Open Access Article
Open Access ArticleA novel square-planar Pt(II) complex as a monomeric and dimeric G-quadruplex DNA binder†
Chun-Qiong Zhou *a,
Zi-Qi Lia,
Ting-Cong Liaoa,
Tian-Zhu Maa,
Shuo-Bin Chen
*a,
Zi-Qi Lia,
Ting-Cong Liaoa,
Tian-Zhu Maa,
Shuo-Bin Chen b and
Yuan-Yuan Lianga
b and
Yuan-Yuan Lianga
aGuangdong Provincial Key Laboratory of New Drug Screening, School of Pharmaceutical Sciences, Southern Medical University, Guangzhou 510515, China. E-mail: zcqlg@smu.edu.cn; Fax: +86 20 61648549
bSchool of Pharmaceutical Sciences, Sun Yat-sen University, Guangzhou 51006, China
First published on 26th June 2018
Abstract
A novel phenanthroimidazole ethylenediamine Pt(II) complex with coumarin derivative (1) was synthesized and showed higher affinity, selectivity and thermal stabilization for mixed-type dimeric G-quadruplexes (G2T1) over monomeric G-quadruplexes (G1) and duplex DNA. Complex 1 could bind to G-quadruplexes via end-stacking and external-binding modes.
G-quadruplex DNA, as a noncanonical secondary DNA structure, is formed by G-rich sequences widespread in biologically important regions of the human genome. Its stabilization at telomeric regions can inhibit telomerase activity and interfere with telomere biology, which makes it a potential target for the development of new anticancer therapies.1,2 However, bioinformatic studies have shown that over 700, 000 DNA sequences within the human genome have potential to form G-quadruplex structures.3 So it is crucial to selectively bind the different sequences and conformations of G-quadruplexes. The ca. 200 bases of the single-stranded overhang of telomeric DNA can potentially fold into multimeric telomeric G-quadruplexes consisting of several consecutive G-quadruplex units linked by TTA spacers.4,5 Moreover, multimeric G-quadruplexes could be formed by telomeric DNA and r(GGGGCC)n RNA repeats, being relevant in amyotrophic lateral sclerosis.6,7 Thus it is significant to design some binders for selectively binding and stabilizing multimeric G-quadruplexes.
Many square-planar Pt(II) complexes, such as square-planar platinum(II) phenanthroline complexes, have been reported as good binders of G-quadruplexes for possessing a large electron deficient π-aromatic surface, positively charged substituents and a positively charged center.8–12 Some Pt(II) complexes have been modified with a pendant cyclic amine or pyridine side arm and exhibited high affinity for human telomeric G-quadruplexes.8–10 Other structurally analogous Pt(II) complexes, including ones with phenanthroimidazol,11 dipyridophenazine,12 and C-coordinated phenylpyridine ligands,12 have exhibited considerably stronger interactions with G-quadruplex DNA for possessing an extended π-surface. Though a few small molecules have been studied to specifically bind multimeric G-quadruplex structures,13–18 most square-planar Pt(II) phenanthroline complexes have been discussed to selectively bind monomeric G-quadruplexes rather than multimeric G-quadruplexes.
It has been reported that an excellent binder of monomeric G-quadruplexes, such as TMPyP4 and azatrux, could also show high binding properties toward multimeric G-quadruplexes, and even result in the significant differences of the binding properties toward multimeric G-quadruplexes and monomeric G-quadruplexes.19 With this thought in mind, together that the coumarin derivatives possess a π-surface and an amino substituent, we chose phenanthroimidazole ethylenediamine with coumarin derivative L previously reported20 as a ligand, synthesized its Pt(II) complex 1 (Scheme 1), and systematically studied its binding affinities, selectivities and thermal stabilization towards human telomeric dimeric quadruplexes G2T1 and monomeric quadruplexes G1.
