Open Access Article
Open Access ArticleThe self-assembly of monosubstituted BODIPY and HFBI-RGD†
Fengnan Suna,
Guang Yangab,
Qian Zhanga,
Zhongbo Xuea,
Chengzhi Guc,
Zhuozhi Chend,
Boying Yane,
Yaqing Fengab,
Zefang Wang*d and
Shuxian Meng *a
*a
aSchool of Chemical Engineering and Technology, Tianjin University, Tianjin 300050, China. E-mail: msxmail@tju.edu.cn
bCollaborative Innovation Center of Chemical Science and Engineering, Tianjin 300072, China
cSchool of Chemical Engineering and Technology, Shihezi University, Xinjiang 832000, China
dSchool of Life Sciences, Tianjin University, Tianjin 300072, China. E-mail: zefangwang@tju.edu.cn
eGeneral Hospital of Tianjin Medical University, Tianjin 300052, China
First published on 12th June 2018
Abstract
A novel fluorescent probe was constructed by the self-assembly of monosubstituted BODIPY and a novel targeted hydrophobin named hereafter as HFBI-RGD. Optical measurements and theoretical calculations confirmed that the spectral properties of the probe were greatly influenced by the BODIPY structure, the appropriate volume of BODIPY and the cavity of HFBI-RGD. The experiments in vivo and ex vivo demonstrated that the probe had excellent ability for tumor labelling.
1. Introduction
Functional dyes have been applied to a variety of research fields such as fluorescent molecular probes,1,2 dye sensitized solar cells (DSSCs)3,4 and photodynamic therapy (PDT).5,6 As an indispensable tool for modern biomedical research and pre-clinical study, fluorescence imaging has attracted much attention. Dyes emitting in the far-red or near infra-red (NIR) region (650–900 nm) are ideal materials for this application.7 Of all these dyes, 4,4-difluoro-4-bora-3a,4a-diaza-s-indacenes (BODIPYs) are promising for their high quantum yields, large extinction coefficients, and good absorption and emission intensities. However, critical defects including short wavelength (470–530 nm),8 low water solubility and poor biocompatibility are yet to be solved for further applications. Many strategies are used to modify the BODIPY core to tune the emission into red/near infrared region by mainly including a conjugate structure at the 3,5-position,9,10 replacing the meso-carbon atom of the BODIPY framework to form aza-BODIPY,11,12 and displacing the fluoride atom in conventional BODIPY with oxygen (carbon) atom to form O-BODIPY (C-BODIPY).13–15 Hydrophobicity can be modified by introducing an ionized hydrophilic16,17 or neutral hydrophilic group18,19 and self-assembly of amphiphilic block copolymer.20The hydrophobins are small (7–15 kDa) proteins produced by the filamentous fungi during a specific period. Their structures have been discussed in detail previously,21–25 and Fig. 120 shows the three-dimensional structure of one hydrophobin. The hydrophobins possess amphiphilic characteristics, remarkable surface activity26,27 and various biological functions, which makes them fascinating materials across a number of fields. Integrin αvβ3 (ref. 28) can serve as a receptor for a variety of extracellular matrix proteins displaying the RGD tripeptide sequence.29 Due to high expression in endothelial cells during tumor angiogenesis, αvβ3 is considered as a suitable membrane structure for tumor targeting.30,31 In this study, a hydrophobin modified by RGD peptide was prepared by genetic engineering. Considering the specific binding of RGD and αvβ3, we envisaged that this hydrophobin named hereafter as HFBI-RGD can be used as a useful tool for labelling tumors.
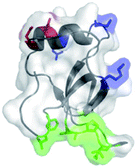 | ||
| Fig. 1 Three-dimensional structure of HFBI. The green part on the surface of HFBI is the hydrophobic patch that can bind to the hydrophobic solid surface. | ||
We designed three monosubstituted BODIPYs (Scheme 1) and prepared HFBI-RGD. Then, the dyes were wrapped into the cavity formed by the self-assembly of HFBI-RGD to produce protein nanocage—fluorescent probes, which not only retained the fluorescence emission of BODIPYs, but also displayed water solubility, biocompatibility and tumor-targeting. We further analysed the main factors which affected the spectral properties of these protein nanocage-fluorescent probes. Optical measurements and theoretical calculations confirmed that the spectral properties were greatly influenced by the BODIPY structure, the appropriate volume of BODIPY and the cavity of HFBI-RGD. Last, using live animal imaging experiments, the probes were verified to have the excellent capability of targeting tumors.
