Open Access Article
Open Access ArticleNon-targeted metabolite profiling and specific targeted discrimination strategy for quality evaluation of Cortex Phellodendri from different varieties†
Guoxue Zhua and
Fang Feng *abc
*abc
aDepartment of Pharmaceutical Analysis, China Pharmaceutical University, Nanjing 210009, China
bKey Laboratory of Drug Quality Control and Pharmacovigilance (Ministry of Education), China Pharmaceutical University, Nanjing 210009, China
cState Key Laboratory of Natural Medicine, China Pharmaceutical University, Nanjing 210009, China. E-mail: fengfangl@hotmail.com
First published on 15th June 2018
Abstract
Cortex Phellodendri is derived from two species of Phellodendron amurense Rupr. and Phellodendron chinense Schneid. Traditionally, the two species are utilized interchangeably under the name of “huangbo” in the clinic because they are believed to share the same clinical efficacy. However, the chemical analysis in vitro couldn't directly reflect the pharmacological effects. Therefore, whether the constituents could be absorbed into the blood becomes the uppermost problem to account for the clinical efficacy differences of the two species. Therefore, a rapid and sensitive approach to differentiate the two species of Cortex Phellodendri based on non-targeted metabolite profiling and the specific targeted discrimination strategy was first established. Samples from different cultivars were clearly discriminated by principal component analysis and orthogonal partial least squares discriminant analysis. 17 prototype compounds and 22 metabolites contributing to the group separation were identified and tentatively characterized, three of which were found for the first time. Moreover, six of them were screened out as the chemical markers which contribute most to the differences between the two species. Taken together, the application of the non-targeted metabolite profiling and specific targeted discrimination strategy is suitable for the assessment of Cortex Phellodendri.
1 Introduction
Traditional Chinese medicine (TCM) has been widely utilized in the prevention and treatment of human diseases and has an indispensable role in the healthcare system in China.1 Up to now, TCM has garnered increasing attention, not only because of its inspiring and reliable clinical efficacy, but also its outstanding sources of bioactive compounds in the search for new drugs.2–5 Nevertheless, in the procedure of “modernization” and “globalization” of TCM, a pivotal problem is the consistency and controllability of the quality of TCM. Cortex Phellodendri (huangbo in TCM), derived from the dried bark of Phellodendron chinensis Schneid. and Phellodendron amurense Rupr., is one of the 50 fundamental herbs of TCM and is utilized as an essential part of herbal medicine in curing diarrhea, jaundice and other syndromes.6 The two species of herbs are known as Cortex Phellodendri chinensis (HB, chuanhuangbo in China) and Cortex Phellodendri amurensis (GHB, guanhuangbo in China) respectively, which are usually utilized interchangeably in clinical applications because they are believed to have similar pharmacological effects. Accordingly, they are included in the Chinese Pharmacopoeia as official drugs in a single monograph.7 However, some researchers discovered that the constituents existing among these two species are significant differences. And the doubt has been begun on whether their clinical efficacy is similar. Therefore, it is essential and urgent to search for a rapid and reliable analytical method for identify and control the quality of Cortex Phellodendri.Nowadays, liquid chromatography coupled with tandem mass spectrometry (LC-MS) lays a foundation for the analysis of herbal constituents in vivo.8 It has already become a powerful technique for rapid identification of constituents in TCM and biological samples, which can be utilized to clarify underlying mechanisms. However, content change of trace constituents between species may not be detected but largely affect the health of human beings. Multivariate statistical analysis such as principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) is concerned with the analysis and explication of intricate data types built up by numerous and highly related variables.9–11 Using the LC-MS coupled with multivariate statistical analysis approaches, the constituents can be filtered easily with no previous study of the compound structure.
