 Open Access Article
Open Access ArticleSurface modification and pattern formation by nucleobases and their coordination complexes
R. Kamal Saravanana,
Ilesha Avasthia,
Rajneesh Kumar Prajapatiab and
Sandeep Verma *ab
*ab
aDepartment of Chemistry, Indian Institute of Technology Kanpur, Kanpur, 208016, UP, India. E-mail: sverma@iitk.ac.in
bCentre for Nanoscience, Indian Institute of Technology Kanpur, Kanpur 208016, UP, India
First published on 6th July 2018
Abstract
This review presents recent progress concerning the organization of nucleobases on highly ordered pyrolytic graphite (HOPG), mica, Cu(110) and Au(111) surfaces, followed by their studies using microscopy methods such as atomic force microscopy (AFM), scanning tunneling microscopy (STM) and transmission electron microscopy (TEM). Interesting research prospects related to surface patterning by nucleobases, nucleobase-functionalized carbon nanotubes (CNTs) and metal–nucleobase coordination polymers are also discussed, which offer a wide array of functional molecules for advanced applications. Nucleobases and their analogs are able to invoke non-covalent interactions such as π–π stacking and hydrogen bonding, and possess the required framework to coordinate metal ions, giving rise to fascinating supramolecular architectures. The latter could be transferred to conductive substrates, such as HOPG and gold, for assessment by high-end tunneling microscopy under various conditions. Clear understanding of the principles governing nucleobase self-assembly and metal ion complexation, and precise control over generation of functional architectures, might lead to custom assemblies for targeted nanotechnological and nanomaterial applications.
Introduction
Adsorption and growth of ordered and self-assembled molecular layers on various surfaces such as highly ordered pyrolytic graphite (HOPG), mica, gold, copper, silicon, etc. lead to interesting topological features, mirroring their own organizational arrangement, and permit access to new properties of these molecules when patterned on surfaces. Surface modification by means of covalent or non-covalent interactions results in 1- or 2D networks on the surface, which have applications in the field of sensing, catalysis, optics, semiconductors, functional nanomaterials and many more. Deposition of small molecules and polymers requires surface optimization based on the nature of the molecule, deposition method, solvent used and surface annealing temperature as well as the other experimental conditions. An entire range of molecular systems such as organic molecules,1–16 biological molecules,17–22 discrete metal complexes, 1-, 2- and 3D coordination polymers,23–31 metal organic frameworks32–43 etc. have been studied on substrates using different microscopy techniques, in order to explore their surface properties.In this review, we focus on surface organization of nucleobases, their derivatives and related functionalized systems, on different metallic and non-metallic substrates. The inherent hydrogen-bonding ability of nucleobases has also been exploited efficiently to get desired morphologies and nanostructures on surfaces. These supramolecular assemblies could be tuned further by chemical derivatization of nucleobases to allow for stable deposition. Also, the presence of heteroatoms in the core skeleton of nucleobases allows metal ions to interact, in order to achieve various binding patterns on the surface. The choice of substituent, metal salt used for complexation, solvent and substrate is critical for surface patterning.44,45
This review focuses on recent advancements in an exciting area of surface modifications by nucleobases, which has applications in many nanotechnological applications. Nucleobases are useful building blocks, modulators or templates for nanomaterial synthesis and our ability to further fine-tune surface-patterning could provide entry to novel applications in functional materials science.45–51,74,77 Here, subsequent sections describe the surface patterns of unmodified nucleobases, their derivatives, metal-complexes and their functionalized forms. Considerable attention has been invested toward patterning and high-resolution imaging of custom surfaces.45,52–61,105 This review exclusively focuses on fundamental and biologically important five canonical nucleobases, namely adenine, guanine, thymine, cytosine and uracil, surface morphologies created by them alone, followed by coadsorption with peptides, metal–nucleobase complexes and nucleobase-modified carbon nanotubes (CNTs).
Surface modification by adenine (A) and its analogs
Nucleobase chemisorption and surface immobilization are primarily guided by operating dispersion forces. In addition to microscopy analyses, many a times these investigations are also supported by density functional theory (DFT) and post-Hartree–Fock calculations. For example, the creation of (modified) adenine monolayers has been subjected to classical molecular dynamics (MD) simulations and it is articulated that interaction of adenine with surfaces is closely governed by varying degree of base–base aromatic interactions.Chi and coworkers illustrated in situ STM imaging of 9-icosyladenine physisorbed on HOPG surface revealing two types of temperature-dependent self-assembled structures: α- and β-phase (Fig. 1). Formation of these on surface phases was driven by H-bonding or aromatic interactions between nucleobases, which was further augmented by van der Waals interactions operating between proximal alkyl chains.62
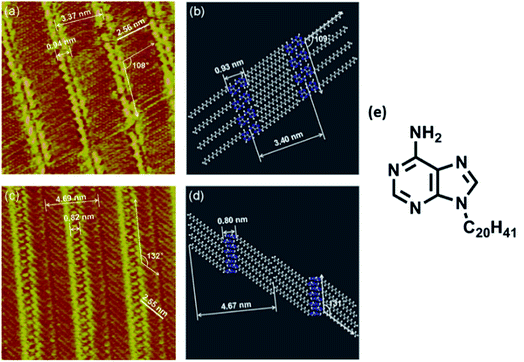 | ||
| Fig. 1 (a) STM image of 9-icosyladenine on HOPG surface; (c) high-resolution STM image of (a); (b) and (d) display model structures with scales relatable to (a) and (c) respectively; (e) structure of 9-icosyladenine. (Reproduced with permission from ref. 62 © 2013, American Chemical Society). | ||
Similar investigations involving a C22 alkyl chain at adenine N9 position revealed two structural phases on the graphite surface, albeit different compared to the previous example, where the deposition pattern was ascribed to the vacuum conditions used for sample preparation (Fig. 2). These highly resolved images helped conclude that substrate temperature and rate of deposition could influence domination of the α phase.63
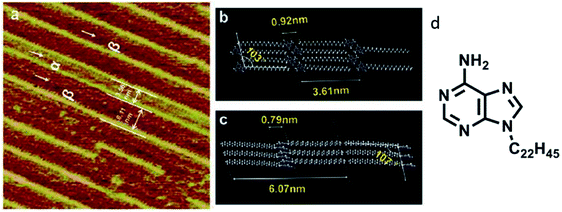 | ||
| Fig. 2 (a) High-resolution STM image of adenine-C22 on a graphite surface indicating two phases; (b)and (c) comprise of model representations demonstrating intermolecular distances in the two phases; (d) structure of adenine-C22. (Reproduced from ref. 63 with permission from The Royal Society of Chemistry). | ||
Ordered growth of adenine epitaxial monolayers on graphite were reported by Allen et al.64 Two dimensional (2D) networks of adenine dimers, stabilized by H-bonding interactions, were observed at 1-octanol/graphite interface by STM imaging.65 Crystalline nature of 2D films, prepared by molecular-beam deposition method, was established using STM and low energy electron diffraction (LEED) techniques,66 while the coexistence of both homochiral and heterochiral domains was identified with high-resolution STM at 1-octanol/solid interface, at room temperature.67
Tao and Shi subjected monolayers of adenine, in aqueous NaCl solution, to AFM and STM investigations. Both of these techniques suggested that adenine arranged in a hydrogen bonded rectangular lattice on HOPG surface, where molecular packing was independent of the potential, but the electronic states exhibited sensitivity to the substrate potential, as judged by STM.68 Notably, the coexistence of homo/hetero chiral domains of self-assembled adenine molecules on various surfaces open up pathways to develop novel surface architectures, which might find useful applications in chiral recognition, chiral separation, heterogeneous catalysis and bioactive surfaces.
Although HOPG is a common substrate for nucleobase organization studies, surfaces such as Au(111), Cu(110) and others are also used to address specific questions. For example, adenine exhibited formation of well-ordered 2D islands on Au(111) revealing two discerning structural possibilities: one of the coexisting phases displayed two adenines per unit cell, while the other presented four adenine molecules, when annealed at 370 K (Fig. 3).69,70 On the contrary, high resolution STM imaging of adenine on Cu(110), at elevated temperature, revealed 1D dimeric chains stabilized by hydrogen bonds, in addition to direct bonding between exocyclic NH2 group and the substrate.71 Interestingly, flat orientation of adenine is observed on flat Au(111) and graphite surfaces, whereas non-planar deposition on Cu(110) could be ascribed to its corrugated surface and favorable interactions of exocyclic amino group with copper.
 | ||
| Fig. 3 (a) Ordered 2D structures of adenine on Au(111) as observed through STM; (b) island size of several tens of molecules. Two different structures exist with four [(a) and (c)] and two [(b) and (d)] molecules in the unit cell, respectively. The unit cell is indicated by the corresponding lattice vectors d1 and d2; (e) structure of adenine. (Reproduced with permission from ref. 69 © 2009 American Institute of Physics). | ||
Surface modification by thymine (T)
Physisorbed thymine at 1-octanol/graphite surface affords zig-zag chain-like structure, as viewed through STM, where each thymine is associated with two neighboring molecules, supported by hydrogen bonds.65 Interestingly, thymine on Au(111) substrate leads to randomly oriented 1D filaments, when viewed at low surface coverage, which gets morphed to 2D island-like growth patterns on increasing the coverage area, supported by weak van der Waals forces. The 2D islands could be reverted to 1D filaments through STM tip perturbation, suggesting that 2D islands were loosely held compared to more stable filaments (Fig. 4).72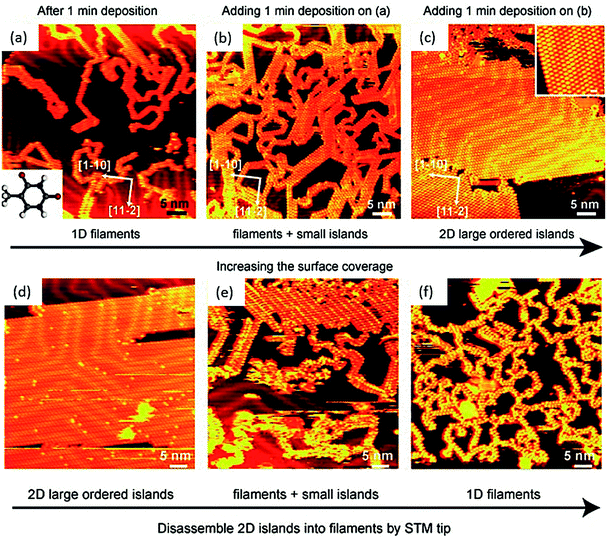 | ||
| Fig. 4 STM images of: (a) 1D thymine filaments on Au(111) surface at low surface coverage, inset: structure of thymine; (b) 1D filaments and small islands; (c) extended 2D islands. Inset: magnified STM image of 2D island structure. (d–f) Images of STM tip manipulation experiment. (Reproduced with permission from ref. 72 © 2007 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim). | ||
2D island formation by thymine was also observed on Cu(111) surface.73 On the contrary, surface patterning investigations on Cu(110) surface revealed parallel aligned 1D chains consisting of oval-shaped structures. These structures were found preferentially oriented along the [001] direction of the substrate. The role of weak dispersion forces in stabilizing thymine molecules was determined by DFT calculations. Desorption studies suggested possible interactions of copper with carbonyl groups as well as with deprotonated N3 of thymine.74 Such disassembly of self-organized molecular structures into higher-order superstructures might be helpful in deciphering the nature of intermolecular interactions.