2. Results and discussion
2.1. Synthesis and optical characterization of 3a–3c
Our initial efforts involved the synthesis and optical measurement of BODIPYs. In this study, we reported three new asymmetric structures with flexible and semi-rigid triarylamino groups at the 3-position of the framework (Scheme 1). The dyes were characterized by 1H NMR, 13C NMR and HRMS. The optical results demonstrated that the mono-substituted structures possessed good spectral properties, especially with the fluorescence emission in the visible region. 3a–3c were found to show similar profiles with the maximum absorption wavelength (λmax) of approximately 600 nm and an intense emission peak at around 650 nm (Fig. 2 and S2 in ESI†). In consistent with their fluorescence quantum yields (Φ3a = 0.38, Φ3b = 0.73, and Φ3c = 0.46), which might be ascribed to the triarylamino groups at the C3 position. The dye 3a had the lowest fluorescence intensity, which was caused by the free rotation of benzene rings in the triarylated amine group.32,33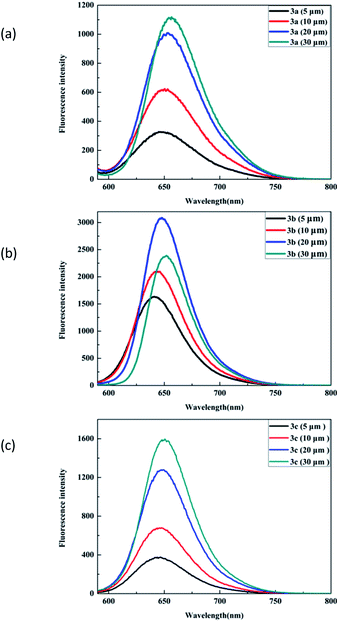 | ||
| Fig. 2 Fluorescence spectra of dyes 3a, 3b and 3c in CH2Cl2 with different concentrations. Condition: excitation wavelength 560 nm. | ||
The dyes 3b and 3c with rigid triarylamino groups exhibited higher emission peaks, especially compound 3b. The absorption and emission spectral behaviors of the dyes in other solvents are also presented (Fig. S3 in ESI†). As indicated by the plots, the fluorescence intensity decreased with the increase in polarity, which could be due to a positive solvent kinetic effect.34,35
Because of the presence of π–π interactions between molecules, the reduced HOMO–LUMO energy gap resulted in a slight bathochromic shift.36,37 Thus, a slight red-shift appeared for all these dyes with the increase in concentration. Among the three BODIPY dyes, dye 3b exhibited the best co-planarity; on the one hand, this property limited free rotation, resulting in the least non-radiative energy loss but on the other hand, it greatly contributed to intermolecular π–π aggregation compared to the observations for dye 3a and dye 3c. Dye 3b showed the strongest emission and exhibited a sharp drop in fluorescence intensity at high concentration.