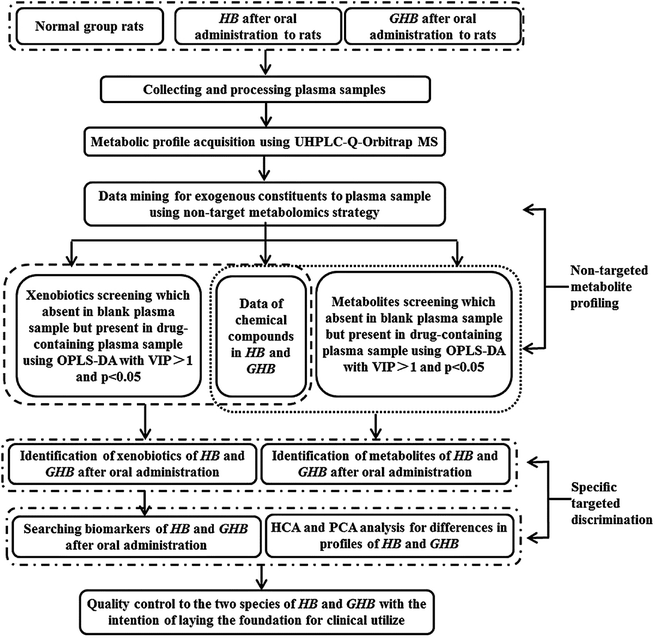
Several analytical methods on chemical analysis have been utilized to evaluate TCM quality, such as fingerprint analysis and quantitative determination of HPLC et al.12–14 However, whether these alleged bioactive constituents can be absorbed into blood to produce pharmacological function is still uncertain. In this study, quality evaluation and species discrimination of Cortex Phellodendri from different varieties was performed to deliver the strategy based on both non-targeted and targeted analysis. The schematic flow is illustrated in Fig. 1. In the research, non-targeted metabolite profiling method based on multivariate statistical analysis was developed and fully validated to screen out the constituents absorbed into blood for confirming the xenobiotic structures. For the purpose of finding the potential chemical markers, specific targeted discrimination approach was employed to analyze the discrimination of two species of Cortex Phellodendri. In general, a systematic strategy was established for comprehensive quality evaluation of Cortex Phellodendri from different varieties.
2 Experimental
2.1 Chemicals and reagent
Acetonitrile and formic acid of HPLC grade were purchased from Merck (Darmstadt, Germany), and HPLC grade methanol was obtained from Fisher Scientific (Pittsburgh, USA). Ultra high purity water was freshly produced by Millipore-Q water purification system (Bedford, USA). All other chemicals and reagents were of analytical grade and commercially available.2.2 Plant material
Five batches of Cortex Phellodendri chinensis and five batches of Cortex Phellodendri amurensis were purchased from Kangmei pharmaceutical Co., Ltd. and authenticated by Professor Minjian Qin (Department of Chinese Materia Medica, China Pharmaceutical University, Nanjing, China). The quality of these crude drugs was controlled and processed according to the Chinese Pharmacopoeia (2015).2.3 Sample preparation
All crude drugs were ground into powder before use. HB and GHB were percolated in water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, w/v) for 0.5 h and decocted by boiling for 1 h. The extracted solution was then decanted, filtered through six layer gauze. The procedure was repeated and the ratio of the total weight of the herbs and the volume of water for the second were 1
10, w/v) for 0.5 h and decocted by boiling for 1 h. The extracted solution was then decanted, filtered through six layer gauze. The procedure was repeated and the ratio of the total weight of the herbs and the volume of water for the second were 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8. The extracts were combined and evaporated to dry under reduced pressure, and then the solution was freeze-dried and stored in a vacuum desiccator before use. The final ratio of powder to raw herb of HB and GHB were 30.31% and 26.02%, respectively. Before given to rats, appropriate amounts of the freeze-dried powders were dispersed in normal saline.
8. The extracts were combined and evaporated to dry under reduced pressure, and then the solution was freeze-dried and stored in a vacuum desiccator before use. The final ratio of powder to raw herb of HB and GHB were 30.31% and 26.02%, respectively. Before given to rats, appropriate amounts of the freeze-dried powders were dispersed in normal saline.