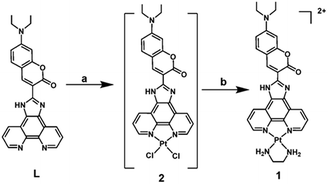 | ||
| Scheme 1 Synthesis of complex 1. Reagents and conditions: (a) K2PtCl4, aqueous DMSO, 140 °C, 2 h; (b) ethylenediamine, EtOH, 80 °C, 12 h. | ||
Complex 1 was synthesized according to the synthetic route in Scheme 1. Phenanthroimidazole with coumarin derivative L reacted with K2PtCl4 in aqueous DMSO and got a red solid of Pt(II) complex 2. Complex 2 reacted with ethylenediamine in ethanol and got the crude product, which was washed with CHCl3 to afford complex 1 as a red solid in 39% yield. Complex 1 was fully characterized by 1H NMR, MS (LR and HR), IR and elemental analysis (seeing ESI†).‡
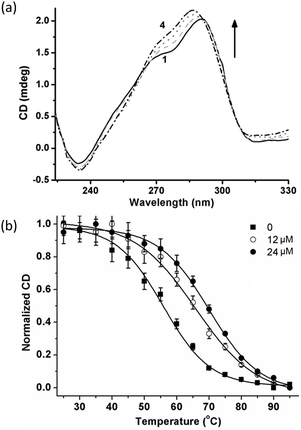
The binding effect of complex 1 on the structures of dimeric G-quadruplexes G2T1 was investigated by circular dichroism (CD) spectra. At first, addition of complex 1 led no significant changes in the ellipticity of antiparallel G2T1, and only induced minor changes in the negative ellipticity at 265 nm (Fig. S3†). These results suggest that complex 1 brought about no structural changes and low binding affinity toward antiparallel G2T1. Subsequently, for mixed-type G2T1, addition of complex 1 led to the increasement of the band with maximum at 291 nm and the shoulder at 270 nm and the shift of the maximum band from 291 nm to 287 nm (Fig. 1a). These results show that complex 1 strongly bound with mixed-type G2T1.
Thermal stabilization of complex 1 towards mixed-type G2T1 was further assessed by CD-melting assays (Fig. 1b and S4†). Complex 1 displayed a ΔTm value being 9.0 °C at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex-to-G2T1 ratio. And the values of ΔTm increased with the increasing amounts of complex 1. A higher thermal stabilization (ΔTm = 11.5 °C) was observed at 8
1 complex-to-G2T1 ratio. And the values of ΔTm increased with the increasing amounts of complex 1. A higher thermal stabilization (ΔTm = 11.5 °C) was observed at 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex-to-G2T1 ratio, which suggests that complex 1 exhibited comparable thermal stabilization with those mixed-type G2T1 binders reported in the literature.16,17,19,21 In contrast, complex 1 had negligible thermal stabilization towards monomeric quadruplexes G1 (ΔTm = 1.2 °C, Fig. S5a†) and double-stranded (ds) DNA (ΔTm = −2.9 °C, Fig. S5b†). These results show that complex 1 had the preferential thermal stabilization towards mixed-type G2T1 over G1 and ds DNA.
1 complex-to-G2T1 ratio, which suggests that complex 1 exhibited comparable thermal stabilization with those mixed-type G2T1 binders reported in the literature.16,17,19,21 In contrast, complex 1 had negligible thermal stabilization towards monomeric quadruplexes G1 (ΔTm = 1.2 °C, Fig. S5a†) and double-stranded (ds) DNA (ΔTm = −2.9 °C, Fig. S5b†). These results show that complex 1 had the preferential thermal stabilization towards mixed-type G2T1 over G1 and ds DNA.