Surface modification by guanine (G) and its analogs
High resolution studies of guanine self-organization have been carried out on surfaces such as HOPG, Cu(111), and Au(111).75 These studies have ranged from tunneling microscopy of simple guanine monolayers on HOPG76 to self-assembled N9-alkyl derivatives of guanine at 1,2,3-trichlorobenzene/HOPG interface. Self-assembled monolayers of single/dimeric molecules were obtained for C6–C10 alkyl guanines, while linear ribbon-like structures were observed when alkyl chains were longer than C12 carbons (Fig. 5). These guanine-mediated self-assembled supramolecular architectures could be used to create responsive nanopatterned surfaces, possessing desired motifs with organized electrically/optically active groups for applications in electronics and optical devices.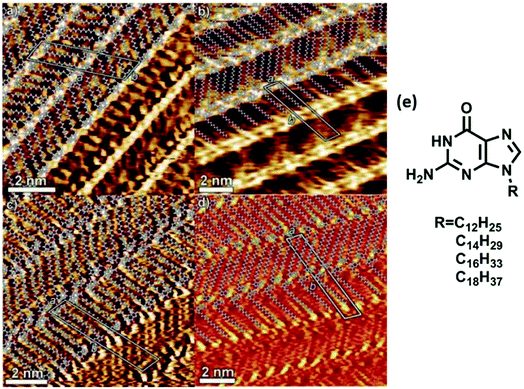 | ||
| Fig. 5 Hydrogen bonded ribbons obtained from (a) C12, (b) C14, (c) C16, and (d) C18 alkyl analogs of guanine by STM imaging; (e) structure of alkyl analogs of guanine. (Reproduced from ref. 77 with permission from The Royal Society of Chemistry). | ||
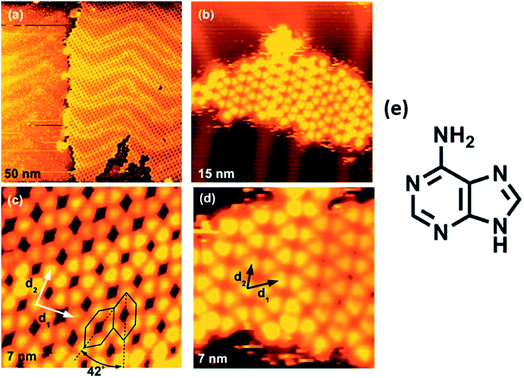
High resolution STM of guanine on Au(111) showed appearance of triangularly protruded 2D islands, which on magnification revealed a stable G-quartet like arrangement. Interestingly, an increase in the temperature resulted in appearance of anti-parallel molecular ribbon-like chains, without altering the local environment of guanine (Fig. 6).78 Guanine forms close-packed 2D square lattices on Cu(111) surface,73 but an ordered arrangement of guanine revealing 1D worm-like structures was also illustrated by Kawai et. al.79 Notably, guanine adsorption was also investigated on a less commonly used Ge(100) surface where detailed STM analyses confirmed that the adsorption occurred via “N(1)–H dissociation through an O dative bonded structure” mechanism.80
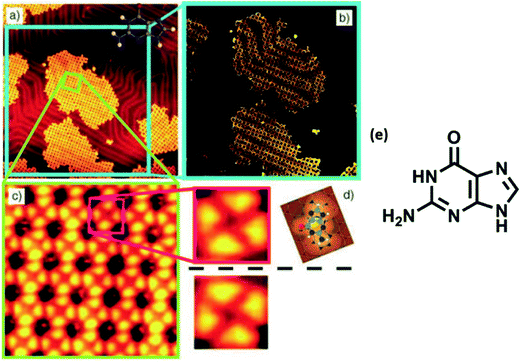 | ||
| Fig. 6 (a) STM image of self-assembled G islands on Au(111); (b) magnified image of (a); (c) STM image of network of guanine assembly; (d) magnified image of (c); (e) structure of guanine. (Reproduced with permission from ref. 78 © 2005 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim). | ||
Surface modification by cytosine (C)
There are limited studies pertaining to interaction of cytosine with surfaces. In one study, cytosine was used as a model nonsymmetrical molecule for deposition on Au(111) surface. STM studies revealed formation of a disordered cytosine network with the identification of three structural possibilities viz. zigzag filaments and five- and six-membered rings, supported by DFT calculations.81 In another example, deposition of cytosine on gold surfaces was used to assess the orientation of adsorption and ensuing electronic structure.82 It adsorbed in two different ways: the vacuum deposited sample displayed a flat state where interaction through N3 nitrogen dominated, whereas deposition from an aqueous solution involved interaction through N3 and exocyclic amino group. Such studies provide insights into interesting behavior of nucleobase interaction with surfaces and the possibility of studying medium-range order in amorphous and glassy systems.Co-adsorption of different nucleobases
Mixing of adenine and thymine followed by co-deposition led to 2D nanoassemblies comprised of reverse Hoogsteen A–T–A–T quartets, separated by homochiral rows of hydrogen bonded adenine dimers, at 1-octanol/HOPG interface. It was suggested that such structures might offer applications in biocompatible nanopatterned surfaces and for targeted drug delivery.65 Co-adsorption of guanine and cytosine also resulted in ordered assemblies at 1-octanol/graphite interface, where one of the phases consisted of hydrogen-bonded guanine-cytosine dimers (Fig. 7).83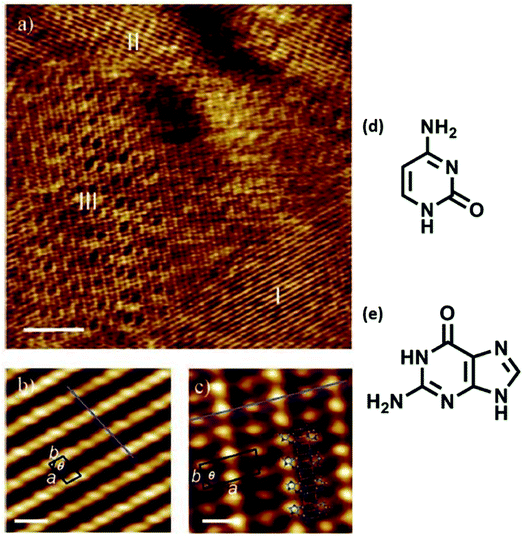 | ||
| Fig. 7 Co-adsorption of cytosine and guanine on HOPG. (a) STM image displaying three distinct domains (labeled I–III), scale bar: 10 nm; (b) zoomed-in view of domain I structure, scale bar: 1 nm; (c) magnified view of domain III revealing an ordered array of high and low protrusions, scale bar: 1 nm; (d) structure of cytosine; (e) structure of guanine. (Reproduced with permission from ref. 83 ©2006, American Chemical Society). | ||
A novel cyclic 2D supramolecular nanostructure was obtained by co-adsorption of guanine and uracil at 1-octanol/HOPG interface, which was different compared to homomolecular layers of guanine and uracil. These cyclic structures could be correlated to GU dimers, arranged in a parallel fashion, to reveal patterns of 1D chains in the translated surface monolayer (Fig. 8).84
 | ||
| Fig. 8 High-resolution STM images of (a) GU-base pairs; (b) large scale STM image; (c) molecular structure from ab initio calculations. GU-cyclic structures are represented by yellow ovals. (Reproduced with permission from ref. 84 © 2008, American Chemical Society). | ||
N-Aryl modified cytosine and guanine were co-deposited on Au(111) surface to afford stable patterns, supported by Watson–Crick hydrogen bonding and van der Waals interactions, which were also validated by DFT calculations.84 In another report, Besenbacher and coworkers elaborated structural details of cytosine deposition and its co-adsorption with adenine and guanine on Au(111) substrate. Cytosine alone gave rise to zig-zag ring-like arrangement, while with A or G on Au(111) surface revealed a significant shift from homomolecular structure. G + C admix afforded ring-like structures, while A + C admix formed large islands and zigzag chains on heating, perhaps suggesting a non-interactive nature of A and C combination (Fig. 9 and 10).85,86
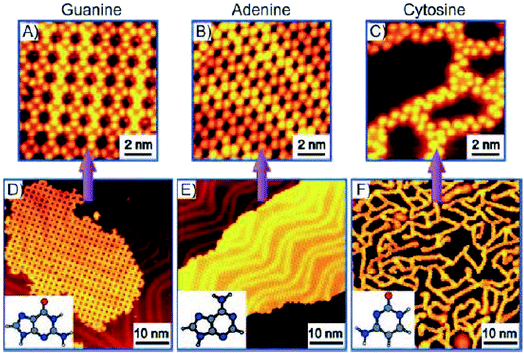 | ||
| Fig. 9 STM images of guanine (A and D) and adenine (B and E) on Au(111) displayed 2D island growth. However, cytosine showed (C and F) 1D filaments consisting of zig-zag branches and ring-like structures. (Reproduced with permission from ref. 86 © 2008 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim). | ||
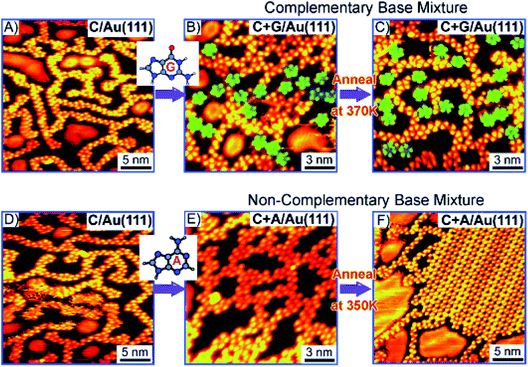 | ||
| Fig. 10 STM images: (A and D) Deposition of C at room temperature; (B and C) co-deposition of complementary C + G; and (E and F) non-complementary C + A bases; (B) deposition of guanine reveals increase in five-fold rings (indicated by the green shading). C + G and C + A mix on co-deposition (B and E). (Reproduced with permission from ref. 86 © 2008 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim). | ||
Surface modification by nucleobase functionalised molecules
Nucleobase terminated, rigid oligo(phenyleneethynylene) molecular rods were deposited on HOPG surface and studied with the help of tunneling microscopy. Adenine–adenine (A–A) dimer displayed a regular arrangement of ordered rectangular patterns, while thymine–thymine (T–T) dimers resulted in an intricate nanostructural pattern. However, an equimolar solution of the two dimers resulted in the structure same as that of homomeric adenine (A–A) system (Fig. 11).87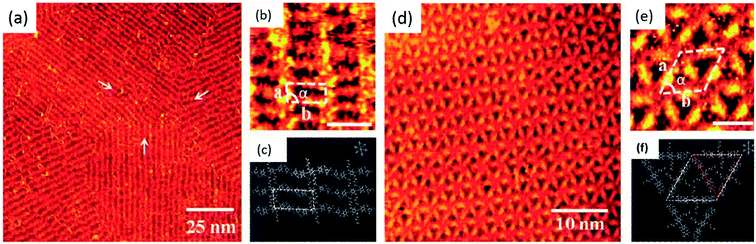 | ||
| Fig. 11 (a and b) STM images of A–A dimer on HOPG surface. Scale bar in (b) is 3 nm; (c) schematic representation of self-assembly; (d) STM images of T–T dimer on HOPG surface. Scale bar in (e) is 3 nm; (f) schematic representation of supramolecular networks. (Reproduced from ref. 87 with permission from The Royal Society of Chemistry). | ||
Chiral and achiral oligo-(p-phenylenevinylene) (OPV) derivatives, functionalized with a diaminotriazine fragment that has hydrogen-bonding sites complementary to thymine, were probed by STM on 1-octanol/HOPG interface, in the presence of added thymine. High resolution micrographs revealed that rosette-like structures of OPV derivatives transformed to supramolecular diastereomeric patterns due to achiral thymine moiety. This study brings a new dimension to tuning of surface chirality through directed chiral–achiral interactions.88
Champness and coworkers reported surface studies with a thymine-modified porphyrin (tetra-TP), which formed uniform self-assembled square lattices on HOPG substrate. Interestingly, a tetra-TP⋯9-propyladenine array was achieved by challenging tetra-TP with 9-propyladenine nucleobase. These studies confirmed concentration dependence and importance of molar ratio of individual components, indicating role of multiple factors in directing surface self-assembly (Fig. 12).89 Such functionalization is also helpful in crafting of well-tuned surface patterns by modifying the size and shape of supramolecular synthons.