2.2. The morphology of BODIPY and HFBI-RGD/BODIPY
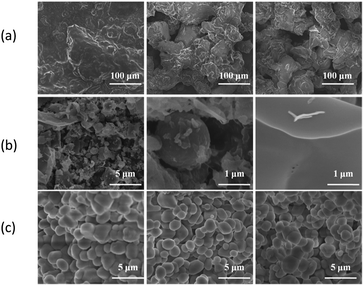
We utilized scanning electron microscopy to characterize the morphological changes between BODIPY and HFBI-RGD/BODIPY. The SEM images suggested that the morphological variation was strongly and closely related to HFBI-RGD. The rightmost image of Fig. 3b shows the membrane structure of the hydrophobic protein modified by RGD. In the self-assembly process, the amphiphilic film entrapped the BODIPY molecule to form an analogous structure to that of a microcapsule, varying the holistic morphology and dispersity of BODIPY. As depicted in Fig. 3c, HFBI-RGD/BODIPY categorically showed much smaller size and regular shape in comparison to BODIPY dyes, which presented massive texture, as shown in Fig. 3a.2.3. Fluorescence properties of the HFBI-RGD/BODIPY fluorescent probe
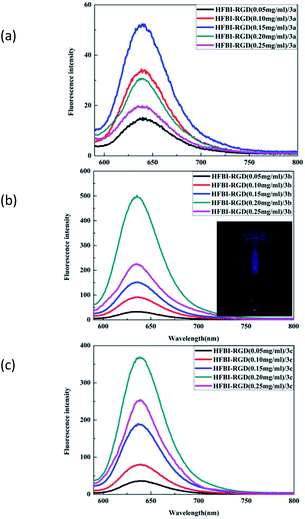
To overcome the hydrophobicity of BODIPY dyes, we encapsulated the dyes with the targeted hydrophobin HFBI-RGD by self-assembly to form a protein nanocage, which exhibited water solubility and excellent spectroscopic properties (Fig. 4), and the protein nanocage also showed stability without aggregation caused by the volatilization of dichloromethane (inset in Fig. 4b). As the figure depicts, the fluorescence intensity increased with the HFBI-RGD concentration in a non-linear manner. The corresponding plot (Fig. S4 in ESI†) clearly indicates the relationship between fluorescence and HFBI-RGD. When the concentration of HFBI-RGD reached a critical value, a decline in the fluorescence intensity was observed. The following presumption might account for this phenomenon: At low concentrations, the hydrophobin molecules in the solution were insufficient to disperse the dyes, whereas the scattered dyes were linked by the aggregation of redundant hydrophobins at high concentrations.20 Another factor that was taken into consideration was the cavity in the structure of hydrophobins. Limited by the size of the cavity, fluorescence intensity for HFBI-RGD/BODIPY was not observed when the BODIPY dyes were mixed in the organic solvents. When the concentration of HFBI-RGD was low, an excess of dye caused aggregation, resulting in the decrease of fluorescence intensity and vice versa. We also found that the critical value of dye 3a was 0.15 mg mL−1, whereas that of each of the other dye was 0.20 mg mL−1, and this divergence was observed due to the different substituents at the 3-position of the central structure. The fluorescence emission wavelengths of the three dyes blue-shifted slightly in close proximity to 645 nm compared to those of the dyes in dichloromethane, which might be caused by the negative solvatochromic effect.38In the process of encapsulating the dyes with HFBI-RGD, we also noticed another interesting phenomenon. The compound 1 and monosubstituted BODIPY molecules 3a–3c could be wrapped by hydrophobins to form protein nanocages with strong fluorescence emissions (Fig. S6 in ESI,† 4), but disubstituted BODIPY molecules could not be wrapped by the hydrophobins. Different concentrations of dyes and hydrophobins were assayed in this self-assembly process, but no fluorescence emission of the disubstituted BODIPYs was observed. We attributed this fluorescence quenching to the difference in volumes of the BODIPYs. The volumes of these dyes are listed in Table S1.† On account of the influence of other substituents, namely, triarylamino groups at the 5-position and steric hindrance, the disubstituted dyes indisputably had bigger volumes than compound 1 and monosubstituted dyes. Based on these evidences, we speculated that hydrophobins with a certain vacuum structure could be prepared by means of ultrasonic agitation. This cavity might affect the fluorescence properties of the dyes of different sizes, which were wrapped into the protein nanocages.