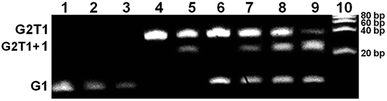
Based on higher thermal stabilization of complex 1 towards mixed-type G2T1 over G1, the binding selectivity of complex 1 was further confirmed for mixed-type G2T1 by gel electrophoresis (Fig. 2). The gel reveals that addition of complex 1 to mixed-type G1 led to no appearance of any new band (lane 2–3). However, the presence of complex 1 increased the mobility of the mixed-type G2T1 (lane 5). These results suggest that complex 1 could form a compact complex with G2T1 rather than G1,14,16 which was further verified by incubating complex 1 with a mixture of G1 and G2T1 and then analysing their gel electrophoresis. Obviously, a mixture of G1 and G2T1 in the absence of complex 1 gave the characteristic bands corresponding to intramolecular G1 and G2T1 (lane 6). When complex 1 added to a mixture of G1 and G2T1, a new band corresponding to the complex of G2T1 with complex 1 (G2T1 + 1) appeared and became more intense with the increasing amounts of complex 1 (lanes 7 to 9). However, no new band appeared corresponding to the complex of G1 and complex 1 (lanes 7 to 9). These results indicate that complex 1 had higher binding selectivity towards G2T1 over G1.
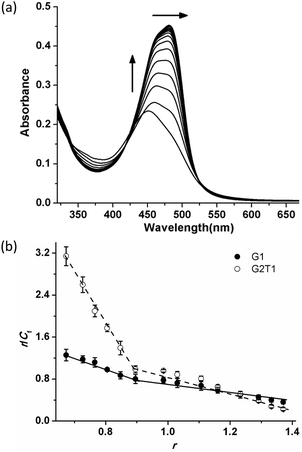
The binding affinities of complex 1 towards mixed-type G2T1 and G1 were determined by UV-Vis titrations (Fig. 3 and S6a†). The gradual addition of G2T1 and G1 to complex 1 resulted in considerable hyperchromicity, a noticeable red-shift at ca. 449 nm (15 nm for G2T1 and 11 nm for G1) and the appearance of a new and strong absorbance peak at ca. 481 nm (Fig. 3a and S6a†), which suggests that complex 1 could interact with G2T1 and G1. Then the data of UV-Vis titrations were also used to calculate the binding constant (K) of complex 1 and the number of binding sites towards G2T1 and G1 by Scatchard eqn (1a):22
| r/Cf = nk − rK | (1a) |
| r = Cb/CDNA | (1b) |
| Cb = Ct (A − A0)/(Amax − A0) | (1c) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for G2T1 and 1.1
1 for G2T1 and 1.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for G1, respectively, when [1]/[DNA] was lower than 1.0. And the binding stoichiometries of complex 1 were 1.5
1 for G1, respectively, when [1]/[DNA] was lower than 1.0. And the binding stoichiometries of complex 1 were 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for G2T1 and 1.7
1 for G2T1 and 1.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for G1, respectively, when [1]/[DNA] was higher than 1.0 (Table 1). Moreover, the binding constants K1 and K2 of complex 1 towards G2T1 had higher than those towards G1. The binding constant of complex 1 towards CT DNA was (0.22 ± 0.04) μM−1 (Fig. S6b†). These results show that complex 1 had higher binding affinity towards G2T1 over G1 and CT DNA. The Ka value of complex 1 towards mixed-type G2T1 was (9.70 ± 0.26) μM−1 (Table 1), which suggests that complex 1 exhibited comparable binding affinity with those mixed-type G2T1 binders reported in the literature.13,19,21,23
1 for G1, respectively, when [1]/[DNA] was higher than 1.0 (Table 1). Moreover, the binding constants K1 and K2 of complex 1 towards G2T1 had higher than those towards G1. The binding constant of complex 1 towards CT DNA was (0.22 ± 0.04) μM−1 (Fig. S6b†). These results show that complex 1 had higher binding affinity towards G2T1 over G1 and CT DNA. The Ka value of complex 1 towards mixed-type G2T1 was (9.70 ± 0.26) μM−1 (Table 1), which suggests that complex 1 exhibited comparable binding affinity with those mixed-type G2T1 binders reported in the literature.13,19,21,23