2.4 Animal experiments
Fifteen male Sprague-Dawley (SD) rats with the weight of 200 ± 20 g were obtained from the Animal Multiplication Center of Qinglong mountain (Permission No. SCXK-ZHE 2014-0001). All animals were adapted for one week under 50 ± 20% humidity and 25 ± 2 °C with a 12 h light/dark cycle circumstances with purified water and standard chow ad libitum before experiment. All rats were fasted for 12 h with free access to water prior to the experiment. All protocols and care of the rats were in accordance with the Guidelines for the Care and Use of Laboratory Animals of China Pharmaceutical University and approved by the Animal Ethics Committee of China Pharmaceutical University. The rats were stochastically divided into the HB-dosed group, GHB-dosed group and control group (n = 5 for each group). HB and GHB were dissolved in normal saline and intragastrically administered to rats at a dose of 20 g kg−1 d−1 for consecutive three days. While the rats serving as control received normal saline with equivalent volume. After three days, blood samples (approximately 0.5 mL) were collected from the fosse orbital vein using heparinized 1.5 mL polythene tubes after dosing at 0.5, 0.75, and 1 h, which were selected by the T max reported of related compounds and preliminary experiment, then centrifuged immediately at 12,000 rpm for 10 min at 4 °C to obtain plasma. The plasma samples at different time points of a single rat were mixed, then labeled and stored at −80 °C until analysis.2.5 Sample preparation
One milliliter of thawed serum samples and 3 mL of acetonitrile were transferred to a polypropylene tube, and the solution was then mixed stand for 20 min at 4 °C before use. The mixture was centrifuged at 3500 rpm for 10 min at 4 °C and evaporated to dryness at 30 °C under nitrogen gas. The residues were re-dissolved in 200 μL of 50% methanol, centrifuged at 12000 rpm for 10 min at 4 °C and then filtered through a 0.22 μm membrane to obtain the injection samples.2.6 Instrumentation and analytical conditions
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 full-width at half-maximum (FWHM) at m/z = 200, which provided approximately three full scan-ddMS2 (data points) per second.
000 full-width at half-maximum (FWHM) at m/z = 200, which provided approximately three full scan-ddMS2 (data points) per second.2.7 Data processing
3 Results and discussion
3.1 Technical reproducibility of the analytical approach
Preliminary preparation treatment including organic solvent precipitation and C18 solid-phase extraction (SPE) were compared to confirm the appropriate method for different plasma samples. In the result, organic solvent precipitation could obtain more integrated information of exogenous constituents and the interference of endogenous constituents was acceptable. Among the method of organic solvent precipitation, methanol and acetonitrile were compared and the result showed that acetonitrile has lower endogenous interference.Meanwhile, different mobile phases were screened for the purpose of obtain LC chromatograms with better peak shape and separation. Finally, 0.1% formic acid in acetonitrile and 0.1% formic acid in water were selected as the most suitable eluting solvent system. The majorization of MS spectra which associated with baseline stability and higher ionization efficiency, including capillary voltage, nebulizer pressure, fragmentor voltage, drying gas flow rate and temperature were realized. And the typical global profiling about HB-containing plasma sample in positive and negative ion modes was showed in Fig. S1 (ESI).†