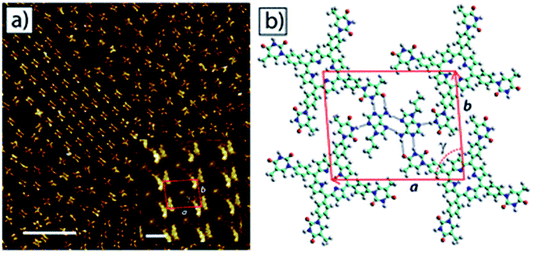 | ||
| Fig. 12 STM image of (a) self-assembled network of 9-propyladenine and tetra-TP on HOPG surface. Inset: high resolution image of the network. Scale bar = 20 nm, inset: 1.6 nm; (b) molecular model of tetra-TP network. (Reproduced from ref. 89 with permission from The Royal Society of Chemistry). | ||
Surface modification by nucleobase functionalised polymers
Long and coworkers reported copolymerisation of n-butylacrylate with novel adenine and thymine acrylate monomers, and the resulting adenine functionalized copolymer afforded formation of highly-ordered nanostructures due to facile stacking interactions in adenine.90 Notably, thymine did not exhibit special surface features. In a related study, synthesis of adenine/thymine functionalized acrylic ABA triblock copolymers was carried out via modified chain transfer polymerization method. This triblock copolymer self-organized to afford microphase-separated structures, supported by stacking and hydrogen bonding interactions, as observed by AFM studies.91Another report by Long and coworkers described synthesis of cytosine and ureido-cytosine functionalized diacrylic copolymers, where cytosine-modified copolymer revealed irregular shapes, while ureido-cytosine analog formed nanofibrillar structures. This self-organization behavior accounted for improved thermal and mechanical properties making them promising candidates for adhesives and thermoplastic elastomers (Fig. 13).92
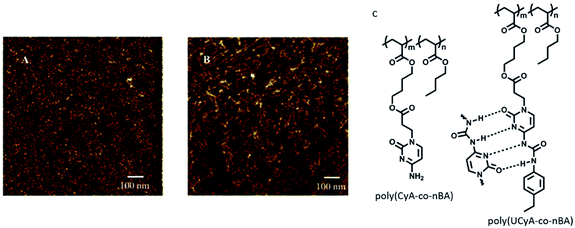 | ||
| Fig. 13 AFM phase images for (A) poly(CyA-co-nBA); (B) poly(UCyA-co-nBA) films; (C) hydrogen bonding interactions in polymers. (Reproduced from ref. 92 with permission from The Royal Society of Chemistry). | ||
Novel biocompatible nucleobase-grafted polycaprolactones consisting of adenine and uracil were prepared and their morphologies were analyzed using AFM and TEM. Both copolymers were studied separately and as a blend, to ascertain favorable adenine–uracil interactions. Nucleobase functionalization enabled transformation of star-like morphology of caprolactone to nanospherical shape of the blend, perhaps due to cooperative hydrogen bonding interactions.93
Investigation concerning self-assembly of guanine-/guanosine-capped oligo(p-phenylene-vinylene) oligomers revealed that oligomer formation and ensuing nanoassemblies were organized primarily through the formation of G-quartets, mediated by monovalent cations. These nanoassemblies were found refractory towards concentration and temperature modifications and stable towards disaggregation (Fig. 14).94
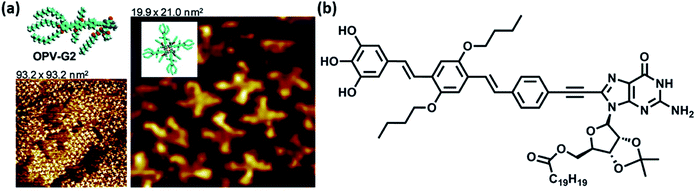 | ||
| Fig. 14 (a) High resolution STM images of OPV-G2 on HOPG surface. Inset: tentative models of various structural features in STM images; (b) structure of OPV-G2. (Reproduced with permission from ref. 94 © 2010, American Chemical Society). | ||
Co-adsorption of nucleobase and peptides
Amino acid sidechains in peptide also have the ability to interact with nucleobases through the Hoogsteen face. Stability of two dimensional structures obtained by co-adsorption of adenine with L-serine (L-Ser) and L-tyrosine (L-Tyr), revealed that adenine–adenine networks are energetically stronger compared to Ser- and Tyr-adenine interactions. This was ascertained by co-deposition of adenine with amino acids, which resulted in morphologies that could be ascribed to homoadenine networks suggesting preponderance of self-sorting was in favor of homoadenine system (Fig. 15).95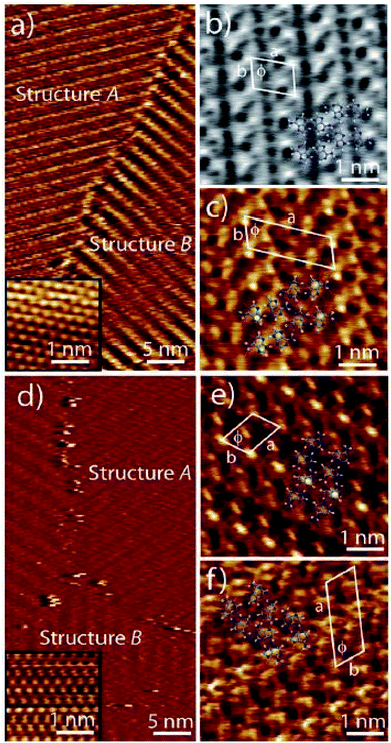 | ||
| Fig. 15 Co-adsorption of adenine with L-serine (a–c) and with L-tyrosine (d–f) revealing pure adenine domains by STM imaging. (Reproduced from ref. 95 with permission from The Royal Society of Chemistry). | ||
Sleiman and coworkers reported nucleobase–peptide amphiphiles based on dityrosine peptide and evaluated their solution-phase self-organization by microscopy methods. Growth of morphologies ranging from rod-like or helical micelles to the formation of nanoribbons was observed.96 The potential of peptide assembly and amphiphilic character led to large-scale scaffolds embellished with addressable nucleobases for recognition.