| DNA | K1 (μM−1) | n1 | K2 (μM−1) | n2 |
|---|---|---|---|---|
| a K1 and K2 are the binding affinities of complex 1 for the strong and weak binding sites in the G-quadruplexes, respectively. They are the average ± SD of three independent measurements. n1 and n2 are the numbers of strong and weak binding sites, respectively. | ||||
| G2T1 | 9.70 ± 0.26 | 1.0 | 2.10 ± 0.32 | 1.5 |
| G1 | 2.34 ± 0.14 | 1.1 | 0.98 ± 0.09 | 1.7 |
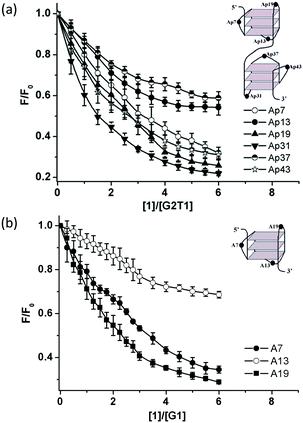
Furthermore, the binding modes of complex 1 towards G2T1 and G1 were discussed by emission spectroscopic assays with G-quadruplexes modified with 2-aminopurine (Ap).15,17 These G2T1 structures with a single Ap base at positions 7, 13, 19, 31, 37 and 43 were selected and named as Ap7, Ap13, Ap19, Ap31, Ap37 and Ap43, respectively (Table S1† and Fig. 4a). As shown in Fig. 4a and S7,† addition of complex 1 significantly decreased the fluorescence intensities of Ap7, Ap19, Ap31 and Ap43, which suggests that complex 1 had strong contact with two propeller loops (Ap7 and Ap43) and two G-tetrads (Ap19 and Ap31). However, addition of complex 1 led to a little decrease on the fluorescence intensities of Ap13 and Ap37 at quadrplex grooves, which excludes the pocket-intercalation mode. At the same conditions, these G1 structures with a single Ap base at positions 7, 13 and 19 (named as A7, A13 and A19, respectively, Table S1†) were selected and discussed the binding mode of complex 1 towards G1. In contrast, complex 1 displayed strong binding with the propeller loop (A7) and 5′-G-tetrads (A19), and slightly weak binding with 3′-G-tetrads (A13-labelled G1) (Fig. 4b and S8†).
To further elucidate the binding mode, molecular docking was performed using complex 1 with hybrid-1 G1 (PDB code: 2MB3, a hybrid-type NMR G-quadruplex structure). The docking results reveal that square-planar Pt(II) section stacked with the guanine residue at the 5′-terminal by a π–π stacking interaction, and the section of coumarin derivative bound with the propeller loop by external binding (Fig. 5a). The docking results for complex 1 bound with the 3′-terminal were also shown in Fig. S9.† The docking score for complex 1 bound with the 5′-terminal was lower than that for complex 1 bound with the 3′-terminal (−3.37 vs. −2.53), which suggests the stronger binding ability at the 5′-terminal than at the 3′-terminal and is consistent with our above experimental data (Fig. 4b).
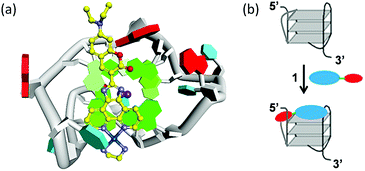 | ||
| Fig. 5 (a) Docked model of complex 1 with G1 (hybrid-1); (b) proposed binding mode of complex 1 with mixed-type G1. | ||
Based on the results of UV-Vis titrations, fluorescence studies with Ap-labelled G2T1 and G1 and molecular modeling studies, complex 1 strongly bound to G2T1 and G1 with the stoichiometry of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 by the external-binding and end-stacking modes. However, their binding modes were different for their different structures of G-quadruplexes (a monomeric model hybrid-1 and a dimeric model hybrid-12 formed by a hybrid-1 G-quadruplex at 5′-end and a hybrid-2 G-quadruplex at 3′-end13). For G1, complex 1 bound to 5′-G-tetrad and the propeller loop of G1 (Fig. 5b). For G2T1, complex 1 possibly bound to 5′-G-tetrad, 3′-G-tetrad and two propeller loops of G2T1. For the binding mode of complex 1 with G2T1 was more complicated, more insights will need to be carried out in future work by molecular modeling.