3.2 Metabolic profile identification based on non-targeted metabolic profile strategy
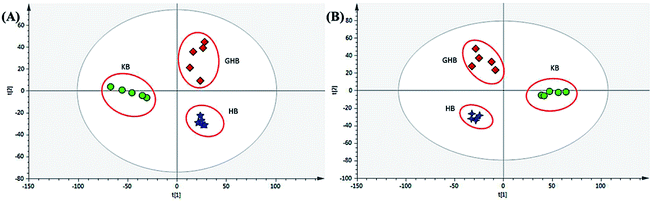
After Pareto scaling with mean-centering, the filtered data were analyzed by PCA and OPLS-DA. PCA, an unsupervised analysis, was firstly employed to observe the classification trends. In the score plots of PCA, it was found that the blank plasma sample (green dots) and HB-containing plasma (blue 4-point star) and GHB-containing plasma (red diamond) were showed a quite clear discrimination (Fig. 2A and B). Afterwards, supervised OPLS-DA model was investigated to identify potential markers accounting for the group separation. And the prototype compounds and metabolites of HB and GHB which absent in blank plasma sample but present in drug-containing plasma sample were obtained by using S-plot analysis. It can be seen from Fig. S2A to D,† in the green 4-point star S-plot, variables that significantly contributed to the discrimination were screened according to a threshold of variable importance in the projection (VIP) values (VIP > 1.0) and p-value (cross-validated predictive residuals, CV-ANOVA, p < 0.05). Finally, 39 compounds, including 17 prototype constituents and 22 metabolites, were filtered out and their fragment pathways have been tentatively identified by searching their accurate masses and MS fragmentation behaviors compared with an online biochemical database service METLIN.3.3 Specific targeted discrimination of HB and GHB in rat
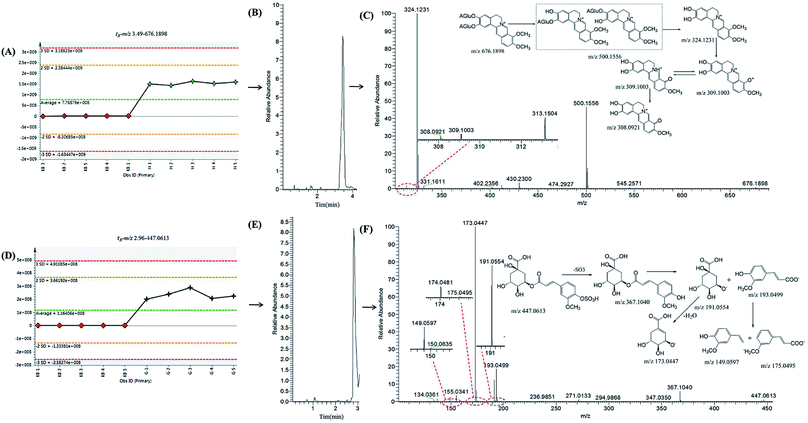
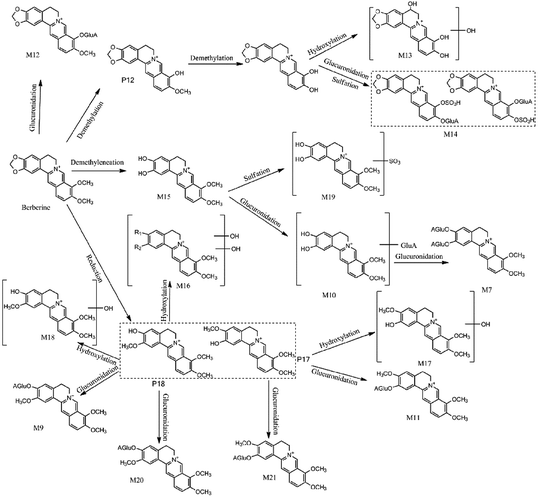
Metabolite M14 showed quasi-molecular ion [M]+ at m/z 592.1170 and calculated molecular formula was C26H26NO13S of which yielded the daughter ions at 416.0801 [M − GluA]+ and 336.1233 [M − GluA–SO3]+. And the successive fragmentation ions at m/z 321.1005 [M − GluA–SO3–CH3]+, 306.0764 [M − GluA–SO3–2CH3]+, 278.0815 [M − GluA–SO3–2CH3–CO]+, all of which were same with berberine. Therefore, M14 was tentatively designated as berberine-O-sulfate-O-glucuronide.
M2 was eluted at 1.47 min and had an [M]+ ion at m/z 518.2007 (C26H32NO10), which was 176 Da higher than that of phellodendrine, demonstrating that the constituents were combined with glucuronic acid. In its MS/MS spectra, the [M]+ ion produced fragment ions at 342.1688 [M − GluA]+,192.1012 [M − GluA–C9H11O2]+, 177.0762 [M − GluA–C9H11O2–CH3]+, which was similar with phellodendrine. Therefore, M2 was identified as phellodendrine-O-glucuronide. The metabolites M4, M9, M11 also exhibited fragment ion [M − 176]+, and notable fragmentation pathways also similar with magnoflorine, jatrorrhizine and columbamine which present at HB and GHB samples. According to the retention time and the previous report,15 they could tentatively be identified as magnoflorine-O-glucuronide, jatrorrhizine-3-O-β-D-glucuronide, columbamine-2-O-β-D-glucuronide, respectively.
