, 5, 7, 10, 12, 15, 17, 20, 23, and 25% (w/v)] were added to minimal medium and the pH was adjusted to 8.0 with HCl/NaOH before sterilization. Growth performance was monitored by measuring the optical density at 600 nm with a UV-vis spectrophotometer. Growth curves were performed using minimal medium and TSB medium supplemented with different NaCl concentrations (5%, 10% and 15%, w/v). All assays were carried out at least in triplicate.2.3 Genomic sequencing and annotation
Genomic DNA of strain HNA-14 was isolated as previously described by Li et al.28 To quantitate and check contamination of the extracted DNA, a NanoDrop2000 (Thermo Fisher) and 16S rRNA gene sequencing (ABI 3730XL, Applied Biosystems) were used. The genome of strain HNA-14 was sequenced using Illumina Miseq technology at the Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China). Reads were assembled using SOAP denovo v2.04, after which the low-quality reads were filtered out. Gene prediction was carried out using Glimmer 3.02. The assembled contigs were submitted to the Prokaryotic Genome Automatic Annotation Pipeline (PGAAP) provided by the National Center for Biotechnology Information (NCBI) for subsystem classification and functional annotation. Gene ontology (GO) functional annotation of genes was carried out using the blast2go algorithm. Clusters of Orthologous Groups of proteins (COG) annotation was carried out using the NCBI String database (v 9.05) with blastp (BLAST 2.2.28+).2.4 Transcriptome analysis
For transcriptomic comparisons, strain HNA-14 was cultivated at different NaCl concentrations (5% and 15%, m/v). Bacterial cells were grown in minimal medium and harvested during the exponential growth stage by centrifugation at 3500 rpm for 10 min at 4 °C. Total RNA was isolated using Trizol Reagent (Invitrogen Life Technologies). Quality and integrity were determined using a NanoDrop spectrophotometer (Thermo Scientific) and a Bioanalyzer 2100 system (Agilent). For mRNA sequencing, the Ribo-Zero rRNA Removal Kit (Illumina, San Diego, CA, USA) was used. Random oligonucleotides and SuperScript III were used to synthesize the first strand cDNA. Second strand cDNA synthesis was subsequently performed using DNA polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities and the enzymes were removed. Following adenylation of the 3′ ends of the DNA fragments, Illumina PE adapter oligonucleotides were ligated to prepare for hybridization. cDNA fragments of the preferred 300 bp in length, were size selected using the AMPure XP beads (Beckman Coulter, Beverly, CA, USA). DNA fragments with ligated adaptor molecules on both ends were selectively enriched using the Illumina PCR Primer Cocktail in a 15 cycle PCR reaction. Products were purified (AMPure XP beads) and quantified using the Agilent high sensitivity DNA assay on a Bioanalyzer 2100 system (Agilent). The sequencing library was then sequenced on a NextSeq 500 platform (Illumina) by Shanghai Personal Biotechnology Cp. Ltd.2.5 Metabolome analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min at 4 °C, after which the supernatants were analyzed by LC-MS.
000 rpm for 10 min at 4 °C, after which the supernatants were analyzed by LC-MS.2.6 Fatty acids analysis
The isolate was cultured using different media (minimal medium and TSB medium) supplemented with different NaCl concentrations (5%, 10% and 15%) at 30 °C until the exponential growth phase. The extraction of bacterial lipids and the preparation of fatty acid methyl esters (FAMEs) were carried out according to the method of Wu et al.31 Briefly, cells were collected by centrifugation (4000 rpm at 4 °C for 10 min). The cell pellets were washed twice with NaCl solution that was isosmotic to the growth medium, then mixed with 1 mL NaOH in a methanol distilled water solution (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, w/v/v) and heated at 100 °C for 30 min. After rapid cooling in an ice bath, FAMEs were extracted with 1.25 mL methyl tertiary butyl ether–hexane (1
10, w/v/v) and heated at 100 °C for 30 min. After rapid cooling in an ice bath, FAMEs were extracted with 1.25 mL methyl tertiary butyl ether–hexane (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) for 10 min and washed with 3 mL 0.33 mol L−1 NaOH. The organic phase (top layer) was transferred and prepared for injecting. FAMEs were analyzed on a Trace GC Ultra gas chromatograph-DSQ II mass spectrometer (Thermo Electron Corporation, Boston, Mass., U.S.A.) equipped with a HP-INNOWAX capillary column (30.0 m × 0.25 mm i.d., 0.25 μm film thickness; Agilent Technology) and a flame ionization detector (FID). Highly purified helium carrier gas (99.999%) was used at a constant flow rate of 1 mL min−1. FAMEs were identified by their mass spectra compared against a database of known compounds. The relative amount of FAMEs was calculated from peak areas.
1, v/v) for 10 min and washed with 3 mL 0.33 mol L−1 NaOH. The organic phase (top layer) was transferred and prepared for injecting. FAMEs were analyzed on a Trace GC Ultra gas chromatograph-DSQ II mass spectrometer (Thermo Electron Corporation, Boston, Mass., U.S.A.) equipped with a HP-INNOWAX capillary column (30.0 m × 0.25 mm i.d., 0.25 μm film thickness; Agilent Technology) and a flame ionization detector (FID). Highly purified helium carrier gas (99.999%) was used at a constant flow rate of 1 mL min−1. FAMEs were identified by their mass spectra compared against a database of known compounds. The relative amount of FAMEs was calculated from peak areas.