1 by the external-binding and end-stacking modes. However, their binding modes were different for their different structures of G-quadruplexes (a monomeric model hybrid-1 and a dimeric model hybrid-12 formed by a hybrid-1 G-quadruplex at 5′-end and a hybrid-2 G-quadruplex at 3′-end13). For G1, complex 1 bound to 5′-G-tetrad and the propeller loop of G1 (Fig. 5b). For G2T1, complex 1 possibly bound to 5′-G-tetrad, 3′-G-tetrad and two propeller loops of G2T1. For the binding mode of complex 1 with G2T1 was more complicated, more insights will need to be carried out in future work by molecular modeling.
Conclusions
In conclusion, a novel square-planar Pt(II) complex with coumarin derivative 1 was synthesized and investigated for its potential as monomeric and dimeric G-quadruplex binder. Complex 1 displayed higher binding affinity, selectivity and thermal stabilization towards dimeric quadruplex G2T1 over G1 and ds DNA. It could bind with mixed-type quadruplexes via end-stacking and external-binding modes. This work provides some new sights to design novel dimeric G-quadruplex binders.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21402085), Guangdong Science and Technology Department of China (2017A050501018 and 2014A050503042), Guangdong Provincial Undergraduate Training Programs for Innovation and Entrepreneurship of China (201612121125) and Southern Medical University (YD2017N001).Notes and references
- D. Sun, B. Thompson, B. E. Cathers, M. Salazar, S. M. Kerwin, J. O. Trent, T. C. Jenkins, S. Neidle and L. H. Hurley, J. Med. Chem., 1997, 40, 2113 CrossRef PubMed.
- S. Neidle, J. Med. Chem., 2016, 59, 5987 CrossRef PubMed; S. Neidle, Nat. Rev. Chem., 2017, 1, 10 CrossRef.
- V. S. Chambers, G. Marsico, J. M. Boutell, M. D. Antonio, G. P. Smith and S. Balasubramanian, Nat. Biotechnol., 2015, 33, 877 CrossRef PubMed.
- C. Saintomé, S. Amrane, J.-L. Mergny and P. Alberti, Nucleic Acids Res., 2016, 44, 2926 CrossRef PubMed.
- J. A. Punnoose, Y. Cui, D. Koirala, P. M. Yangyuoru, C. Ghimire, P. Shrestha and H. Mao, J. Am. Chem. Soc., 2014, 136, 18062 CrossRef PubMed.
- J. G. O'Rourke, L. Bogdanik, A. Yáőez, A. J. Wolf, A. K. M. G. Muhammad, R. Ho, S. Carmona and J. P. Vit, Science, 2016, 351, 1324 CrossRef PubMed.
- J. Brčić and J. Plavec, Nucleic Acids Res., 2015, 43, 8590 CrossRef PubMed.
- J. E. Reed, S. Neidle and R. Vilar, Chem. Commun., 2007, 4366 RSC.
- B. Zamiri, M. Mirceta, K. Bomsztyk, J. R. B. Macgregor and C. E. Pearson, Nucleic Acids Res., 2015, 43, 10055 Search PubMed.
- S. Ghosh, O. Mendoza, L. Cuboo, F. Rosu, V. Gabelica, A. J. P. White and R. Vilar, Chem.–Eur. J., 2014, 20, 4772 CrossRef PubMed.
- P. V. Grebe, K. Suntharalingam, R. Vilar, P. J. S. Miguel, S. Herres-Pawlis and B. Lippert, Chem.–Eur. J., 2013, 19, 11429 CrossRef PubMed.
- K. Suntharalingam, A. Łęczkowska, M. A. Furrer, Y. Wu, M. K. Kuimova, B. Therrien, A. J. P. White and R. Vilar, Chem.–Eur. J., 2012, 18, 16277 CrossRef PubMed.
- M. H. Hu, S. B. Chen, B. Wang, T.-M. Ou, L.-Q. Gu, J.-H. Tan and Z.-S. Huang, Nucleic Acids Res., 2017, 45, 1606 CrossRef PubMed.