Metabolites M1 (tR = 1.35 min) and M3 (tR = 1.88 min) exhibited the [M]+ ion at m/z 474.1764 and 474.1746, which calculated molecular formula was C24H28NO9. The [M]+ ion showed the fragment ion at 298.1439 [M − GluA]+, which was 16 Da lower than armepavine, and the other notable fragmentation pathways are similar. According to the retention time and above-mentioned information as well as literature report,16 M1 and M3 were tentatively determined as hydroxymethyl armepavine-O-glucuronide, and M8 was tentatively identified as metabolites of armepavine.
Metabolite M15 had an [M]+ ion at m/z 324.0773 and was eluted at 8.30 min. The molecular formula was C19H18NO4, which was 14 Da lower than that of columbamine and jatrorrhizine. In its MS/MS information, the [M]+ ion exhibited fragment ions at 308.1785 [M − CH4]+, 293.0962 [M − CH4–CH3]+, 266.0475 [M − CH4–CH3–CO]+, all of which were the same as the previous report.17 Compared with the reference standard, metabolite M15 was confirmed as demethyleneberberine. M10 had the molecular ion at m/z 500.1556, which was 176 Da higher than M15 and showed the fragment ion at 324.1231 [M − GluA]+. The MS spectra of M7 (tR = 1.35 min) exhibited the [M]+ ion at m/z 676.1898, and a successive fragmentation ions the same as M10, such as 500.1556 [M − GluA]+,324.1223 [M − 2GluA]+,309.1003 [M − 2GluA–CH3]+, 308.0921 [M − 2GluA–CH4]+. Therefore, M7 was designated as diglucuronide conjugate of demethyleneberberine, owing to the loss of two glucuronic acid units in the [M − 2GluA]+ ion at m/z 324.1231. Thus, M10 and M7 were assigned as demethyleneberberine-O-glucuronide and 2,3-di-O-glucuronide demethyleneberberine, respectively. M13 showed [M]+ ion at m/z 340.1535, which was 16 Da heavier than M15 and was tentatively identified as hydroxyldemethyleneberberine. It was evidenced by fragment ions at m/z 307.0475 [M − H2O–CH3]+, 279.0536 [M − H2O–CH3–CO]+.
The MS spectra of M16 showed [M]+ ion at m/z 370.1288 which was 16 Da higher than columbamine and jatrorrhizine. It was exhibited fragment ions at m/z 352.1179 [M − H2O]+, 337.0941 [M − H2O–CH3]+, 308.0917 [M − H2O–CH3–CO]+, 293.1202 [M − H2O–2CH3–CO]+. Therefore, M16 was assgined as dihydroxyjatrorrhizine or dihydroxycolumbamine. Equally, according to the literature reports18 and MS/MS information, M17, M18 and M12 were designated as hydroxycolumbamine, hydroxyljatrorrhizine and berberrubine-9-O-β-D-glucuronide, respectively.
Metabolites M5 (tR = 2.76 min) and M6 (tR = 2.96 min), had an quasimolecular ion [M − H]− at m/z 447.0618 and 447.0613 (C17H20O12S), which were 80 Da higher than that of feruloylquinic acid. The fragmentation pathway of which is similar with 3-O-feruloylquinic acid and 5-O-feruloylquinic acid. It was confirmed with fragment ions at m/z 367.1035 [M − SO3–H]−, 193.0498 [M − SO3–H–C7H11O6]−, 191.0554 [M − SO3–H–C10H9O4]−, 175.0232 [M − SO3–H–C7H11O6–H2O]−, 173.0447 [M − SO3–H–C10H9O4–H2O]−. Therefore, M5 and M6 were tentatively identified as sulfated 3-O-feruloylquinic acid and sulfated 5-O-feruloylquinic acid, which were found for the first time. And M19, M20, M21 were tentatively identified as dimethyleneberberine-2-O-sulfate, jatrorrhizine-3-O-sulfate and columbamine-2-O-sulfate, respectively.