Liu and coworkers demonstrated chirality induction to achiral Thioflavin T (ThT) dye from self-assembled, fiber-like helical structures arising from the interaction of Fmoc-glutamic acid and achiral guanine or adenine nucleobases. Interestingly, ThT fluorescence changed from transparent to yellow and cyan, when added to gel fibers. This property was further confirmed by circular polarised fluorescence studies (Fig. 16).22
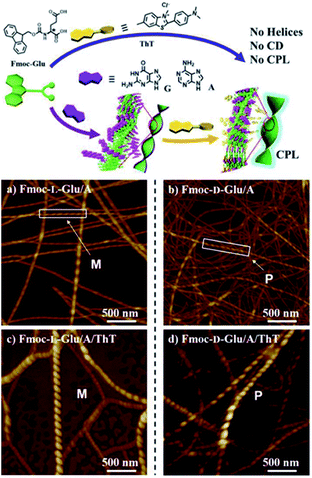 | ||
| Fig. 16 Helical supramolecular fibres depicted by AFM images of (a) Fmoc-L-Glu/adenine; (b) Fmoc-D-Glu/adenine; (c) Fmoc-L-Glu/adenine/ThT (d) Fmoc-D-Glu/adenine/ThT. (Reproduced from ref. 22 with permission from © 2016 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim). | ||
Surface modification by nucleobase-CNT hybrids
Carbon nanotubes (CNTs) are sparingly soluble in most solvents, which restricts their widespread use in certain applications. Covalent functionalization strategies are commonly used to improve upon physicochemical characteristics of CNTs, which includes functionalization at end tips and CNT sidewalls.97,98 Consequently, suitably altered functionalised CNTs are attractive candidates for biomedical applications as well as material development approaches.99–102 There are numerous examples CNTs functionalised with small organic molecules,103 polymers,104 peptides,105 peptide nucleic acids,106 nucleic acids107 and nucleobases.108–115Inherent hydrogen bond-mediated assembly of nucleobases provided the impetus to explore self-assembly of guanine-single walled CNT hybrids on HOPG surface. Covalently functionalised guanine–SWCNT (SWNT: single walled carbon nanotubes) 1 appeared as small bundles on TEM analysis, which in the presence of K+ ions (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), indirectly revealed a G-quartet type of self-assembled pattern on HOPG/mica surface (Fig. 17a), thereby aligning CNTs on surface.108
1), indirectly revealed a G-quartet type of self-assembled pattern on HOPG/mica surface (Fig. 17a), thereby aligning CNTs on surface.108
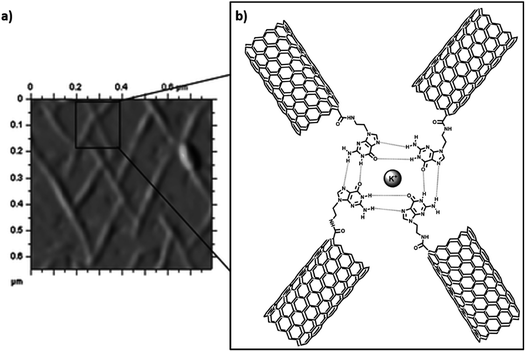 | ||
| Fig. 17 (a) AFM image of Guanine-SWCNT 1 with potassium ions on mica surface; (b) proposed model CNT alignment. (Reproduced with permission from ref. 108 The Royal Society of Chemistry). | ||
In another example, self-organisation of adenine-SWCNT -triethyleneglycol (TEG) linker and its interaction with silver ions was studied on HOPG with the help of AFM and TEM. Adenine-SWCNT 3 showed horizontally aligned nanotube on HOPG surface (Fig. 18b), which was ascribed to parallel-oriented assembly. On the contrary, AFM images of adenine-SWCNT 2, without TEG chain, revealed the occurrence of a fibrous morphology, primarily supported by van der Waals interactions. On interaction with silver ions, hybrid adenine-SWCNT 2 resulted in deposition of silver nanoparticles over nanotube surface (Fig. 18c).109 It was surmised that such nucleobase-carbon nanotubes hybrids, along with metal coordination, could provide a new platform for biosensors and heterogeneous catalysis.
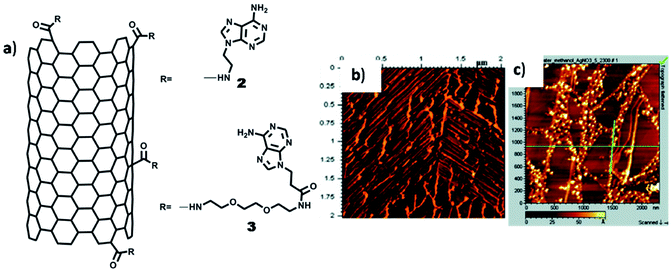 | ||
| Fig. 18 (a) Chemical structures of adenine-SWCNTs; (b) AFM image of adenine-SWCNT 3 showing alignment of nanotubes on HOPG surface; (c) AFM image of Ag(I) complex of adenine-SWCNT 2. (Reproduced with permission from ref. 109 © 2009 American Chemical Society). | ||
Silver coated adenine-CNT hybrids were employed as heterogeneous catalyst for the conversion of 2-methylhydroquinone to 2-methylbenzoquinone. Three multiwalled CNT (MWCNT) derivatives (4–6) were prepared (Fig. 19a), followed by their interaction with silver ions as judged by TEM analysis (Fig. 19b–d). Catalytic activity of 4/Ag hybrid was demonstrated by achieving oxidation of 2-methylhydroquinone to 2-methylbenzoquinone, with successful catalyst recycling without appreciable loss of catalytic activity (Fig. 19e).110
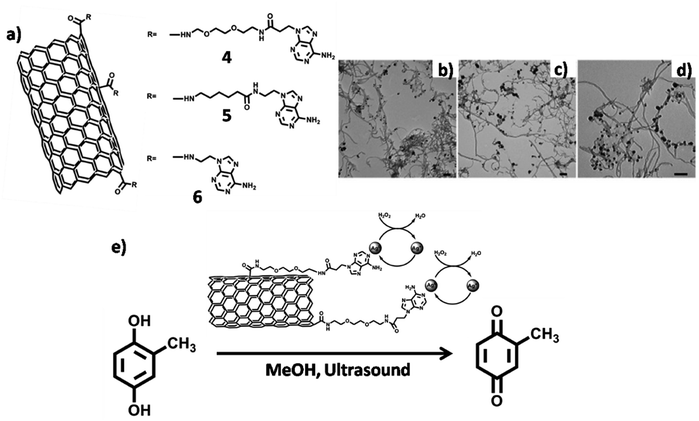 | ||
| Fig. 19 (a) Chemical structures of adenine-MWCNTs 4–6. TEM micrographs of adenine-MWCNT hybrids (b) 4 (c) 5 and (d) 6 complexed with silver ions. Scale bars = 100 nm; (e) silver complexed adenine-MWCNT 4 hybrid in catalytic conversion. (Reproduced with permission from ref. 110 © 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim). | ||
Self-organisation of covalently functionalized uracil-SWCNT 7 on freshly cleaved HOPG surface afforded formation of nanoring structures (cross-section: ∼50–70 nm; height: ∼20–25 nm), as observed by AFM analysis (Fig. 20b). Nanoring formation could be ascribed to surface adsorption of SWCNT, followed by hydrogen bonding between uracil–uracil base pair. Notably, uracil-SWCNT 8, devoid of the TEG chain, afforded fibrils or tubes on HOPG surface.111
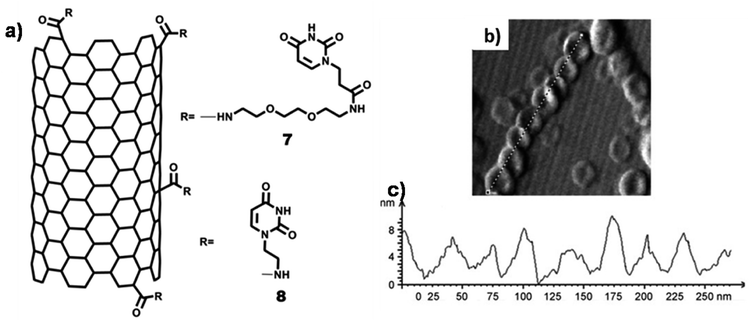 | ||
| Fig. 20 (a) Chemical structures of uracil-SWCNTs 7 and uracil-SWCNT 8; (b) AFM image of uracil-SWCNT 7 on HOPG showing nanorings forming helix-type structure; (c) height–diameter profile of nanorings. (Reproduced with permission from ref. 111 © 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim). | ||
H-bond driven self-assembly of thymine-functionalized CNTs in organic solvents has also been investigated. TEM analysis revealed dispersion of thymine-CNTs in dimethyl formamide (DMF), a solvent that could interact with nucleobase moiety (Fig. 21c) and dichloromethane (DCM), a non-competitive solvent, affording formation of diverse superstructures directed by thymine–thymine interaction (Fig. 21d).114
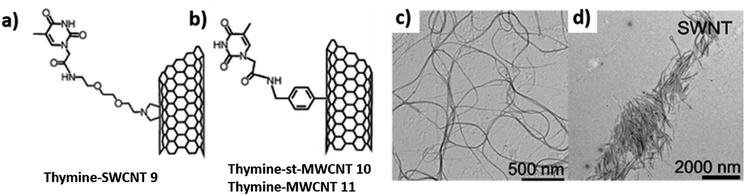 | ||
| Fig. 21 Chemical structures of thymine-functionalised CNTs formed by: (a) 1,3-dipolar cycloaddition (b) arylation reaction. TEM images (c) 9 in DMF (d) 9 in DCM. (Reproduced with permission from ref. 114 The Royal Society of Chemistry). | ||
Selective deposition of thymine-functionalized MWCNTs was achieved on patterned polystyrene matrix using a combination of “top-down” as well as “bottom-up” approaches. Modified MWCNTs were able to interact with the polystyrene matrix having 2,6-di(acetylamino)-4-pyridyl moieties, through complementary three-point hydrogen bonding, as studied by various microscopy techniques (Fig. 22b–g).115
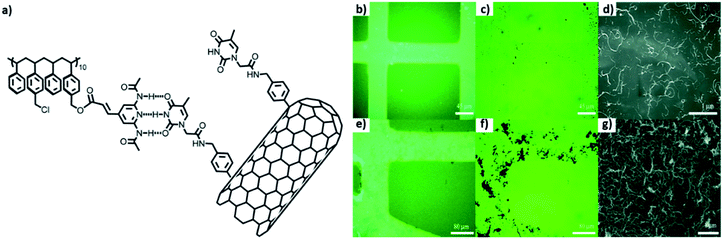 | ||
| Fig. 22 (a) Chemical structure of PS1, thymine-MWCNTs; images of patterned surface by (b and e) fluorescence, (c and f) optical and (d and g) scanning electron microscopy, after deposition of a dispersion of Thymine-MWCNTs in DMF (b–d) and CHCl3 (e–g).(Reproduced with permission from ref. 115 © 2012 American Chemical Society). | ||
Surface modification by metal–nucleobase complexes
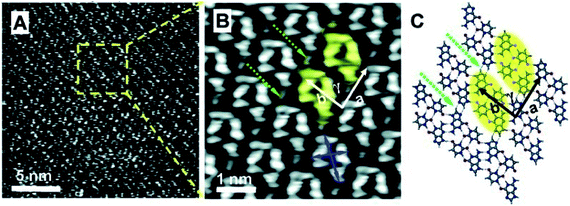
Predisposed metal coordination sites present of nucleobases could be used to construct tailored metal–nucleobase frameworks for designed applications.116 For example, adenine interacts efficiently with metal ions to create a vast number of coordination polymers in the solid state, with a distinct possibility of surface patterning,117–120 in addition to such custom surfaces being amenable to further processing.121–123In one example, 9-allyladenine afforded silver-mediated metallamacrocyclic quartet 1, where all the imino nitrogens of adenine coordinated to silver ions. Complex 1 was transferred on HOPG surface and it showed a well-ordered arrangement consisting of a repetitive zig-zag assembly (Fig. 23b), which resembled the crystallographic signature in the crystal lattice.117
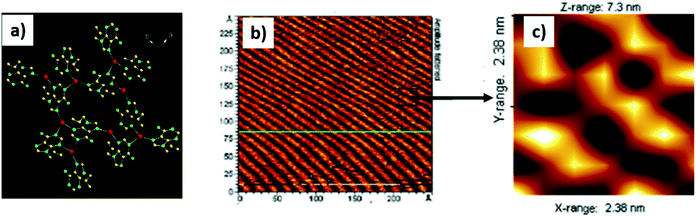 | ||
| Fig. 23 (a) Adenine metallamacrocycle 1. (b) AFM image of complex 1 on HOPG surface; (c) magnified image of (b) showing zig-zag assembly. (Reproduced with permission from ref. 117 © 2006 American Chemical Society). | ||
Similarly, crystals of an adenine-silver helicate (2), obtained by the interaction of 9-allyladenine with ammoniacal silver nitrate, were deposited on HOPG surface. Non-contact mode AFM images revealed persistent silver–adenine interaction via pattered deposition that closely resembled crystal structure of 2 (Fig. 24a–b).118 Such patterning was not formed when 2 was deposited on Si(100) wafer or mica, suggesting the importance of favourable surface properties in stabilizing patterns.