3. Results
In our previous study, we isolated a novel moderately halophilic bacteria, Bacillus shacheensis HNA-14, from saline-alkaline soil samples.27 In order to better understand the physiological resistance of HNA-14 to osmotic pressure at different salt concentrations, genomic, transcriptomic and metabolomic analyses of the strain were performed.3.1 Growth characteristics of HNA-14 in different media and NaCl stress
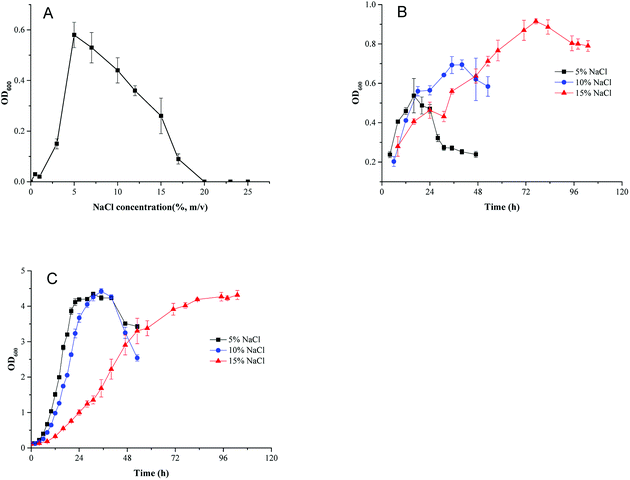
To investigate the effect of osmotic pressure on cell growth, HNA-14 cells were inoculated into minimal medium containing different NaCl concentrations. As shown in Fig. 1A, HNA-14 grew best under 5% (w/v) NaCl concentration, cells did not grow in NaCl concentrations higher than 17%. And no growth of HNA-14 cells was observed when NaCl concentration was lower than 3%. To determine the effects of different culture medium and salt concentration on cell growth, the growth curves of HNA-14 cultured on the minimal medium and TSB medium supplemented with different NaCl concentrations (5%, 10% and 15%, w/v) were performed. The results indicated that HNA-14 grew well on minimal medium and TSB medium containing 5–15% (w/v) NaCl (Fig. 1B and C), the maximum growth rate of HNA-14 cells in the presence of 5% (w/v) sodium chloride was observed, with μmax reaching 0.13 ± 0.011 h−1 (minimal medium) and 0.28 ± 0.029 h−1 (TSB medium), respectively. Lower growth rates were observed with increasing salt concentrations, 0.12 ± 0.014 h−1 (minimal medium) and 0.26 ± 0.011 h−1 (TSB medium), 0.046 ± 0.0028 h−1 (minimal medium) and 0.14 ± 0.001 h−1 (TSB medium) for 10% and 15% NaCl concentration respectively. The results indicated that the growth was better in TSB medium than minimal medium, and the salt concentration can significantly affect the cell growth in both the two medium.3.2 Genome analysis
The draft genome sequence of HNA-14 comprises 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 963
963![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 841 bases with an average G + C content of 47.1%. The assembled genome consists of 98 contigs (N50, 164
841 bases with an average G + C content of 47.1%. The assembled genome consists of 98 contigs (N50, 164![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 238 bp) and 86 scaffolds (N50, 164
238 bp) and 86 scaffolds (N50, 164![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 238 bp). Annotation of the genome identified a total of 3977 genes, of which 3783 have been annotated as coding DNA sequences (CDS), 112 as pseudo genes, 6 as rRNAs, 71 as tRNAs and 5 as ncRNAs. The Bacillus shacheensis HNA-14 whole genome shotgun (WGS) project has been deposited in DDBJ/ENA/GenBank under the accession number LLYY00000000 (Table 1).
238 bp). Annotation of the genome identified a total of 3977 genes, of which 3783 have been annotated as coding DNA sequences (CDS), 112 as pseudo genes, 6 as rRNAs, 71 as tRNAs and 5 as ncRNAs. The Bacillus shacheensis HNA-14 whole genome shotgun (WGS) project has been deposited in DDBJ/ENA/GenBank under the accession number LLYY00000000 (Table 1).
| Features | Genome |
|---|---|
| Genome size | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 963 963![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 841 bp 841 bp |
| G + C contents (%) | 47.1% |
| Genome coverage | 232× |
| No. of all contigs | 98 |
| Contigs N50 | 164![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 238 bp 238 bp |
| No. of all scaffolds | 86 |
| Scaffolds N50 | 164![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 238 bp 238 bp |
| tRNA genes | 71 |
| rRNA genes | 6 |
| ncRNAs | 5 |
| Pseudo genes | 112 |
| Bioproject ID | PRJNA299368 |
| Biosample ID | SAMN04207873 |
| Genome accession | LLYY00000000 |
At present, the salt-in-cytoplasm mechanism and the organic-osmolyte mechanism, as well as an increase in cell membrane fluidity, are known as salt tolerance mechanisms.32 In order to clearly determine the salt-tolerant mechanism of strain HNA-14, we analyzed the genome and briefly summarized genes related to salt tolerance, such as Na+ efflux and K+ uptake genes, compatible solutes synthesis and transport genes, and fatty acid synthesis genes. In prokaryotes, common osmotic protection systems associated with Na+ efflux and K+ uptake include TrK-type systems, Kdp-type systems, Kef-type systems and a voltage-gated potassium channel. We found several genes that encode channel related proteins, such as TRKA/TRKB/TRKD (TrK-type system), KdpD (Kdp-type system), KefA (Kef-type), VGSC (K+ uptake) and MnhA/NhaC (ABC transporter family related to Na+ efflux) (Table S1†). The genes related to common osmoprotectants found in HNA-14 were mainly transport-related genes, which can help the strain to take up compatible solutes present in the environment in order to resist high osmotic pressures. Only a small part of relevant synthesis pathways of small molecules, such as sugars, proline and ectoine, were found. Proline is a common compatible solute that can be synthesized in Bacillus, and genes for the synthesis-related proteins ProB–ProA–ProC (located in the same operon), PutA–ProC and PutA–ProDH, were also found (Fig. S1-A†). These putative pathways are similar to the two pathways ProB–ProA–ProI and ProJ–ProA–ProH in Bacillus subtilis,32 where the former pathway is crucial for normal physiological functions, and the latter plays an important role in resisting osmotic pressure. It is also highly probable that one or more of the three pathways would be activated to resist pressure and clear out toxins when the cells are exposed to a high osmotic environment. Hydroxyectoine is synthesized from aspartic acid by the enzymes Lysc–Asd–EctB–EctA–EctC–EctD (Fig. S1-B†).