2.4. Biological properties
Finally, we examined HFBI-RGD/BODIPY as a fluorescent probe for targeting the tumor tissue. First, the cytotoxicity of the HFBI-RGD/BODIPY fluorescent probe to glioma cells was studied. The glioma cells were incubated with HFBI-RGD/BODIPY at different concentration gradients (0 μM, 0.1 μM, 1 μM, 3 μM, 5 μM, 10 μM, 20 μM, 60 μM, 100 μM, and 200 μM) for 48 h. The data showed cell survival of approximately 80 percent at 100 μM and 56 percent at 200 μM. The probes exhibited weak even negligible cytotoxicity to the U-87 cells at low concentration (Fig. 5) and thus, we used a low concentration (5 μM) in the follow-up experiment.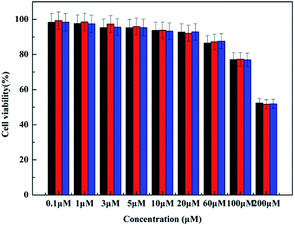 | ||
| Fig. 5 Cytotoxicity of HFBI-RGD/BODIPY fluorescent probe by MTT assay. Black, cytotoxicity of HFBI-RGD/3a; red, cytotoxicity of HFBI-RGD/3b; blue, cytotoxicity of HFBI-RGD/3c. (n = 3, *P < 0.05). | ||
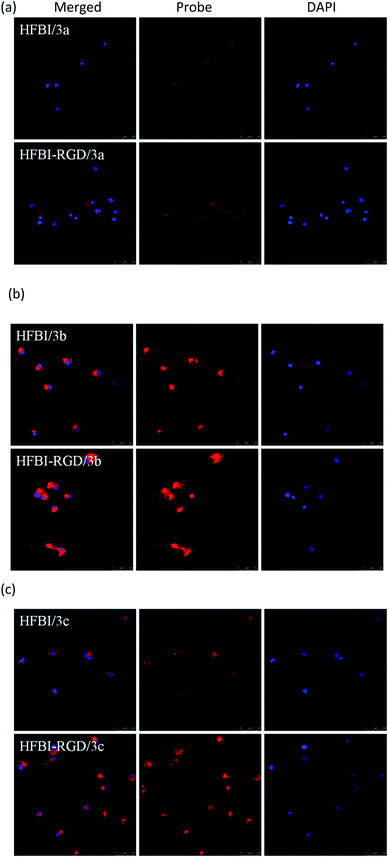
Next, we tested the capability of tumor-targeting by confocal fluorescence microscopy. U-87 cells treated with probes displayed red fluorescence under a confocal fluorescence microscope (Fig. 6). A bright red fluorescence emission was observed clearly when the cells were treated with HFBI-RGD/3b. By contrast, the cells incubated with HFBI-RGD/3a and HFBI-RGD/3c showed indistinct red fluorescence emissions. These changes were consistent with the previous optical data wherein HFBI-RGD/3b displayed better fluorescence intensity than 3a and 3c. It was worth noting that when the cells were treated with HFBI-RGD/(3a–3c), more probes could enter into the cells, which was concluded from the area under the curves of fluorescence spectra in the graphs. The results indicated that HFBI-RGD/BODIPY possessed the ability of targeting the cancer cells to some extent.
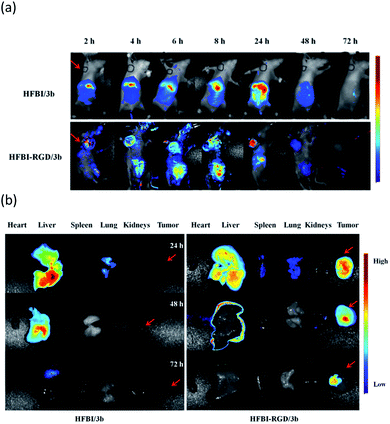
Nude mice bearing U-87-derived glioma were injected with HFBI-RGD/3b or HFBI/3b fluorescent probes (5 μM) via the tail vein. As shown in the real time-dependent fluorescence images of the living tumor-bearing nude mice (Fig. 7a), fuzzy fluorescence at the tumor site was observed at wavelengths close to 645 nm, which may not be completely devoid of interference from the normal tissues. The tumor tissues could be distinguished from the surrounding normal tissues with a certain degree of clarity. Weak fluorescence was observed at the tumor site after 2 h. But as time elapsed, more fluorescent probes gathered around the tumor tissues, and this was accompanied with higher fluorescence intensity, which reached a peak after 24 h. Negligible fluorescence emission was observed after 72 h because of the rapid clearance of the fluorescence probe. To further characterize the tumor-targeting property of the probe, tumor tissues and main organs were harvested at certain times post labelling. The tumors exhibited persistent and strong fluorescence emission without the interference of autofluorescence from the background (Fig. 7b). The results of the control experiment showed that no fluorescence was observed in the tumor tissue whether in the real time-dependent fluorescence images or anatomic images (Fig. 7). These results confirmed the capability of HFBI-RGD/BODIPY fluorescent probes to target the tumors.