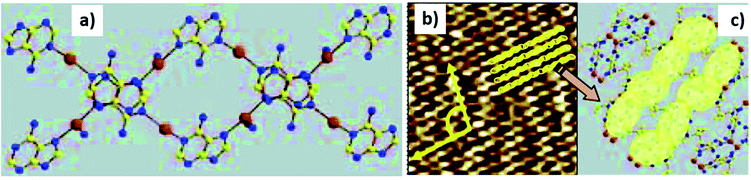 | ||
| Fig. 24 (a) Double-stranded coordination helicate 2. (b) AFM image of 2 on HOPG surface. Image size is 400 × 400 Å2; (c) proposed model of adenine-silver helicate deposition. (Reproduced with permission from ref. 118 © 2007 American Chemical Society). | ||
A C3 symmetric hexaadenine ligand interacted with AgClO4 to form silver complex 3, which showed energetically stable and high abundant 1D needle-like features over HOPG surface (Fig. 25a). A strong interaction between ligand and metal ions was evident from the chain growth over surface kinks and monoatomic steps. Notably, 2D growth was observed with increase in the thickness of monolayer (Fig. 25b and c), where formation of the proposed adlayer was assisted by bridging counterions (Fig. 25d).120
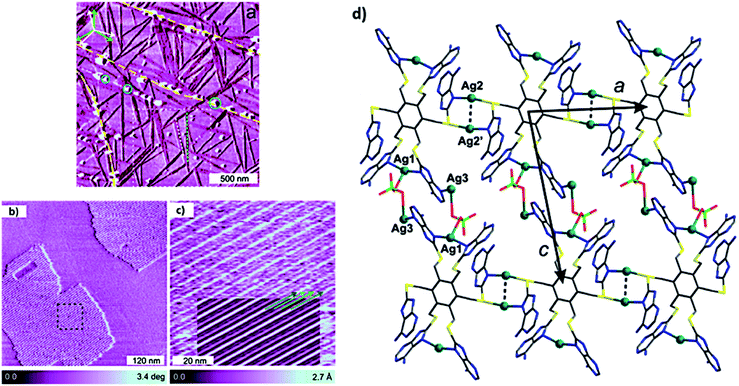 | ||
| Fig. 25 (a) AFM phase image of ultrathin film of hexapodal silver complex 3 on HOPG surface. Graphite multiatomic terrace edges are shown in yellow dashed lines and with growth direction of rotational and mirror domains of 1D chains; (b) AFM constant height phase image of 2D domains; (c) high-resolution AFM topography image of black square region in (b); (d) plane containing Ag-coordinated polymer. (Reproduced with permission from ref. 120 © 2017 American Chemical Society). | ||
Additional metal coordinating sites in adenine was orchestrated by introducing a thiol substitution at the C8 position. Consequently, 8-mercapto-N9-propyladenine afforded a highly porous copper adenine framework upon interaction with cuprous iodide. Notably, time-dependent solution phase growth studies revealed conversion of seed-like particles of complex 4 at 0 h to a cuboidal morphology after 48 h, finally culminating into homogeneous cuboidal morphology after 30 days (Fig. 26a–f).124
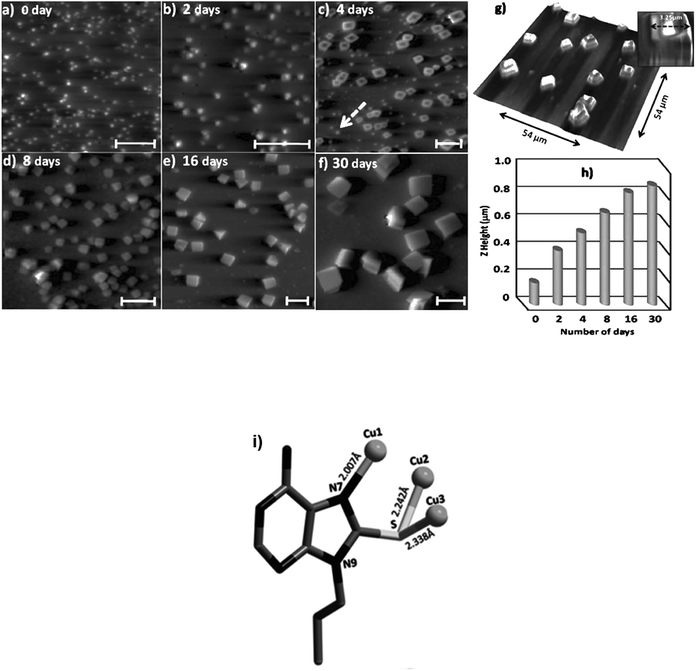 | ||
| Fig. 26 (a–f) 2D AFM image of time-dependent solution phase growth of copper complex on glass surface. Scale bars: 5 μm; (g) 3D AFM image of cuboidal aggregate; (h) periodic increase in Z-height over 0 to 30 days; (i) Crystal structure of copper complex 4 (Reproduced with permission from ref. 124 © 2014 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim). | ||
The effect of substitution at N9 position was investigated in 1,N6-ethenoadenine derivatives where N9 icosyl substitution afforded a well-ordered, uniform lamellar structure on HOPG surface, with alternate bright and dark stripes, where the dark ones represented alkyl chains and the bright ones the aromatic moieties. Notably, Ag(I) complexation with this ligand demonstrated a similar lamellar pattern, albeit with a slight difference in observed patterns.125
Samori and coworkers synthesized a platinum–isocytosine complex 5, followed by its surface study on 1-phenyloctane/HOPG interface. Interestingly, polygonal discrete cyclic motifs were observed, which were stabilized by hydrogen bonding between the isocytosine groups. Notably, formation of trimers, tetramers, pentamers, hexamers and half-hexamers were also surmised as explained by theoretical calculations (Fig. 27).126
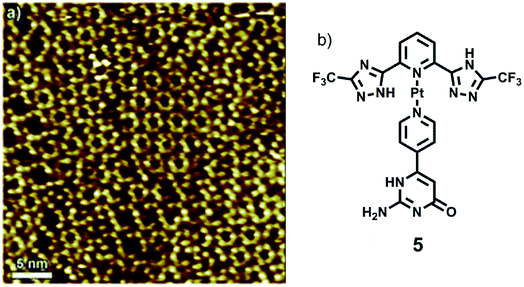 | ||
| Fig. 27 (a) STM image of Pt-complex 5 on HOPG; (b) structure of complex 5. (Reproduced from ref. 126 with permission from The Royal Society of Chemistry). | ||
Zamora and coworkers synthesized 6-mercaptopurine and complexed it with Cd(II) salt to obtain a discrete cadmium complex 6 or a polymeric structure 7, along with the formation of several interconvertible intermediates. AFM was used to study these reactions on HOPG surface, including an in situ deprotonation event, which was analyzed on the mica surface (Fig. 28).127
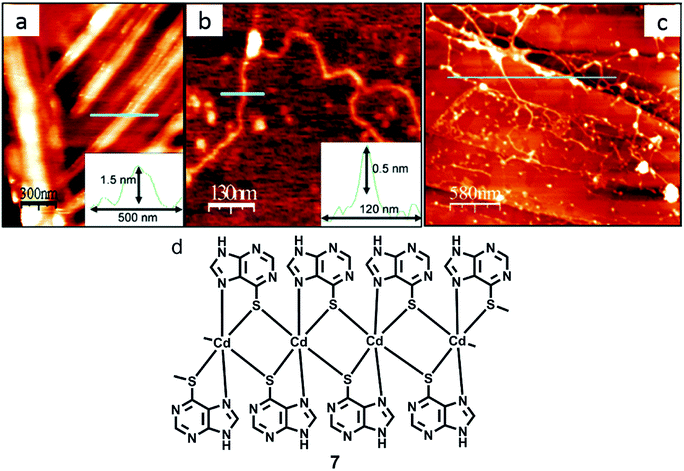 | ||
| Fig. 28 (a) AFM image of crystals acquired by treating 7 with NaOH; (b) AFM image of a single polyanion; (c) AFM micrograph of fibers acquired by in situ reaction on HOPG; (d) structure of 7. (Reproduced with permission from ref. 127 © 2006, American Chemical Society). | ||
In another report, Amo-Ochoa and coworkers reported reaction of thymine-1-acetic acid, copper nitrate and 4,4′-bipyridine in aqueous medium, resulting in the formation of coordination polymer 8 as crystalline nanoribbons. The latter exhibited selectivity for single-stranded adenine-based oligonucleotides on the basis of adenine–thymine interactions. Remarkably, these nanoassemblies show low cytotoxicity and exhibited potential as oligonucleotide nanocarriers for cellular delivery (Fig. 29).128
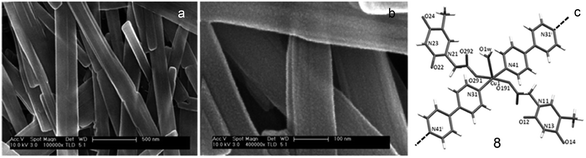 | ||
| Fig. 29 (a) SEM image of nanoribbons of 8; (b) enlarged view of (a); (c) structure of 8. (Reproduced from ref. 128 with permission from © 2017 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim). | ||
Although it is impossible to review the entire literature available in this area, we however present an overview to reiterate the potential of nucleobases and their conjugates in achieving desired metal ion interaction and their natural ability to interact with surfaces to afford facile patterning for advanced applications. The following table gives a snap-shot of model compounds, the preferred substrate for interaction studies and type of topography obtained subsequent to surface adsorption (Table 1).