In minimal medium, the composition of the medium is defined and there are no compatible solutes that could be taken up by the cells, the cells can only uptake some ions or synthesis compatible solutes to balance the internal and external osmotic pressure, which may inevitably lead to a change in central carbon metabolism. In TSB medium, compatible solutes are present in the environment and can help cells to cope with high osmotic pressures, meaning that their metabolism must be different from that in minimal medium, which needs to be explored further using transcriptome and metabolome analyses.
3.3 Transcriptome analysis
According to the salt tolerance results of HNA-14, we chose 5% and 15% (w/v) NaCl concentration as the culture condition for transcriptional analysis. Whole transcriptome analysis was performed using an Illumina NextSeq 500 sequencing platform. Reads were mapped onto the reference genome and RPKM values were compared. Differently expressed genes (DEGs) were identified in the case of the FDR < 0.001 and the |log2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) O/P| > 0.3.
O/P| > 0.3.
According to transcriptomic data, we performed hierarchical clustering of the significantly differentially expressed genes of HNA-14 cultured in 5% and 15% NaCl concentration, the heat map illustrated obvious differences in gene expression between different osmotic pressure (Fig. 2, Table S2†). The changes found by KO enrichment analysis were mainly those genes related to biosynthetic process, translation, transport, cytosol, intracellular, and RNA binding. The KEEG enrichment analysis showed that a few genes involved in the translation, folding and degradation of the protein have obvious changes, while other genes did not have significant changes.
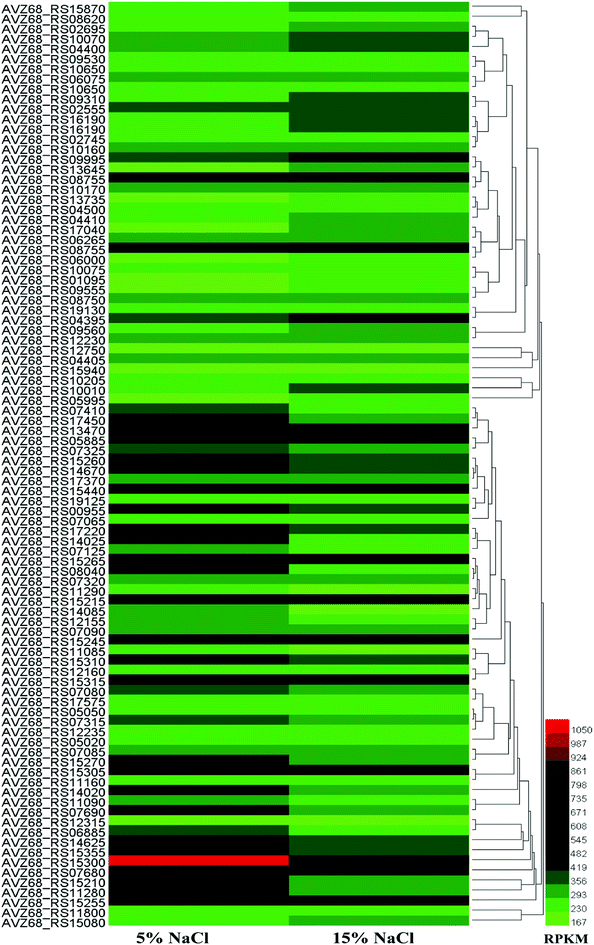 | ||
| Fig. 2 Hierarchical clustering of features, of the whole transcriptome of HNA-14 cultured in 5% and 15% NaCl concentration. The heat map represents the expression of genes in Table S2.† | ||
We further analysis the data and found the expression of a large number of ribosomal proteins showed tended to be down-regulated at higher salt concentrations (16 of 30s RNA and 20 of 50s RNA ribosomal protein), while translation-related initiation factors and elongation factors were also decreased, the reason might be the cell growth become slower under higher salt stress. In addition, most amino acid synthesis related genes did not change significantly. The proline synthesis genes (proB–proA–proC) were up-regulated to some extent at higher salt concentrations (15% NaCl, w/v), while another gene, putA in the putA–proDH, operon was slightly up-regulated. ProDH was down-regulated. According to the results, when under higher salt stress the genes proB–proA–proC and putA–proC were activated, while the putA–proDH genes were inhibited, which was similarly to the previous study.32 The glutamate synthase genes glnA and gltD were up-regulated, and glutamine synthase may be attributed to an increase in its precursors. Enzymes in the valine and leucine synthetic pathways were activated to some extent under higher salt conditions, which was consistent with a previous study.32 Enzymes in the aspartate synthetic pathway were not up-regulated, but were slightly inhibited by higher salt concentration. It is worth mentioning that the compatible solute ectoine related genes lysC–asd–ectB–ectA–ectC–ectD were also inhibited at higher salt concentrations, and enzymes related to this pathway also decreased, which was different from most of other relevant studies. In general, the genes involved in amino acid synthesis pathways were not activated or repressed too much due to increasing salt concentrations, and were not significantly altered at the transcriptional level.