3. Conclusions
In summary, we have shown the construction of a novel fluorescent probe—HFBI-RGD/BODIPY. Three new asymmetric structures with flexible and semi-rigid triarylamino groups at the 3-position of the framework were designed and synthesized. A novel hydrophobin modified by peptides RGD was prepared by genetic engineering. Next, HFBI-RGD was exploited to encapsulate the dyes to form protein nanocages by self-assembly. The protein nanocages not only retained the fluorescence emission of BODIPYs, but also showed water solubility, biocompatibility and tumor-specific targeting. Subsequently, we turned our attention to the main factors which affected the spectral properties of the protein nanocages. Optical measurements and theoretical calculations confirmed that the spectral properties were greatly influenced by the BODIPY structure, the appropriate volume of BODIPY and the cavity of HFBI-RGD. The experiments in vivo and ex vivo ascertained that the fluorescent probe had a powerful tumor-targeting property.4. Experimental section
4.1 Materials and instruments
All solvents and starting materials were commercially available and used without further purification (unless specified). The synthetic routes of 2b and 2c were described in our lab's previous publications.39 HFBI-RGD was purified by our laboratory. 1H NMR and 13C NMR spectra were recorded on a Bruker AV400 MHz spectrometer. HRMS results were obtained with a Bruker microTof-QII instrument. Absorption spectra were measured using Shimadzu UV-1800. Fluorescence spectra were determined on a Hitachi F-2500 spectrophotometer. The morphology was obtained with a scanning electron microscope (S-4800, Hitachi, Japan). The cell viability was analysed using a microplate reader (Perkin Elmer Enspire). Fluorescent images of cells were obtained with a confocal microscope (Ultra VIEM Vox, PerkinElmer). In vivo images were captured with a Cri Maestro In vivo imaging system (Cri, Woburn, MA).4.2 Synthesis
3a. 13% yield. 1H NMR (400 MHz, CDCl3, δ): 8.18 (d, 2H, J = 8.4 Hz, Ar–H); 7.72–7.69 (m, 1H, Ar–H); 7.57–7.52 (m, 3H, Ar–H); 7.44 (t, 4H, J = 8.6 Hz, Ar–H); 7.14–7.12 (m, 4H, Ar–H); 7.07–7.06 (m, 2H, Ar–H); 7.02 (d, 2H, J = 8.8 Hz, Ar–H); 6.66 (s, 1H, pyrrole–H); 5.99 (s, 1H, pyrrole–H); 3.98 (s, 3H, –COOCH3); 2.58 (s, 3H, pyrrole–CH3); 1.41 (s, 3H, pyrrole–CH3); 1.37 (s, 3H, pyrrole–CH3). 13C NMR (101 MHz, CDCl3, δ): 167.73, 154.85, 153.89, 148.77, 147.09, 140.08, 136.51, 132.44, 132.36, 132.30, 131.97, 130.89, 130.86, 130.27, 128.82, 128.78, 128.70, 128.62, 125.03, 123.62, 122.39, 121.22, 117.86, 114.73, 52.37, 14.05, 13.72, 10.95. HRMS (ESI) calcd for C40H34BF2N3O2 637.2618, found 637.2612.
3b. 17% yield. 1H NMR (400 MHz, CDCl3, δ): 8.16 (d, 2H, J = 8.4 Hz, Ar–H); 7.52–7.39 (m, 11H, Ar–H); 7.12 (d, 1H, J = 15.6 Hz, –CH
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHTrans–); 6.85 (s, 2H, –CH
CHTrans–); 6.85 (s, 2H, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHsyn–); 6.55 (s, 1H, pyrrole–H); 6.28 (d, 2H, J = 8.4 Hz, Ar–H); 5.95 (s, 1H, pyrrole–H); 3.97 (s, 3H, –COOCH3); 2.54 (s, 3H, pyrrole–CH3); 1.37 (s, 3H, pyrrole–CH3); 1.34 (s, 3H, pyrrole–CH3). 13C NMR (101 MHz, CDCl3, δ): 167.73, 149.95, 142.42, 140.26, 137.62, 137.49, 136.08, 132.44, 132.31, 130.90, 130.86, 130.62, 130.50, 130.41, 130.20, 129.94, 129.79, 128.83, 128.79, 128.60, 127.36, 126.69, 120.74, 117.90, 115.04, 112.17, 52.35, 19.16, 14.05, 10.96. HRMS (ESI) calcd for C42H34BF2N3O2 661.2615, found 661.2606.