| S. no. | Compound | Substrate | Topography | Ref. |
|---|---|---|---|---|
| Adenine (A) and its analogs | ||||
| 1 | 9-Icosyladenine | HOPG | Two different phases: α- and β-phase | 62 |
| 2 | Adenine-C22 | HOPG | Two different phases: α- and β-phase | 63 |
| 3 | Adenine | HOPG | 2D networks of adenine dimers | 65 |
| 4 | Adenine | Au(111) | Two different types of islands with four and two molecules per unit cell | 69 |
| 5 | Adenine | Cu(110) | 1D dimeric chains | 71 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Thymine (T) | ||||
| 6 | Thymine | HOPG | Zig-zag chain like structure | 65 |
| 7 | Thymine | Au(111) | Randomly oriented 1D filaments (low coverage) and 2D island-like growth patterns (high coverage) | 72 |
| 8 | Thymine | Cu(111) | 2D island formation | 73 |
| 9 | Thymine | Cu(110) | Parallel aligned 1D chains consisting of oval-shaped structures | 74 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Guanine (G) and its analogs | ||||
| 10 | Guanine | HOPG | Monolayers | 76 |
| 11 | C6–C10 alkyl guanines | HOPG | Self-assembled monolayers of single/dimeric molecules | 77 |
| 12 | C12 or higher alkyl guanines | HOPG | Linear ribbon-like structures | 77 |
| 13 | Guanine | Au(111) | 2D islands with G-quartet like arrangement | 78 |
| 14 | Guanine | Cu(111) | Close-packed 2D square lattices | 73 |
| 15 | Guanine | Au(111) | Self-assembled guanine islands | 78 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Cytosine (C) | ||||
| 16 | Cytosine | Au(111) | Zigzag filaments and five- and six-membered rings | 81 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Co-adsorption of different nucleobases | ||||
| 17 | A + T | HOPG | Well-ordered 2D supramolecular nanoassemblies | 65 |
| 18 | G + C | HOPG | Ordered nanoassemblies with three distinct domains | 83 |
| 19 | G + U | HOPG | 2D cyclic supramolecular nanostructure | 84 |
| 20 | N-aryl modified cytosine and guanine | Au(111) | Porous supramolecular assembly | 84 |
| 21 | C + G | Au(111) | Dense ring-like structures | 86 |
| 22 | C + A | Au(111) | Large islands and zigzag chains | 86 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Nucleobase functionalised molecules | ||||
| 23 | A–A dimer functionalized by oligo(phenyleneethynylene) | HOPG | Ordered rectangular pattern | 87 |
| 24 | T–T dimer functionalized by oligo(phenyleneethynylene) | HOPG | Sparse nanostructural pattern | 87 |
| 25 | Mixture of functionalized A–A and T–T dimers | HOPG | Ordered rectangular pattern | 87 |
| 26 | 9-Propyladenine and tetra-TP | HOPG | Self-assembled network | 89 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Nucleobase functionalised polymers | ||||
| 27 | OPV-G2 | HOPG | Nanoassemblies | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Nucleobase and peptides | ||||
| 28 | adenine + L-Ser | HOPG | Homoadenine networks | 95 |
| 29 | adenine + L-Tyr | HOPG | Homoadenine networks | 95 |
| 30 | Fmoc-L-Glu/adenine | Mica | Fibre-like helical supramolecular structures | 22 |
| 31 | Fmoc-D-Glu/adenine | Mica | Fibre-like helical supramolecular structures | 22 |
| 32 | Fmoc-L-Glu/adenine/ThT | Mica | Fmoc-adenine served as a matrix to transfer chirality to ThT | 22 |
| 33 | Fmoc-D-Glu/adenine/ThT | Mica | Fmoc-adenine served as a matrix to transfer chirality to ThT | 22 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Nucleobase-CNT hybrids | ||||
| 34 | Guanine–SWCNT1 | HOPG/mica | G-quartet type of self-assembled pattern | 108 |
| 35 | Adenine-SWCNT 3 | HOPG | Horizontal alignment of nanotubes | 109 |
| 36 | Silver(I) complexed with adenine-MWCNTs 4, 5 and 6 | TEM grid | Uniform distribution of silver, Ag(I) and Ag(II) ions over nanotubes | 110 |
| 37 | Uracil-SWCNT7 | HOPG | Nanorings forming helix type structure | 111 |
| 38 | Thymine-SWCNT 9 | TEM grid | Formation of superstructure | 113 |
| 39 | PS1,Thymine-MWCNT | Glass/Si | Selective deposition of Thymine-MWCNTs on patterned polystyrene matrix | 114 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Metal–nucleobase complexes | ||||
| 40 | Complex1 | HOPG | Repetitive zig-zag assembly resembling crystal structure | 116 |
| 41 | Complex 2 | HOPG | Patterned deposition resembling crystal structure | 117 |
| 42 | Complex 3 | HOPG | Long and thin 1D needle like features | 119 |
| 43 | MOF 4 | Glass | Homogeneously-sized nanocuboidal morphology after 30 days | 123 |
| 44 | Complex 5 | HOPG | Polygonal discrete cyclic motifs | 125 |
| 45 | Complex 7 | HOPG | Fibre-like structures | |
| 46 | Coordination polymer 8 | SiO2 | Uniform crystalline nanoribbons | 127 |
Conclusions
Surface modifications achieved by heterocyclic natural nucleobases, nucleobase-functionalised carbon nanotubes (CNTs) and metal–nucleobase coordination polymers, are promising candidates for novel materials and for transferring interesting properties onto custom surfaces.44 Multicomponent discrete complexes, mixtures and nucleobase-terminated entities further enrich the repertoire and enhance the possibilities of using these systems, consisting of naturally evolved hydrogen bonding sites, for constructing hierarchically complex supramolecular networks and nanoarchitectures. Additionally, immobilization of nucleobases and their derivatives (including metal adducts) on (conducting) surfaces also presents interesting paradigm for fast expanding biochip technology efforts, necessary for rapid identification and unequivocal detection of important analytes. It could be envisaged that several other areas of custom surface design and applied platforms for detection and device fabrication will also benefit from the continued progress with nucleobase assemblies and transfer of their properties to custom surfaces.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge CSIR for SPM fellowship (RKS), MHRD for pre-doctoral fellowship (IA) and Center for Nanoscience, IIT Kanpur, for support (RKP). SV thanks DST for J. C. Bose National Fellowship and the ongoing work in his laboratory is also supported by DST Nano Mission.References
- M. Moradi, L. G. Tulli, J. Nowakowski, M. Baljozovic, T. A. Jung and P. Shahgaldian, Angew. Chem., Int. Ed., 2017, 56, 14395–14399 CrossRef PubMed.
- J. Lee, A. J. Kalin, T. Yuan, M. Al-Hashimi and L. Fang, Chem. Sci., 2017, 8, 2503–2521 RSC.
- T. Nijs, F. J. Malzner, S. Fatayer, A. Wäckerlin, S. Nowakowska, E. C. Constable, C. E. Housecroft and T. A. Jung, Chem. Commun., 2015, 51, 12297–12300 RSC.
- J. R. Eskelsen, K. J. Phillips, K. W. Hipps and U. Mazur, Chem. Commun., 2015, 51, 10–13 RSC.
- X.-Y. Zhu, B. Tu, G.-J. Hu, Q.-J. Fang, J.-J. Qi, X.-W. Xiao, Y.-F. Geng and Q.-D. Zeng, Phys. Chem. Chem. Phys., 2018, 20, 6383–6389 RSC.
- Y. Yu, J. Lin and S. Lei, RSC Adv., 2017, 7, 11496–11502 RSC.
- A. Eberle, A. Nosek, J. Büttner, T. Markert and F. Trixler, CrystEngComm, 2017, 19, 1417–1426 RSC.
- R. Raval, Faraday Discuss., 2017, 204, 9–33 RSC.
- A. Della Pia, D. Luo, R. Blackwell, G. Costantini and N. Martsinovich, Faraday Discuss., 2017, 204, 191–213 RSC.
- H. Yang, L. Huang, K. Sun, K. Niu, Z. Cui, H. Zhang, Z. Wang, D. Yan and L. Chi, J. Phys. Chem. C, 2017, 121, 25043–25051 CrossRef.
- Z. Li, H. Van Gorp, P. Walke, T. H. Phan, Y. Fujita, J. Greenwood, O. Ivasenko, K. Tahara, Y. Tobe, H. Uji-i, S. F. L. Mertens and S. De Feyter, Nanoscale, 2017, 9, 5188–5193 RSC.
- H. Cao, A. Minoia, I. De Cat, J. Seibel, D. Waghray, Z. Li, D. Cornil, K. S. Mali, R. Lazzaroni, W. Dehaen and S. De Feyter, Nanoscale, 2017, 9, 18075–18080 RSC.
- L. Zhang, J. Li, S. Qiu, X. Huang and Z. Zeng, New J. Chem., 2017, 41, 3260–3264 RSC.
- C. Guo, J. D. Xue, L. X. Cheng, R. C. Liu, S. Z. Kang, Q. D. Zeng and M. Li, Phys. Chem. Chem. Phys., 2017, 19, 16213–16218 RSC.
- Y. Hu, K. Miao, L. Xu, B. Zha, M. Long, X. Miao and W. Deng, Phys. Chem. Chem. Phys., 2017, 19, 19205–19216 RSC.
- M. Dong, K. Miao, Y. Hu, J. Wu, J. Li, P. Pang, X. Miao and W. Deng, Phys. Chem. Chem. Phys., 2017, 19, 31113–31120 RSC.