3.4 Screening chemical markers by specific targeted discrimination strategy
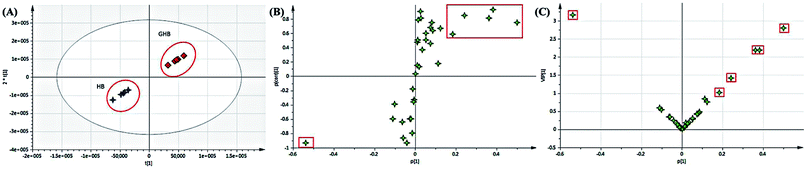
To further study the chemical disparity between two species of drug-containing plasma sample with the purpose of better veritable and effectively investigate the original dissimilarity. The three-dimensional data (retention time vs. m/z vs. response) in positive mode were subjected to PCA and OPLS-DA analysis. As the result, the PCA scores plots exhibited a good separation among the two species of HB and GHB (Fig. 5A). Supervised OPLS-DA was employed to divide the different groups of decoctions and aid the screening of potential marker chemicals (Fig. 5B). The model statistics, R2X = 0.820, R2Y = 0.966, and Q2 = 0.955, indicate that the models are robust and not the result of statistical overfitting.19,20 Moreover, the VIP value plot was utilized to structurally identify these chemical markers (Fig. 5C). In this study, a VIP value above 1.0 scale out by a red box was utilized as a filter standard to select potential chemical markers. Using the method of constituents screening, six ions at the corner of “S” contributed most to the differences between two species drug-containing plasma samples. Thereinto, the concentration levels of berberine, jatrorrhizine, columbamine, berberine-O-sulfate-O-glucuronide detected in HB-containing plasma samples obviously higher than GHB and the compound of 2,3-di-O-glucuronide demethyleneberberine only found in HB-containing plasma samples. In addition, the other compound of magnoflorine has higher content in GHB-containing plasma samples. The intensities difference of 6 potential chemical markers among the HB-containing plasma and GHB-containing plasma were shown in Fig. 6.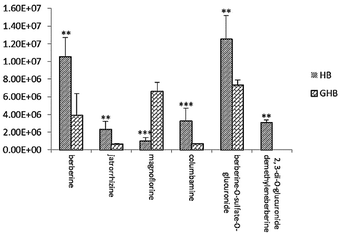 | ||
| Fig. 6 Changes in the intensities of 6 potential chemical markers in plasma sample. **P < 0.01,***P < 0.001 compared with GHB-dosed group (n = 5 for each group). | ||
3.5 Quality assessment of Cortex Phellodendri by hierarchical cluster analysis after metabolic
Hierarchical cluster analysis (HCA) is one of the most common utilized methods in cluster analysis. The principle of HCA is based on the n samples and each of them is classified as one class. Firstly, calculate the similarities of every two elements and combine it into a class which smallest different samples. Then, calculate the distance of this class with the remained n−2 samples. In the procedure of combination, each step is the smallest distance until all samples classified as one class.21,22 In this study, the result showed that the samples can clearly clustered into three groups. It was demonstrated that HB-containing plasma and GHB-containing plasma sample had significant difference so as to indicate that the clinical efficacy is obvious disparity between HB and GHB. HCA results for HB-containing plasma and GHB-containing plasma sample in positive and negative mode were shown in Fig. 7A and B.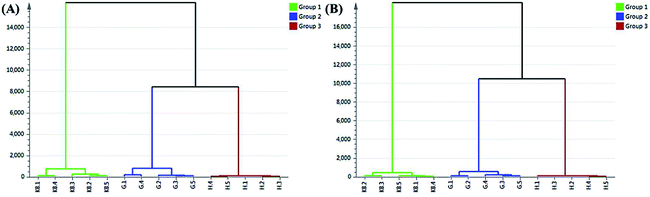 | ||
| Fig. 7 Dendrogram of HCA for blank plasma samples (green), HB-containing plasma samples (brown) and GHB-containing plasma samples (blue) in positive-ion mode (A) and negative-ion mode (B). | ||
4 Conclusions