CHsyn–); 6.55 (s, 1H, pyrrole–H); 6.28 (d, 2H, J = 8.4 Hz, Ar–H); 5.95 (s, 1H, pyrrole–H); 3.97 (s, 3H, –COOCH3); 2.54 (s, 3H, pyrrole–CH3); 1.37 (s, 3H, pyrrole–CH3); 1.34 (s, 3H, pyrrole–CH3). 13C NMR (101 MHz, CDCl3, δ): 167.73, 149.95, 142.42, 140.26, 137.62, 137.49, 136.08, 132.44, 132.31, 130.90, 130.86, 130.62, 130.50, 130.41, 130.20, 129.94, 129.79, 128.83, 128.79, 128.60, 127.36, 126.69, 120.74, 117.90, 115.04, 112.17, 52.35, 19.16, 14.05, 10.96. HRMS (ESI) calcd for C42H34BF2N3O2 661.2615, found 661.2606.
3c. 16% yield. 1H NMR (400 MHz, CDCl3, δ): 8.17 (d, 2H, J = 8.4 Hz, Ar–H); 7.72–7.69 (m, 11H, Ar–H); 7.54–7.52 (m, 11H, Ar–H); 7.43–7.36 (m, 7H, Ar–H); 7.29–7.15 (m, 5H, Ar–H); 7.17 (d, 1H, J = 16 Hz, –CH
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHTrans–); 6.60–6.57 (m, 3H, pyrrole–H and Ar–H); 5.96 (s, 1H, pyrrole–H); 3.97 (s, 3H, –COOCH3); 3.00 (s, 4H, –CH2CH2–); 2.56 (s, 3H, pyrrole–CH3); 1.39 (s, 3H, pyrrole–CH3); 1.35 (s, 3H, pyrrole–CH3). 13C NMR (101 MHz, CDCl3, δ): 167.89, 150.11, 142.58, 140.42, 137.78, 136.24, 132.60, 131.06, 131.02, 130.78, 130.66, 130.57, 130.36, 130.10, 129.95, 128.99, 128.95, 128.76, 127.52, 126.85, 120.90, 118.06, 115.20, 112.33, 52.51, 23.90, 23.14, 19.32, 14.21, 11.12. HRMS (APCI) calcd for C42H36BF2N3O2 663.2290, found 663.2298.
CHTrans–); 6.60–6.57 (m, 3H, pyrrole–H and Ar–H); 5.96 (s, 1H, pyrrole–H); 3.97 (s, 3H, –COOCH3); 3.00 (s, 4H, –CH2CH2–); 2.56 (s, 3H, pyrrole–CH3); 1.39 (s, 3H, pyrrole–CH3); 1.35 (s, 3H, pyrrole–CH3). 13C NMR (101 MHz, CDCl3, δ): 167.89, 150.11, 142.58, 140.42, 137.78, 136.24, 132.60, 131.06, 131.02, 130.78, 130.66, 130.57, 130.36, 130.10, 129.95, 128.99, 128.95, 128.76, 127.52, 126.85, 120.90, 118.06, 115.20, 112.33, 52.51, 23.90, 23.14, 19.32, 14.21, 11.12. HRMS (APCI) calcd for C42H36BF2N3O2 663.2290, found 663.2298.
4.3 Preparation of HFBI-RGD/BODIPY solution
3a–3c were dissolved in dichloromethane to obtain a certain concentration. Phosphate buffer (pH 7.4) was used to prepare five concentrations of HFBI-RGD solution (50, 100, 150, 200, 250 μg mL−1). Subsequently, 30 μL BODIPY-dichloromethane solution was added to five concentrations of HFBI-RGD (200 μL). The layered solutions were self-assembled by ultrasonic agitation for 1 h below 25 °C. The upper transparent solution was separated for further tests.4.4 General biology experiments
Here, V% is the percent of cell viability, Aexperimental is the absorbance of the treated cells, and Acontrol is the absorbance of the untreated cells.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
S. X. Meng acknowledges the support of the National Natural Science Foundation of China (No. 21676187, No. 21476162), and China International Science and Technology Project (No. 2012DFG41980, 2016YFE0114900), National Key R&D Program of China (21761132007). Z. F. Wang acknowledges the support of the National Natural Science Foundation of China (No. 81601593). We also appreciate the favor of Xiaoyan Zhang and Professor Ding from Nankai University and thank Bowei Lu and others for offering help.Notes and references
- S. Osati, H. Ali, F. Marques, M. Paquette, S. Beaudoin, B. Guerin, J. V. Leyton and J. E. van Lier, Bioorg. Med. Chem. Lett., 2017, 27, 443–446 CrossRef PubMed.