Three genes related to central carbon metabolic pathways (glycolysis, TCA, PPP) were up-regulated to some extent at the transcriptional level. In the glycolysis pathway, except for the first step enzyme PTS-Glc-EIIA, which catalyzes the transformation of glucose to glucose-6-phosphate and was up-regulated, pyruvate kinase (PK) was down-regulated, but other enzymes did not change significantly. This may be due to the fact that although the metabolism was slowed down at high salt concentration, cells need to uptake more glucose to synthesis small molecules to resist salt stress. In TCA cycle citrate synthase and fumarate hydratase were slightly up-regulated, while other genes were down-regulated, which is consistent with the previous study.32 PPP pathway related enzymes showed a general downward trend.
In addition to ribosomal proteins, the most varied genes at the transcriptional level were various transport proteins, especially ABC family proteins, of which 12 were down-regulated and 27 were up-regulated (Fig. 3). Among the 12 genes, two were related to manganese and zinc ion transport, two were amino acid transporters, two were saccharide transporters, two were glycine betaine transporters, and one was a transporter for other ions. Small molecule transporters, ion transporters, and amino acid transporters were mainly up-regulated.
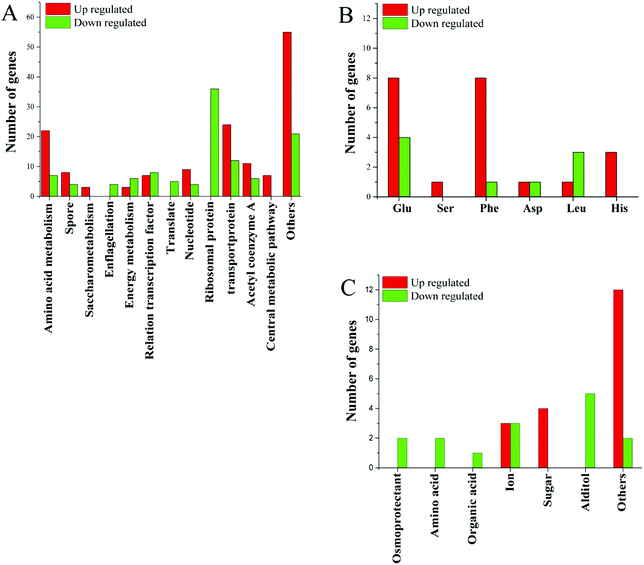 | ||
| Fig. 3 (A) Changes in gene expression profile; (B) changes in gene expression related to amino acids; (C) changes in gene expression related to transporters. | ||
Acetyl-CoA is an important cellular metabolite that plays an important role in the synthesis of fatty acids, the TCA pathway and amino acid metabolism. Seventeen functional genes related to acetyl-CoA were altered at the transcriptional level, of which 6 were down-regulated and 11 were up-regulated (Fig. 3). Of genes related to the energy-producing oxidative phosphorylation pathway, 6 genes related to the synthesis of cytochrome C were down-regulated, the ATP synthase subunit I was up-regulated. Two of the genes involved in fatty acid degradation were down-regulated. This is consistent with the down-regulated of genes involved in protein synthesis and the slower cell growth. The changes in gene expression associated with the energy metabolism pathway may be to respond to different energy needs in different environments.
Several genes related to sulfur metabolism, signal transduction, FE-S proteins, RNA degradation and transcripts, DNA repair and DNA binding proteins, as well as several other genes were altered at the transcriptional level in higher salt concentrations. Changes in many unknown hypothetical proteins were also observed (Fig. 3). These unknown hypothetical proteins may play an important role in bacterial adaptation and survival in high salt environments.
3.4 Metabolomic analysis of strain HNA-14
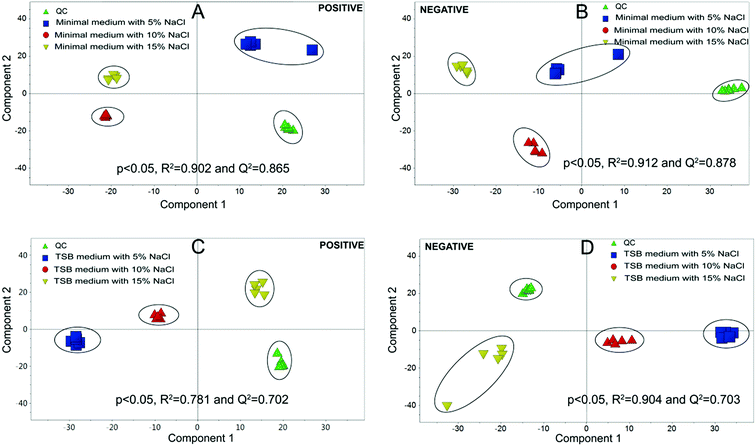
Metabolomics experiments were divided into two groups, (i) minimal medium with glucose as a sole carbon source; (ii) TSB medium, which is a richer medium with abundant nutrients.Multivariate statistical analysis was carried out with PCA and PLS-DA score plots after data processing and normalization in order to evaluate changes in the metabolite profiles under salt stress. As shown in Fig. 4, the quintuplicate of fermentation treatments are generally placed close to each other for all the samples, which indicated good reproducibility of the fermentation treatments and analytical method. The PCA plots in Fig. 4 demonstrated a clear effect of NaCl concentration on the level of intracellular metabolites. PCA on the different treatments revealed a significant separation among the cells with different salt stress, which demonstrated metabolic differences between the cells under different NaCl concentration. All the PLS-DA score plots were cross-validated with permutation tests that showed P < 0.05. The results indicated that significant metabolic differences were detected when strain HNA-14 was cultured in different media and salt concentrations.