- J. Dash, A. J. Patil, R. N. Das, F. L. Dowdall and S. Mann, Soft Matter, 2011, 7, 8120 RSC.
- X. Li, G. Wang, X. Ding, Y. Chen, Y. Gou and Y. Lu, Phys. Chem. Chem. Phys., 2013, 15, 12800 RSC.
- J. T. Davis and G. P. Spada, Chem. Soc. Rev., 2007, 36, 296–313 RSC.
- M. El Garah, R. C. Perone, A. S. Bonilla, S. Haar, M. Campitiello, R. Gutierrez, G. Cuniberti, S. Masiero, A. Ciesielski and P. Samorì, Chem. Commun., 2015, 51, 11677–11680 RSC.
- C. Xing, H. Yuan, S. Xu, H. An, R. Niu and Y. Zhan, ACS Appl. Mater. Interfaces, 2014, 6, 9601–9607 CrossRef PubMed.
- M. Deng, L. Zhang, Y. Jiang and M. Liu, Angew. Chem., Int. Ed., 2016, 55, 15062–15066 CrossRef PubMed.
- H. Ozawa, N. Katori, T. Kita, S. Oka and M. A. Haga, Langmuir, 2017, 33, 11901–11910 CrossRef PubMed.
- S. Zhang, Y. Geng, Y. Fan, W. Duan, K. Deng, D. Zhao and Q. Zeng, Phys. Chem. Chem. Phys., 2017, 19, 31284–31289 RSC.
- G. Nandi, B. Chilukuri, K. W. Hipps and U. Mazur, Phys. Chem. Chem. Phys., 2016, 18, 20819–20820 RSC.
- M. Kohmoto, H. Ozawa, L. Yang, T. Hagio, M. Matsunaga and M. A. Haga, Langmuir, 2016, 32, 4141–4152 CrossRef PubMed.
- M. E. Garah, A. S. Bonilla, A. Ciesielski, A. Gualandi, L. Mengozzi, A. Fiorani, M. Iurlo, M. Marcaccio, R. Gutierrez, S. Rapino, M. Calvaresi, F. Zerbetto, G. Cuniberti, P. G. Cozzi, F. Paolucci and P. Samorì, Nanoscale, 2016, 8, 13678–13686 RSC.
- E. Moreno Pineda, T. Komeda, K. Katoh, M. Yamashita and M. Ruben, Dalton Trans., 2016, 45, 18417–18433 RSC.
- J. Teyssandier, S. De Feyter and K. S. Mali, Chem. Commun., 2016, 52, 11465–11487 RSC.
- S. B. Kim, R. D. Pike, J. S. D'Acchioli, B. J. Walder, G. B. Carpenter and D. A. Sweigart, Angew. Chem., Int. Ed., 2009, 48, 1762–1765 CrossRef PubMed.
- F. Zamora, M. Pilar Amo-Ochoa, P. J. Sanz Miguel and O. Castillo, Inorg. Chim. Acta, 2009, 362, 691–706 CrossRef.
- Z. Xiang, D. Cao and L. Dai, Polym. Chem., 2015, 6, 1896–1911 RSC.
- Z. Li and H. C. Zeng, J. Am. Chem. Soc., 2014, 136, 5631–5639 CrossRef PubMed.
- D. Cui, J. M. MacLeod, M. Ebrahimi and F. Rosei, CrystEngComm, 2017, 19, 4927–4932 RSC.
- R. Wagia, I. Strashnov, M. W. Anderson and M. P. Attfield, Cryst. Growth Des., 2018, 18, 695–700 CrossRef.
- C. Hermosa, B. R. Horrocks, J. I. Martínez, F. Liscio, J. Gómez-Herrero and F. Zamora, Chem. Sci., 2015, 6, 2553–2558 RSC.
- A. Dazzi and C. B. Prater, Chem. Rev., 2017, 117, 5146–5173 CrossRef PubMed.
- R. Sakamoto, K. Takada, T. Pal, H. Maeda, T. Kambe and H. Nishihara, Chem. Commun., 2017, 53, 5781–5801 RSC.
- L. Yu, Z.-B. Li and D. Wang, Chem. Commun., 2016, 52, 13771–13774 RSC.
- Y. Hu, N. Goodeal, Y. Chen, A. M. Ganose, R. G. Palgrave, H. Bronstein and M. O. Blunt, Chem. Commun., 2016, 52, 9941–9944 RSC.
- J.-Y. Yue, X.-H. Liu, B. Sun and D. Wang, Chem. Commun., 2015, 51, 14318–14321 RSC.
- M. El Garah, A. Ciesielski, N. Marets, V. Bulach, M. W. Hosseini and P. Samorì, Chem. Commun., 2014, 50, 12250–12253 RSC.
- Y. Sun, Z. Hu, D. Zhao and K. Zeng, ACS Appl. Mater. Interfaces, 2017, 9, 32202–32210 CrossRef PubMed.
- A. Ciesielski, M. El Garah, S. Masiero and P. Samorì, Small, 2016, 12, 83–95 CrossRef PubMed.
- F. Pu, J. Ren and X. Qu, Chem. Soc. Rev., 2018, 47, 1285–1306 RSC.
- K. Zhang, G. B. Fahs, M. Aiba, R. B. Moore and T. E. Long, Chem. Commun., 2014, 50, 9145–9148 RSC.
- G. Beobide, O. Castillo, A. Luque and S. P. Yáñez, CrystEngComm, 2015, 17, 3051–3059 RSC.
- R. B. Zerdan, P. Cohn, E. Puodziukynaite, M. B. Baker, M. Voisin, C. Sarun and R. K. Castellano, J. Org. Chem., 2015, 80, 1828–1840 CrossRef PubMed.
- Z. Hua, R. Keogh, Z. Li, T. R. Wilks, G. Chen and R. K. O'Reilly, Macromolecules, 2017, 50, 3662–3670 CrossRef PubMed.
- Q. Lin, X. Zou, G. Zhou, R. Liu, J. Wu, J. Li and W. Duan, Phys. Chem. Chem. Phys., 2011, 13, 12225–12230 RSC.
- M. Sharma, A. Kumarb and P. K. Ahluwalia, RSC Adv., 2016, 6, 60223–60230 RSC.
- J. A. A. W. Elemans, S. Lei and S. D. Feyter, Angew. Chem., Int. Ed., 2009, 48, 7298–7332 CrossRef PubMed.
- D. R. San-Miguel, P. Amo-Ochoaa and F. Zamora, Chem. Commun., 2016, 52, 4113–4127 RSC.
- C. R. Pfeiffer, N. Pearce and N. R. Champness, Chem. Commun., 2016, 52, 11528–11539 Search PubMed.
- J. A. Wytko, R. Ruppert, C. Jeandon and J. Weiss, Chem. Commun., 2018, 54, 1550–1558 RSC.
- K. S. Mali, N. Pearce, S. De Feyter and N. R. Champness, Chem. Soc. Rev., 2017, 46, 2520–2542 RSC.
- R. J. Holmberg and M. Murugesu, J. Mater. Chem. C, 2015, 3, 11986–11998 RSC.
- L. S. Vargas, E. Kim and A. J. Attias, Mater. Horiz., 2017, 4, 570–583 RSC.
- L. P. Xu, Y. Liu and X. Zhang, Nanoscale, 2011, 3, 4901–4915 RSC.
- X. Zhang, Q. Zeng and C. Wang, Nanoscale, 2013, 5, 8269–8287 RSC.
- J. L. Zhang, S. Zhong, J. Q. Zhong, T. C. Niu, W. P. Hu, A. T. S. Weea and W. Chen, Nanoscale, 2015, 7, 4306–4324 RSC.
- Z. Mu, O. Rubner, M. Bamler, T. Blömker, G. Kehr, G. Erker, A. Heuer, H. Fuchs and L. Chi, Langmuir, 2013, 29, 10737–10743 CrossRef PubMed.
- C. Wang, P. K. Jana, H. Zhang, Z. Mu, G. Kehr, T. Blömker, G. Erker, H. Fuchs, A. Heuer and L. Chi, Chem. Commun., 2014, 50, 9192–9195 RSC.
- M. J. Allen, M. Balooch, S. Subbiah, R. J. Tench, R. Balhorn and W. Siekhaus, Ultramicroscopy, 1992, 42, 1049–1053 CrossRef.
- W. Mamdouh, M. Dong, S. Xu, E. Rauls and F. Besenbacher, J. Am. Chem. Soc., 2006, 128, 13305–13311 CrossRef PubMed.
- J. Freund, M. Edelwirth, P. Kröbel and W. Heckl, Phys. Rev. B: Condens. Matter Mater. Phys., 1997, 55, 5394–5397 CrossRef.
- W. Mamdouh, D. Mingdong, R. E. A. Kelly, L. N. Kantorovich and F. Besenbacher, J. Phys. Chem. B, 2007, 111, 12048–12052 CrossRef PubMed.
- N. J. Tao and Z. Shi, J. Phys. Chem., 1994, 98, 1464–1471 CrossRef.
- M. Lukas, R. E. A. Kelly, L. N. Kantorovich, R. Otero, W. Xu, E. Laegsgaard, I. Stensgaard and F. Besenbacher, J. Chem. Phys., 2009, 130, 0247051–0247059 CrossRef PubMed.
- R. E. A. Kelly, W. Xu, M. Lukas, R. Otero, M. Mura, Y. J. Lee, E. Lœgsgaard, I. Stensgaard, L. N. Kantorovich and F. Besenbacher, Small, 2008, 4, 1494–1500 CrossRef PubMed.
- Q. Chen, D. J. Frankel and N. V. Richardson, Langmuir, 2002, 18, 3219–3225 CrossRef.
- W. Xu, R. E. A. Kelly, R. Otero, M. Schöck, E. Lœgsgaard, I. Stensgaard, L. N. Kantorovich and F. Besenbacher, Small, 2007, 3, 2011–2014 CrossRef PubMed.
- H. Tanaka, T. Nakagawa and T. Kawai, Surf. Sci., 1996, 364, L575–L579 CrossRef.
- I. Temprano, G. Thomas, S. Haq, M. S. Dyer, E. G. Latter, G. R. Darling, P. Uvdal and R. Raval, J. Chem. Phys., 2015, 142, 101916 CrossRef PubMed.
- S. J. Sowerby, M. Edelwirth and W. M. Heckl, J. Phys. Chem. B, 1998, 102, 5914–5922 CrossRef.