- Y. Hiruta, H. Koiso, H. Ozawa, H. Sato, K. Hamada, S. Yabushita, D. Citterio and K. Suzuki, Org. Lett., 2015, 17, 3022–3025 CrossRef PubMed.
- M. Mao, Q.-S. Li, X.-L. Zhang, G.-H. Wu, C.-G. Dai, Y. Ding, S.-Y. Dai and Q.-H. Song, Dyes Pigm., 2017, 141, 148–160 CrossRef.
- Y. Ooyama, M. Kanda, T. EnoKi, Y. Adachi and J. Ohshita, RSC Adv., 2017, 7, 13072–13081 RSC.
- G. Durán-Sampedro, N. Epelde-Elezcano, V. Martínez-Martínez, I. Esnal, J. Bañuelos, I. García-Moreno, A. R. Agarrabeitia, S. de la Moya, A. Tabero, A. Lazaro-Carrillo, A. Villanueva, M. J. Ortiz and I. López-Arbeloa, Dyes Pigm., 2017, 142, 77–87 CrossRef.
- B. Kim, B. Sui, X. Yue, S. Tang, M. G. Tichy and K. D. Belfield, Eur. J. Org. Chem., 2017, 25–28 CrossRef.
- L. Yuan, W. Lin, K. Zheng, L. He and W. Huang, Chem. Soc. Rev., 2013, 42, 622–661 RSC.
- J. Xu, L. Zhu, Q. Wang, L. Zeng, X. Hu, B. Fu and Z. Sun, Tetrahedron, 2014, 70, 5800–5805 CrossRef.
- E. Y. Schmidt, N. V. Zorina, M. Y. Dvorko, N. I. Protsuk, K. V. Belyaeva, G. Clavier, R. Meallet-Renault, T. T. Vu, A. I. Mikhaleva and B. A. Trofimov, Chemistry, 2011, 17, 3069–3073 CrossRef PubMed.
- R. Ziessel, S. Rihn and A. Harriman, Chemistry, 2010, 16, 11942–11953 CrossRef PubMed.
- J. Bartelmess, M. Baldrighi, V. Nardone, E. Parisini, D. Buck, L. Echegoyen and S. Giordani, Chem.–Eur. J., 2015, 21, 9727–9732 CrossRef PubMed.
- S. Liu, Z. Shi, W. Xu, H. Yang, N. Xi, X. Liu, Q. Zhao and W. Huang, Dyes Pigm., 2014, 103, 145–153 CrossRef.
- B. Brizet, C. Bernhard, Y. Volkova, Y. Rousselin, P. D. Harvey, C. Goze and F. Denat, Org. Biomol. Chem., 2013, 11, 7729–7737 Search PubMed.
- H. Manzano, I. Esnal, T. Marqués-Matesanz, J. Bañuelos, I. López-Arbeloa, M. J. Ortiz, L. Cerdán, A. Costela, I. García-Moreno and J. L. Chiara, Adv. Funct. Mater., 2016, 26, 2756–2769 CrossRef.
- X.-D. Jiang, Y. Fu, T. Zhang and W. Zhao, Tetrahedron Lett., 2012, 53, 5703–5706 CrossRef.
- M. Kamiya and K. Johnsson, Anal. Chem., 2010, 82, 6472–6479 CrossRef PubMed.
- J. Zhang, X. Bao, J. Zhou, F. Peng, H. Ren, X. Dong and W. Zhao, Biosens. Bioelectron., 2016, 85, 164–170 CrossRef PubMed.
- N. Shivran, M. Tyagi, S. Mula, P. Gupta, B. Saha, B. S. Patro and S. Chattopadhyay, Eur. J. Med. Chem., 2016, 122, 352–365 CrossRef PubMed.
- H. Lee, W. Akers, K. Bhushan, S. Bloch, G. Sudlow, R. Tang and S. Achilefu, Bioconjugate Chem., 2011, 22, 777–784 CrossRef PubMed.
- K. Wang, Y. Xiao, Y. Wang, Y. Feng, C. Chen, J. Zhang, Q. Zhang, S. Meng, Z. Wang and H. Yang, Sci. Rep., 2016, 6, 23061 CrossRef PubMed.