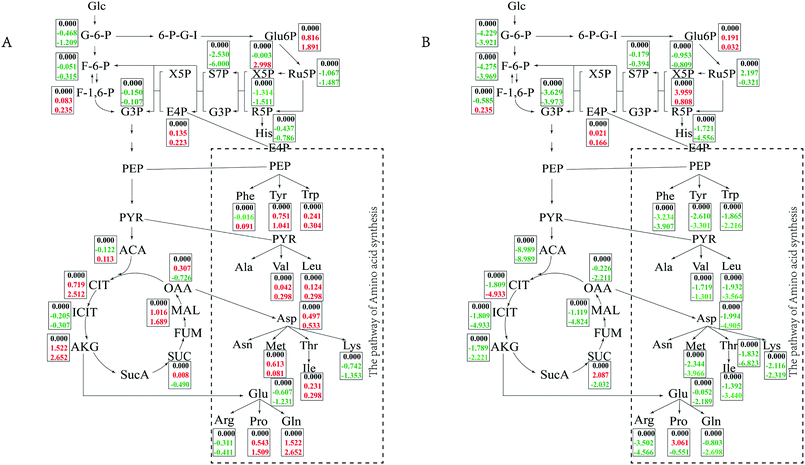
In minimal medium, according to transcriptomic analysis, there was no significant up-regulation of the relevant ion channel proteins, which indicated that the cells did not balance the osmotic pressure through uptake ions. The cells might synthesize compatible solutes de novo instead of taking them up from the environment to resist osmotic pressure. Changes in central carbon metabolism can also reflect this salt tolerance mechanism (Fig. 5). Intracellular accumulation of metabolites in the TCA pathway, glycolysis pathway and PPP pathway were detected. In minimal medium, an accumulation of citrate and malate in the TCA cycle increased with increasing salt concentration, while oxaloacetate and succinate first increased and then decreased when NaCl concentration changed form 5% to 10% and 15%, which was consistent with the transcriptome data. Acetyl-CoA, as the starting material for the TCA cycle, first decreased and then increased with increasing salt concentration. In the glycolysis pathway, except for fructose-1, 6-bisphosphate, which accumulated gradually with increasing salt concentration, the intracellular accumulation of other metabolites gradually decreased. Among metabolites of the PPP pathway, 6-phospho-gluconate and erythrose-4-phosphate accumulated with increasing salt concentration, which were consistent with the up-regulated of the first enzyme in glucose uptake in transcriptome analysis. Whereas xylulose-5-phosphate accumulated at high salt concentrations (15%), increasing up to 3-fold compared to 5% NaCl. Other metabolites of the PPP pathway had reduced concentrations with increasing salt concentration, it was also the same with transcriptome data, the reason might be the cell growth slowed down so that the demand for energy and precursor were reduced.
In TSB medium, intermediate of the TCA cycle and glycolysis pathway tended to decrease with increasing salt concentration, while fructose-1,6-bisphosphate first increased then decreased. 6-Phospho-gluconate, ribulose-5-phosphate and ribose-5-phosphate also showed a trend of first increasing and then decreasing. Erythrose-4-phosphate began to accumulate at high salt concentrations (15% NaCl). The changes of the central metabolic pathway in different media indicated that the bacteria cells take different strategies to resist the high salt stress for long-term survival depending on whether the surrounding nutrients are abundant or not. The cells can synthesize compatible solutes against osmotic pressure themselves when nutrients are scarce, and it can uptake small molecules in the environment when nutrients are rich.
Accumulation of valine and leucine increased slightly in minimal medium, which was consistent with the previous report.32 Aspartate, isoleucine and methionine also accumulated with increasing salt concentration. The concentration of proline increased significantly with increasing salt concentration, and the same phenomenon was observed for glutamine. A large number of intracellular amino acid derivatives, such as nitrotyrosine, pyrrolysine, N2-acetyl-L-ornithine, Nα-acetyl-L-glutamine, accumulated with increasing salt concentration. These phenomena are consistent with the changes cells suffered from salt shock for short time,32 which indicated that there was certain similarities between the mechanism of osmotic pressure resistance in a short time and for long-term survival. In TSB medium, all of the detected amino acids were greatly reduced with increasing salt concentration. Ectoine and 5-hydroxyectoine as common compatible solutes, tended to first increase and then decrease with increasing salt concentration. The content of some substances, such as nucleotides and their derivatives, amides and organic acids, changed with salt concentrations and changing culture media (Tables S3 and S4†).
As a precursor of many other substances, amino acids played an important role in the synthesis of small molecules. In the absence of nutrients, the cells have to accumulate some amino acids to ensure the synthesis of other compatible solutes, and at the same time, they also have the function of balancing osmotic pressure. When the environment is rich in nutrients, cells can uptake small molecules from the environment to resist osmotic pressure, the demand for amino acids becomes smaller, and its accumulation in cells also decreases accordingly, and less synthesis of excess amino acids can make more energy available for maintenance the necessary growth, which also indicated that in this case amino acids are not the main compatible solutes.