- C.-Q. Zhou, T.-C. Liao, Z.-Q. Li, J. Gonzalez-Garcia, M. Reynolds, M. Zou and R. Vilar, Chem.–Eur. J., 2017, 43, 4713 CrossRef PubMed.
- C.-Q. Zhou, J.-W. Yang, C. Dong, Y.-M. Wang, B. Sun, J.-X. Chen, Y.-S. Xu and W.-H. Chen, Org. Biomol. Chem., 2016, 14, 191 RSC.
- Z.-Q. Li, T.-C. Liao, C. Dong, J.-W. Yang, X.-J. Chen, L. Liu, Y.-Y. Liang, W.-H. Chen and C.-Q. Zhou, Org. Biomol. Chem., 2017, 15, 10221 RSC.
- Q. Zhang, Y.-C. Liu, D.-M. Kong and D.-S. Guo, Chem.–Eur. J., 2015, 21, 13253 CrossRef PubMed.
- J. A. Punnoose, Y. Ma, Y. Li, M. Sakuma, S. Mandal, K. Nagasawa and H. Mao, J. Am. Chem. Soc., 2017, 139, 7476 CrossRef PubMed.
- A. Cummaro, I. Fotticchia, M. Franceschin, C. Giancola and L. Petraccone, Biochimie, 2011, 93, 1392 CrossRef PubMed.
- J. Sun, J. Zhao, H. Guo and W. Wu, Chem. Commun., 2012, 48, 4169 RSC; J. Sun, W. Wu, H. Guo and J. Zhao, Eur. J. Inorg. Chem., 2011, 21, 3165 CrossRef.
- X.-X. Huang, L.-N. Zhu, B. Wu, Y.-F. Huo, N.-N. Duan and D.-M. Kong, Nucleic Acids Res., 2014, 42, 8719 CrossRef PubMed.
- L.-N. Zhu, B. Wu and D.-M. Kong, Nucleic Acids Res., 2013, 41, 4324 CrossRef PubMed.
- B. Jin, X. Zhang, W. Zheng, X. Liu, J. Zhou, N. Zhang, F. Wang and D. Shangguan, Anal. Chem., 2014, 86, 7063 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Electronic Supplementary Information (ESI) available: synthesis and characterization of complex 1, and experimental procedures for the measurement of its DNA-binding properties and thermal stabilization. See DOI: 10.1039/c8ra03606c |
‡ Structural data for complex 1: 1H NMR (CD3OD, 400 MHz): δ 8.94 (s, 2H), 8.68 (s, 2H), 8.51 (s, 1H), 8.32 (s, 2H), 8.21 (s, 1H), 7.70 (s, 2H), 7.61 (s, 2H), 6.85 (d, J = 8.0 Hz, 1H), 6.60 (s, 1H), 5.36 (s, 1H), 3.58 (m, 4H), 2.97 (s, 4H), 1.30 (t, J = 6.0 Hz, 6H). Main IR (KBr disc, cm−1): ν 3435 (m, N–H), 3152, 2323, 1633 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1400 (s, C O), 1400 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 683, 654. ESI-MS: m/z 689.2 ([1-2Cl–H]+) and HR-MS for C28H29Cl2N7O2Pt ([1-2Cl]2+) calcd: 345.1010, found: 345.1006. Elemental analysis calcd (%) for C28H29Cl2N7O2 Pt·0.5EDA 0.5CHCl3·6H2O: C 36.97, H 4.68, N 11.69; found: C 36.95, H 4.53, N 11.91. N), 683, 654. ESI-MS: m/z 689.2 ([1-2Cl–H]+) and HR-MS for C28H29Cl2N7O2Pt ([1-2Cl]2+) calcd: 345.1010, found: 345.1006. Elemental analysis calcd (%) for C28H29Cl2N7O2 Pt·0.5EDA 0.5CHCl3·6H2O: C 36.97, H 4.68, N 11.69; found: C 36.95, H 4.53, N 11.91. |
| This journal is © The Royal Society of Chemistry 2018 |