This study provided insights into the chemical profiles of Cortex Phellodendri from different cultivars by a non-targeted metabolite profiling and specific targeted discrimination strategy. Also, it is noted that 17 prototype compounds and 22 metabolites were annotated and tentatively characterized based on their exact mass and predicted or authentic retention times. And only six of them were filtered out as the potential chemical markers which showed the differences of the two varieties. Alkaloids are the major active compounds in HB and GHB.6,23,24 Additionally, among them, berberine is known to have anti-inflammatory, hepatoprotective, anti-oxidant effects.25 Besides berberine, the main protoberberine-type alkaloids also exhibited various biological activities similar with that of berberine.26,27 Recently, considerable attention has been focused on its anticytotoxicity effects with the purpose of developing magnoflorine as a potential agent.28 As the result of Fig. 6, the concentration levels of chemical markers indicated that the pharmacological functions are different among the two varieties. Generally, HB is mainly utilized to anti-diarrheal, anti-bacterial and anti-inflammatory in clinical application. Interestingly, GHB is more potential of the two species to be utilized in traditional medicine as an anti-Alzheimer's disease agent.29Previous studies proved that the content of berberine, jatrorrhizine and magnoflorine in extraction is different in the two species, however, columbamine has nearly the same. Therefore, whether these alleged bioactive constituents can be absorbed into blood to produce pharmacological function is more important and necessary.6,13,30 That is to say, these findings systematically indicate that mix or misuse the two varieties of Cortex Phellodendri are crucial for the safe and effective usage. And in order to better comprehensively explain the reason of diversity, proposed metabolic pathways, taking berberine as an example, have been researched. Taken altogether, the proposed strategy integrating non-targeted and targeted analysis is a promising approach for the quality evaluation of Cortex Phellodendri and the obtained knowledge would be useful for guiding the clinical applications of these herbal drugs.
Conflicts of interest
The authors have declared no conflict of interest.Acknowledgements
We would like to express our gratitude to National Natural Science Foundation of China (no. 30973858) and a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.References
- Y. Ling, Z. X. Li, M. C. Chen, Z. L. Sun, M. S. Fan and C. G. Huang, Analysis of multiple constituents in Cong-Ming-Tang, a Chinese herbal formula for the treatment of amnesia, by high-performance liquid chromatography with quadrupole time-of-flight mass spectrometry, Phytochem. Anal., 2013, 24, 677–688 CrossRef PubMed.
- H. Y. Wang, H. Sun, A. H. Zhang, Y. Li, L. H. Wang, H. Shi and X. J. Wang, Rapid identification and comparative analysis of the chemical constituents and metabolites of Phellodendri amurensis cortex and Zhibai dihuang pill by ultra-performance liquid chromatography with quadrupole TOF-MS, J. Sep. Sci., 2013, 36, 3874–3882 CrossRef PubMed.
- X. J. Wang, H. Sun, A. H. Zhang, G. Z. Jiao, W. J. Sun and Y. Yuan, Pharmacokinetics screening for multi-components absorbed in the rat plasma after oral administration traditional Chinese medicine formula Yin-Chen-Hao-Tang by ultra performance liquid chromatography-electrospray ionization/quadrupole-time-of-flight mass spectrometry combined with pattern recognition methods, Analyst, 2011, 136, 5068–5076 RSC.
- M. Wang, Y. Li, Y. Huang, Y. Tian, F. Xu and Z. Zhang, Chemomic and chemometric approach based on ultra-fast liquid chromatography with ion trap time-of-flight mass spectrometry to reveal the difference in the chemical composition between Da-Cheng-Qi decoction and its three constitutional herbal medicines, J. Sep. Sci., 2014, 37, 1148–1154 CrossRef PubMed.