The cell membrane is the first barrier against osmotic pressure and is primarily composed of membrane lipids, proteins and carbohydrates. In this study, phosphatidylethanolamine (PE), phosphatidylinositol (PI) and sphinganine, which are important components of membrane lipids, increased with increasing salt concentration when strain HNA-14 was cultured in minimal medium. In TSB medium, PE and PI increased while sphinganine showed the opposite result.
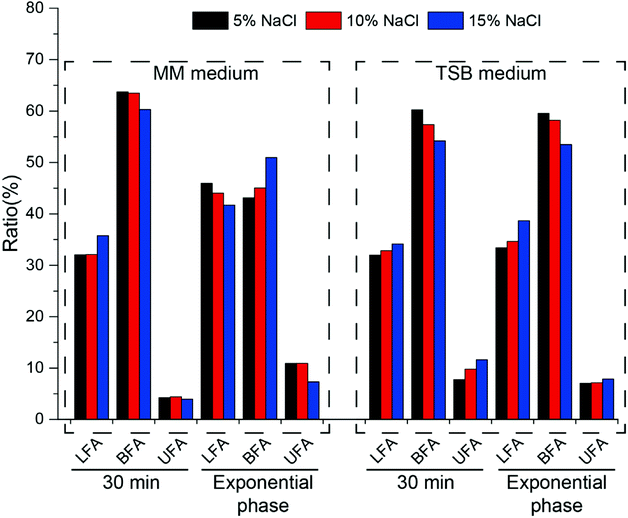
In order to further clarify the changes in cell membrane composition under different culture conditions, fatty acid components were tested (Fig. 6). As an important component of the cell membrane, changes in fatty acids play an important role in resistance to osmotic pressure.
In minimal medium, after increasing the salt concentration for 30 min, the content of linear fatty acids increased, branched fatty acids decreased and unsaturated fatty acids did not change significantly. At exponential phase, linear and unsaturated fatty acids decreased with increasing salt concentration, while branched fatty acids increased. After cells were cultured to exponential phase, the ratio of branched fatty acids (C14:0, C15:0) gradually increased from 43.12% to 50.95% as the salt concentration increased, and the ratio of unsaturated fatty acids gradually decreased (from 10.94% to 7.33%), which is opposite to that reported in the literature.33 In TSB medium, salt shock for 30 min led to an increase in the amount of linear and unsaturated fatty acids and a decrease in the amount of branched fatty acids (Fig. 6). During long-term growth, the same phenomenon was observed. Whether in minimal or TSB medium, the amount of branched fatty acids comprised the majority of the whole fatty acid content, which indicated that halophilic strain HNA-14 possessed good cell membrane fluidity.
4. Discussion
In our previous study, we isolated a moderately halophilic bacterium Bacillus shacheensis HNA-14 from saline alkaline soil samples collected in Shache County, Xinjiang Province, China. In the natural environment, strain HNA-14 has to deal with the double impact of high salt concentrations and nutrient deficiency. In Bacillus, a large number of studies have demonstrated the mechanisms of adaptation to high salt stress, however, most of these have focused on the short period after osmotic shock. When cells are exposed to different osmotic pressures for a long time in the natural environment, is the osmoprotection mechanism consistent with that seen during short time osmotic shock? Therefore, we have performed a multi-omics study to demonstrate the long-term survival of strain HNA-14 under different osmotic pressures, which may help illustrate the adaptive mechanisms of Bacillus when grown in different osmotic and nutrient environments.Genomic analysis, transcriptome analysis at different osmotic pressures, and metabolomic analysis at different osmotic conditions and under different nutrient conditions were performed on strain HNA-14. Based on the previous studies1–4 on the mechanisms of osmotic pressure resistance, we analyzed the genes related to this resistance pathway. Similar to other members of the genus Bacillus,34,35 we did not find genes related to the biosynthetic pathways for common compatible solutes (glycine betaine and trehalose) in the genome sequence of strain HNA-14, except for proline and ectoine. However, a large number of transporters such as those for glycine, betaine and proline, were found. These proteins can help strain HNA-14 to take up compatible solutes from a hypertonic environment against osmotic pressure. Genes related to common K+ uptake systems, such as TrK-type systems, Kdp-type systems and Kef-type systems, Na+ efflux systems and some voltage-gated potassium channels were identified. These systems can help cells cope with sudden changes in osmotic pressure. Although transcriptional changes in these genes were not observed in our study (long-term growth in osmotic environment), there have been many studies1–4 that indicated that these genes are up-regulated to resist osmotic shock. This mechanism may play a role in the early stages of the osmotic shock. However, whether the functional genes still played an important role in cells for long-term survival?