- A.-M. Chiorcea and A. M. Oliveira-Brett, Bioelectrochemistry, 2002, 55, 63–65 CrossRef PubMed.
- A. Ciesielski, R. Perone, S. Pieraccini, G. P. Spada and P. Samorì, Chem. Commun., 2010, 46, 4493–4495 RSC.
- R. Otero, M. Schöck, L. M. Molina, E. Lœgsgaard, I. Stensgaard, B. Hammer and F. Besenbacher, Angew. Chem., Int. Ed., 2005, 44, 2270–2275 CrossRef PubMed.
- M. Furukawa, H. Tanaka and T. Kawai, J. Chem. Phys., 2001, 115, 3419–3423 CrossRef.
- Y.-S. Youn, D. H. Kim, H. J. Lee and S. Kim, Chem. Commun., 2015, 51, 12815–12818 RSC.
- R. Otero, M. Lukas, R. E. A. Kelly, W. Xu, E. Lægsgaard, I. Stensgaard, L. N. Kantorovich and F. Besenbacher, Science, 2008, 319, 312–315 CrossRef PubMed.
- M. Iakhnenko, V. Feyer, N. Tsud, O. Plekan, F. Wang, M. Ahmed, O. V. Slobodyanyuk, R. G. Acres, V. Matolín and K. C. Prince, J. Phys. Chem. C, 2013, 117, 18423–18433 CrossRef.
- S. Xu, M. Dong, E. Rauls, R. Otero, T. R. Linderoth and F. Besenbacher, Nano Lett., 2006, 6, 1434–1438 CrossRef PubMed.
- W. Mamdouh, R. E. A. Kelly, M. Dong, L. N. Kantorovich and F. Besenbacher, J. Am. Chem. Soc., 2008, 130, 695–702 CrossRef PubMed.
- W. Xu, J. G. Wang, M. F. Jacobsen, M. Mura, M. Yu, R. E. A. Kelly, Q. Q. Meng, E. Lægsgaard, I. Stensgaard, T. R. Linderoth, J. Kjems, L. N. Kantorovich, K. V. Gothelf and F. Besenbacher, Angew. Chem., Int. Ed., 2010, 49, 9373–9377 CrossRef PubMed.
- R. Otero, W. Xu, M. Lukas, R. E. A. Kelly, E. Lægsgaard, I. Stensgaard, J. Kjems, L. N. Kantorovich and F. Besenbacher, Angew. Chem., Int. Ed., 2008, 47, 9673–9676 CrossRef PubMed.
- C. Shen, J. R. Cramer, M. F. Jacobsen, L. Liu, S. Zhang, M. Dong, K. V. Gothelf and F. Besenbacher, Chem. Commun., 2013, 49, 508–510 RSC.
- Z. Guo, I. De Cat, B. Van Averbeke, J. Lin, G. Wang, H. Xu, R. Lazzaroni, D. Beljonne, A. P. H. J. Schenning and S. De Feyter, Chem. Commun., 2014, 50, 11903–11906 RSC.
- A. G. Slater, Y. Hu, L. Yang, S. P. Argent, W. Lewis, M. O. Blunt and N. R. Champness, Chem. Sci., 2015, 6, 1562–1569 RSC.
- S. Cheng, M. Zhang, N. Dixit, R. B. Moore and T. E. Long, Macromolecules, 2012, 45, 805–812 CrossRef.
- K. Zhang, M. Aiba, G. B. Fahs, A. G. Hudson, W. D. Chiang, R. B. Moore, M. Ueda and T. E. Long, Polym. Chem., 2015, 6, 2434–2444 RSC.
- K. Zhang, M. Chen, K. J. Drummey, S. J. Talley, L. J. Anderson, R. B. Moore and T. E. Long, Polym. Chem., 2016, 7, 6671–6681 RSC.
- I.-H. Lin, C.-C. Cheng, C.-W. Huang, M.-C. Liang, J.-K. Chen, F.-H. Ko, C.-W. Chu, C.-F. Huang and F.-C. Chang, RSC Adv., 2013, 3, 12598–12603 RSC.
- D. González-Rodríguez, P. G. A. Janssen, R. Martín-Rapún, I. De Cat, S. De Feyter, A. P. H. J. Schenning and E. W. Meijer, J. Am. Chem. Soc., 2010, 132, 4710–4719 CrossRef PubMed.
- B. Ilko, W. Sigrid, M. Xiaojing, P. Xie, S. Ramesh, D. Mingdong, W. Chen, M. Wael, W. Jianguo and F. Besenbacher, Phys. Chem. Chem. Phys., 2010, 12, 3616–3621 RSC.
- C. J. Serpell, M. Barłóg, K. Basu, J. F. Fakhoury, H. S. Bazzi and H. F. Sleiman, Mater. Horiz., 2014, 1, 348–354 RSC.
- P. Singh, S. Campidelli, S. Giordani, D. Bonifazi, A. Bianco and M. Prato, Chem. Soc. Rev., 2009, 38, 2214 RSC.
- N. Karousis, N. Tagmatarchis and D. Tasis, Chem. Rev., 2010, 110, 5366–5397 CrossRef PubMed.
- C. Farrera, F. Torres Andón and N. Feliu, ACS Nano, 2017, 11, 10637–10643 CrossRef PubMed.
- R. Alshehri, A. M. Ilyas, A. Hasan, A. Arnaout, F. Ahmed and A. Memic, J. Med. Chem., 2016, 59, 8149–8167 CrossRef PubMed.
- E. Heister, E. W. Brunner, G. R. Dieckmann, I. Jurewicz and A. B. Dalton, ACS Appl. Mater. Interfaces, 2013, 5, 1870–1891 CrossRef PubMed.
- J. M. Schnorr and T. M. Swager, Chem. Mater., 2011, 23, 646–657 CrossRef.
- R. Soleyman, S. Hirbod and M. Adeli, Biomater. Sci., 2015, 3, 695–711 RSC.
- M. Adeli, R. Soleyman, Z. Beiranvand and F. Madani, Chem. Soc. Rev., 2013, 42, 5231 RSC.
- A. Bianco, K. Kostarelos, C. D. Partidos and M. Prato, Chem. Commun., 2005, 571–577 RSC.
- K. V. Singh, R. R. Pandey, X. Wang, R. Lake, C. S. Ozkan, K. Wang and M. Ozkan, Carbon, 2006, 44, 1730–1739 CrossRef.
- C. Dwyer, M. Guthold, M. Falvo, S. Washburn, R. Superfine and D. Erie, Nanotechnology, 2002, 13, 601–604 CrossRef.
- P. Singh, V. Venkatesh, N. Nagapradeep, S. Verma and A. Bianco, Nanoscale, 2012, 4, 1972–1974 RSC.
- P. Singh, J. Kumar, F. M. Toma, J. Raya, M. Prato, B. Fabre, S. Verma and A. Bianco, J. Am. Chem. Soc., 2009, 131, 13555–13562 CrossRef PubMed.
- P. Singh, G. Lamanna, C. Ménard-Moyon, F. M. Toma, E. Magnano, F. Bondino, M. Prato, S. Verma and A. Bianco, Angew. Chem., Int. Ed., 2011, 50, 9893–9897 CrossRef PubMed.
- P. Singh, F. M. Toma, J. Kumar, V. Venkatesh, J. Raya, M. Prato, S. Verma and A. Bianco, Chem.–Eur. J., 2011, 17, 6772–6780 CrossRef PubMed.
- P. Singh, C. Ménard-Moyon, A. Battigelli, F. M. Toma, J. Raya, J. Kumar, N. Nidamanuri, S. Verma and A. Bianco, Chem.–Asian J., 2013, 8, 1472–1481 CrossRef PubMed.
- P. Singh, C. Ménard-Moyon, J. Kumar, B. Fabre, S. Verma and A. Bianco, Carbon, 2012, 50, 3170–3177 CrossRef.
- M. Quintana and M. Prato, Chem. Commun., 2009, 6005–6007 RSC.
- M. Quintana, H. Traboulsi, A. Llanes-Pallas, R. Marega, D. Bonifazi and M. Prato, ACS Nano, 2012, 6, 23–31 CrossRef PubMed.
- B. Mohapatra, P. Pratibha and S. Verma, Chem. Commun., 2017, 53, 4748–4758 RSC.
- C. S. Purohit and S. Verma, J. Am. Chem. Soc., 2006, 128, 400–401 CrossRef PubMed.
- C. S. Purohit and S. Verma, J. Am. Chem. Soc., 2007, 129, 3488–3489 CrossRef PubMed.
- A. K. Mishra, C. S. Purohit, J. Kumar and S. Verma, Inorg. Chim. Acta, 2009, 362, 855–860 CrossRef.
- R. K. Saravanan, P. Saha, V. Venkatesh, T. G. Gopakumar and S. Verma, Inorg. Chem., 2017, 56, 3976–3982 CrossRef PubMed.
- J. Gómez-Herrero and F. Zamora, Adv. Mater., 2011, 23, 5311–5317 CrossRef.
- R. Mas-Ballesté, J. Gómez-Herrero and F. Zamora, Chem. Soc. Rev., 2010, 39, 4220 RSC.
- J. V. Barth, G. Costantini and K. Kern, Nature, 2005, 437, 671–679 CrossRef PubMed.
- V. Venkatesh, P. Pachfule, R. Banerjee and S. Verma, Chem.–Eur. J., 2014, 20, 12262–12268 CrossRef PubMed.
- S. Mandal, C. Wang, R. K. Prajapati, J. Kösters, S. Verma, L. Chi and J. Müller, Inorg. Chem., 2016, 55, 7041–7050 CrossRef PubMed.
- M. El Garah, S. Sinn, A. Dianat, A. Santana-Bonilla, R. Gutierrez, L. De Cola, G. Cuniberti, A. Ciesielski and P. Samori, Chem. Commun., 2016, 52, 11163–11166 RSC.
- P. Amo-Ochoa, M. I. Rodríguez-Tapiador, O. Castillo, D. Olea, A. Guijarro, S. S. Alexandre, J. Gómez-Herrero and F. Zamora, Inorg. Chem., 2006, 45, 7642–7650 CrossRef PubMed.
- G. Vegas, R. Lorca, A. Latorre, K. Hassanein, J. G. Carlos, O. Castillo, F. Zamora and P. Amo-Ochoa, Angew. Chem., Int. Ed., 2017, 56, 987–991 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2018 |