- M. Lienemann, J.-A. Gandier, J. J. Joensuu, A. Iwanaga, Y. Takatsuji, T. Haruyama, E. Master, M. Tenkanen and M. B. Linder, Appl. Environ. Microbiol., 2013, 79, 5533–5538 CrossRef PubMed.
- A. Paananen, E. Vuorimaa, M. Torkkeli, M. Penttila, M. Kauranen, O. Ikkala, H. Lemmetyinen, R. Serimaa and M. B. Linder, Biochemistry, 2003, 42, 5253–5258 CrossRef PubMed.
- J. H. A. Hakanpää, G. Z. R. Szilvay, H. Kaljunen, M. Maksimainen, M. Linder and J. Rouvinen, Protein Sci., 2006, 15, 2129–2140 CrossRef PubMed.
- A. Magarkar, N. Mele, N. Abdel-Rahman, S. Butcher, M. Torkkeli, R. Serimaa, A. Paananen, M. Linder and A. Bunker, PLoS Comput. Biol., 2014, 10, e1003745 Search PubMed.
- M. B. Linder, G. z. R. Szilvay, T. N. Setälä and M. E. Penttilä, FEMS Microbiol. Rev., 2005, 29, 877–896 CrossRef PubMed.
- H. J. Hektor and K. Scholtmeijer, Curr. Opin. Biotechnol., 2005, 16, 434–439 CrossRef PubMed.
- H. A. B. Wosten and K. Scholtmeijer, Appl. Microbiol. Biotechnol., 2015, 99, 1587–1597 CrossRef PubMed.
- M. Baruah, W. Qin, C. Flors, J. Hofkens, R. A. L. Vallée, D. Beljonne, M. Van der Auweraer, W. M. De Borggraeve and N. Boens, J. Phys. Chem. A, 2006, 110, 5998–6009 CrossRef PubMed.
- Z. Q. Zhao, Y. Yang, W. Fang and S. Liu, Nucl. Med. Biol., 2016, 43, 661–669 CrossRef PubMed.
- W. Xiao, Y. Wang, E. Y. Lau, J. Luo, N. Yao, C. Shi, L. Meza, H. Tseng, Y. Maeda, P. Kumaresan, R. Liu, F. C. Lightstone, Y. Takada and K. S. Lam, Mol. Cancer Ther., 2010, 9, 2714–2723 CrossRef PubMed.
- Q. Zhang, X. Dong, K. Wang, T. Zhu, F. Sun, S. Meng and Y. Feng, Chin. Chem. Lett., 2017, 28, 777–781 CrossRef.
- C. Wang, J. Li, Z. Ning, D. Zhao and Q. Zhang, Dyes Pigm., 2012, 94, 40–48 CrossRef.
- K. Do, D. Kim, N. Cho, S. Paek, K. Song and J. Ko, Org. Lett., 2012, 14, 222–225 CrossRef PubMed.
- L. Gai, H. Lu and B. Zou, et al., RSC Adv., 2012, 2, 8840–8846 RSC.
- Z. Li, Y. Chen and X. Lv, et al., New J. Chem., 2013, 37, 3755–3761 RSC.
- R. Gresser, M. Hummert, H. Hartmann, K. Leo and M. Riede, Chemistry, 2011, 17, 2939–2947 CrossRef PubMed.
- X. Cao, L. Meng, Z. Li, Y. Mao, H. Lan, L. Chen, Y. Fan and T. Yi, Langmuir, 2014, 30, 11753–11760 CrossRef PubMed.
- B. Dhokale, T. Jadhav, S. M. Mobin and R. Misra, Dalton Trans., 2015, 44, 15803–15812 RSC.
- J. Zhang, F. Lu, S. Qi, Y. Zhao, K. Wang, B. Zhang and Y. Feng, Dyes Pigm., 2016, 128, 296–303 CrossRef.
- H. He, D.-W. Li, L.-Y. Yang, L. Fu, X.-J. Zhu, W.-K. Wong, F.-L. Jiang and Y. Liu, Sci. Rep., 2015, 5, 13543 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, 1H NMR, SEM, UV/vis, fluorescence spectrum, computational data of BODIPYs, cytotoxicity, confocal fluorescence images, in vivo experiment. See DOI: 10.1039/c8ra03687j |
| This journal is © The Royal Society of Chemistry 2018 |