In minimal medium, the transcriptome results showed that during long-term growth of cells in a high salt environment, a large number of ribosomal proteins, translation-related initiation factors and elongation factors were down-regulated, which is consistent with previous reports.36 In order to resist osmotic pressure, cells may shift more energy away from basal metabolism. On the other hand, our results showed that most genes in the central carbon metabolic pathways changed minimally or were slightly down-regulated, which also illustrates this point. The enzyme PTS-Glc-EIIA, which catalyzes the synthesis of glucose-6-phosphate from glucose in the central carbon metabolic pathway, was up-regulated, and the intracellular accumulation of glucose-6-phosphate also increased with increasing salt concentration. This may be due to high osmotic pressure that assimilates more substrates for energy and produces compatible solutes to resist the stress. Pyruvate kinase was down-regulated, which may result in a decrease in the accumulation of pyruvate. This was also confirmed by the intracellular metabolite test results (Fig. 5). However, the accumulation of the branched amino acids (Val, Leu) of the pyruvate node increased with increasing salt concentration, which is consistent with a previous study,32 and these played an important role in strain HNA-14. Accordingly, a slight upregulation of the citrate synthase GltA in the TCA cycle leads directly to the accumulation of citrate, which also is consistent with previous reports.32 The up-regulation of fumarate hydratase IDH1 may be to an increase in the rate of oxaloacetate formation and provides conditions for proline synthesis.32,34,37,38 In the proline synthesis pathway, ProB–ProA–ProC and PutA–ProC were activated, while the PutA–ProDH pathway was inhibited to a certain extent. That is, as the salt concentration increased, more proline was synthesized via the ProB–ProA–ProC and PutA–ProC pathways, which indicated that strain HNA-14 can further accumulate proline by regulating enzyme expression to resist osmotic pressure. According to the metabolomics analysis, proline increased gradually with increasing salt concentration, indicating that proline is an important compatible solute for strain HNA-14 to resist osmotic pressure. The transcription of genes in the ectoine-related pathway did not change significantly, and were even slightly inhibited, at high salt concentrations, but in minimal medium ectoine gradually accumulated as the salt concentration increased. Therefore, it was likely that ectoine was accumulated by increasing protein activity or decreasing protein degradation rate to accumulate the glycine betaine. Transcriptional results showed similarities with previous studies,39 in that a large number of transport proteins, especially ABC family proteins, were up-regulated. These primarily included glycine betaine and some other compatible solutes transporters, ATP-binding proteins and ion pumps. Activation of these genes helps to take up compatible molecules from the environment and excrete high concentrations of intracellular ions to maintain normal cellular metabolism and growth. Almost all metabolites in TSB medium decreased with increasing salt concentration and only a small number of metabolites increased. This suggests that strain HNA-14 may use a different mechanism to resist osmotic pressure under nutrient-rich conditions.
The cell membrane is the first barrier against osmotic pressure in Bacillus. According to the metabolomic analysis, an increase in the content of phosphoric fatty acids in either minimal medium or TSB medium can help to quickly change the cell membrane, reduce the passive transport of the membrane and reduce the loss of intracellular water.36,39 Further analysis of fatty acid content results showed that in minimal medium, the content of branched-chain fatty acids and unsaturated fatty acids were the majority of the entire fatty acid content, which indicated that the membrane had great fluidity, which could help to resist the high osmotic pressure.
5. Conclusion
The results indicated that (1) the cells mainly synthesized compatible solutes to resist osmotic pressure when cultured under nutrient deficient conditions; (2) cells can uptake compatible solutes from the environment to balance the internal and external osmotic pressure when cultured under a nutritionally rich environment; (3) cells can change the composition of the cell membrane to improve the fluidity of cell membrane to reduce the damage from high osmotic pressure whether in minimal or TSB medium. The salt-in-cytoplasm mechanism to relieve short-term salt stress did not seem to work in long-term survival when cells were under high osmotic stress.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key Research and Development Program of China (2017YFB0308401); and Chengdu Science and Technology Project (2016HM0100409SF).References
- E. O. Casamayor, R. Massana, S. Benlloch, L. Øvreås, B. Díez, V. J. Goddard, J. M. Gasol, I. Joint, F. Rodríguez-Valera and C. Pedrós-Alió, Environ. Microbiol., 2002, 4(6), 338–348 CrossRef PubMed.
- A. Ventosa, J. J. Nieto and A. Oren, Microbiol. Mol. Biol. Rev., 1998, 62(2), 504–544 Search PubMed.
- K. Johnson, Y. Jiang, R. Kleerebezem, G. Muyzer and M. C. Loosdrecht, Biomacromolecules, 2009, 10(4), 670–676 CrossRef PubMed.
- J. Yin, J. C. Chen, Q. Wu and G. Q. Chen, Biotechnol. Adv., 2015, 33(7), 1433 CrossRef PubMed.
- E. A. Galinski and H. G. Trüper, FEMS Microbiol. Rev., 1994, 15(2–3), 95–108 CrossRef.
- S. Shabala, J. Exp. Bot., 2009, 60, 709–712 CrossRef PubMed.
- I. R. Booth, R. M. Douglas, A. W. Munro, A. J. Lamb, G. Y. Ritchie, S.-P. Koo and G. P. Ferguson, Regulated transport systems in bacteria, in Molecular Mechanisms of Transport, E. Quagliariello and F. Palmieri, 1992, pp. 59–66 Search PubMed.
- I. Ochrombel, L. Ott, R. Krämer, A. Burkovski and K. Marin, Biochim. Biophys. Acta, 2011, 1807(4), 444–450 CrossRef PubMed.
- C. Cui and J. Adler, J. Membr. Biol., 1996, 150(2), 143–152 CrossRef PubMed.
- A. D. Brown, Bacteriol. Rev., 1976, 40, 803–846 Search PubMed.
- A. R. Strom and I. Kaasen, Mol. Microbiol., 1993, 8(2), 205–210 CrossRef PubMed.
- K. Strange, Cellular and molecular physiology of cell volume regulation. CRC Press, 1993 Search PubMed.
- M. T. Record Jr, E. S. Courtenay, D. S. Cayley and H. J. Guttman, Trends Biochem. Sci., 1998, 23(4), 143–148 CrossRef.
- M. T. Record Jr, E. S. Courtenay, D. S. Cayley and H. J. Guttman, Trends Biochem. Sci., 1998, 23(5), 190–194 CrossRef.
- P. Lamosa, L. O. Martins, M. S. da Costa and H. Santos, Appl. Environ. Microbiol., 1998, 64(10), 3591–3598 Search PubMed.
- K. Gouffi and C. Blanco, Int. J. Food Microbiol., 2000, 55, 171–174 CrossRef PubMed.
- T. Shirai, A. Suzuki, T. Yamane, T. Ashida, T. Kobayashi, J. Hitomi and S. Ito, Protein Eng., 1997, 10(6), 627–634 CrossRef PubMed.
- D. Cánovas, C. Vargas, S. Kneip, M. J. Morón, A. Ventosa, E. Bremer and J. J. Nieto, Microbiology, 2000, 146(2), 455–463 CrossRef PubMed.
- Y. Liu, W. Gao, Y. Wang, L. Wu, X. Liu, T. Yan, E. Alm, A. Arkin, D. Thompson, M. Fields and J. Zhou, J. Bacteriol., 2005, 187(7), 2501–2507 CrossRef PubMed.
- Z. J. Gu, L. Wang, D. Le Rudulier, B. Zhang and S. S. Yang, Curr. Microbiol., 2008, 57(4), 306 CrossRef PubMed.
- J. M. Pastor, M. Salvador, M. Argandoña, V. Bernal, M. Reina-Bueno, L. N. Csonka, J. L. Iborra, C. Vargas, J. J. Nieto and M. Cánovas, Biotechnol. Adv., 2010, 28(6), 782–801 CrossRef PubMed.
- M. E. Setati, Afr. J. Biotechnol., 2010, 9(11), 1555–1560 CrossRef.
- S. K. Satpute, I. M. Banat, P. K. Dhakephalkar, A. G. Banpurkar and B. A. Chopade, Biotechnol. Adv., 2010, 28(4), 436–450 CrossRef PubMed.
- K. Marin, Y. Kanesaki, D. A. Los, N. Murata, I. Suzuki and M. Hagemann, Plant Physiol., 2004, 136(2), 3290–3300 CrossRef PubMed.
- S. Kol, M. E. Merlo, R. A. Scheltema, M. de Vries, R. J. Vonk, N. A. Kikkert, L. Dijkhuizen, R. Breitling and E. Takano, Appl. Environ. Microbiol., 2010, 76(8), 2574–2581 CrossRef PubMed.
- N. N. Joghee and G. Jayaraman, Biochimie, 2014, 102, 102–111 CrossRef PubMed.
- Z. Lei, P. Qiu, R. Ye, J. Tian, Y. Liu, L. Wang, S. Tang, W. Li and Y. Tian, Appl. Microbiol., 2014, 60(3), 101–105 Search PubMed.
- W. J. Li, P. Xu, P. Schumann, Y. Q. Zhang, R. Pukall, L. H. Xu, E. Stackebrandt and C. L. Jiang, Int. J. Syst. Evol. Microbiol., 2007, 57, 1424–1428 CrossRef PubMed.
- Q. Teng, W. Huang, T. W. Collette, D. R. Ekman and C. Tan, Metabolomics, 2009, 5(2), 199 CrossRef.
- F. Chen, J. Xue, L. Zhou, S. Wu and Z. Chen, Anal. Bioanal. Chem., 2011, 401(6), 1899 CrossRef PubMed.
- C. Wu, J. Zhang, M. Wang, G. Du and J. Chen, J. Ind. Microbiol. Biotechnol., 2012, 39(7), 1031 CrossRef PubMed.
- M. Kohlstedt, P. K. Sappa, H. Meyer, S. Maaß, A. Zaprasis, T. Hoffmann, J. Becker, L. Steil, M. Hecker, J. M. Dijl and M. Lalk, Environ. Microbiol., 2014, 16(6), 1898–1917 CrossRef PubMed.
- G. He, C. Wu, J. Huang and R. Zhou, Biotechnol. Bioprocess Eng., 2017, 22(4), 366–375 CrossRef.
- A. M. Whatmore, J. A. Chudek and R. H. Reed, Microbiology, 1990, 136(12), 2527–2535 CrossRef PubMed.
- J. Brill, T. Hoffmann, M. Bleisteiner and E. Bremer, J. Bacteriol., 2011, JB-05490 Search PubMed.
- Y. M. Zhang and C. O. Rock, Nat. Rev. Microbiol., 2008, 6(3), 222 CrossRef PubMed.
- T. Hoffmann, A. Wensing, M. Brosius, L. Steil, U. Völker and E. Bremer, J. Bacteriol., 2013, 195(3), 510–522 CrossRef PubMed.
- A. Zaprasis, J. Brill, M. Thüring, G. Wünsche, M. Heun, H. Barzantny, T. Hoffmann and E. Bremer, Appl. Environ. Microbiol., 2013, 79(2), 576–587 CrossRef PubMed.
- J. I. Khudyakov, P. D'haeseleer, S. E. Borglin, K. M. DeAngelis, H. Woo, E. A. Lindquist, T. C. Hazen, B. A. Simmons and M. P. Thelen, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(32), E2173–E2182 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra05472j |
| This journal is © The Royal Society of Chemistry 2018 |