- Y. F. Li, S. Qiu, L. J. Gao and A. H. Zhang, Metabolomic estimation of the diagnosis of hepatocellular carcinoma based on ultrahigh performance liquid chromatography coupled with time-of-flight mass spectrometry, RSC Adv., 2018, 8, 9375–9382 RSC.
- Y. Zhang, Z. P. Zhang, H. T. Liu, B. G. Zhang, Y. H. Liao and Z. Zhang, Quantitative and chemical fingerprint analysis for quality evaluation of the dried bark of wild Phellodendron amurense Rupr. based on HPLC-DAD-MS combined with chemometrics methods, Anal. Methods, 2015, 7, 2041–2049 RSC.
- Chinese Pharmacopoeia Commission, Pharmacopoeia of the People's Republic of China, Beijing, 2015, pp. 146–147 Search PubMed.
- H. H. Li, J. L. Pan, S. Hui, X. W. Ma, Z. L. Wang, H. X. Yao, J. F. Wang and H. Li, High-throughput metabolomics identifies serum metabolic signatures in acute kidney injury using LC-MS combined with pattern recognition approach, RSC Adv., 2018, 8, 14838–14847 RSC.
- H. Gika, C. Zisi, G. Theodoridis and I. Wilson, Protocol for quality control in metabolic profiling of biological fluids by U(H)PLC-MS, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2016, 1008, 15–25 CrossRef PubMed.
- L. M. Wei, Y. Wang, L. Lin, L. Zhang, Y. Shi, P. Xiang, S. J. Cao, M. Shen and P. Y. Yang, Identification of potential serum biomarkers of acute paraquat poisoning in humans using an iTRAQ quantitative proteomic, RSC Adv., 2018, 8, 10598–10609 RSC.
- J. Kambeitz, C. Cabral, M. D. Sacchet, I. H. Gotlib, R. Zahn, M. H. Serpa, M. Walter, P. Falkai and N. Koutsouleris, Detecting Neuroimaging Biomarkers for Depression: A Meta-analysis of Multivariate Pattern Recognition Studies, Biol. Psychiatry, 2017, 82, 330–338 CrossRef PubMed.
- Z. P. Zhang, Y. Zhang, Z. Zhang, H. Yao, H. T. Liu, B. G. Zhang and Y. H. Liao, Comparative Analysis of DNA Barcoding and HPLC Fingerprint to Trace Species of Phellodendri Cortex, an Important Traditional Chinese Medicine from Multiple Sources, Biol. Pharm. Bull., 2016, 39, 1325–1330 CrossRef PubMed.
- H. Sun, H. Y. Wang, A. H. Zhang, G. L. Yan, Y. Han, Y. Li, X. H. Wu, X. C. Meng and X. J. Wang, Chemical discrimination of Cortex Phellodendri amurensis and Cortex Phellodendri chinensis by Multivariate Analysis Approach, Pharmacogn. Mag., 2016, 12, 41–49 CrossRef PubMed.
- L. Zhao, Z. X. Zhang, M. M. Zhou, X. J. Gou, Z. Zeng, J. Song, W. N. Ma and Y. Xu, A urinary metabolomics (GC-MS) strategy to evaluate the antidepressant-like effect of chlorogenic acid in adrenocorticotropic hormone-treated rats, RSC Adv., 2018, 8, 9141–9151 RSC.
- K. Wang, L. W. Chai, X. C. Feng, Z. B. Liu, H. X. Liu, L. Q. Ding and F. Qiu, Metabolites identification of berberine in rats using ultra-high performance liquid chromatography/quadrupole time-of-flight mass spectrometry, J. Pharm. Biomed. Anal., 2017, 139, 73–86 CrossRef PubMed.
- H. Y. Wang, G. L. Yan, A. H. Zhang, Y. Li, Y. Y. Wang, H. Sun, X. H. Wu and X. J. Wang, Rapid discovery and global characterization of chemical constituents and rats metabolites of Phellodendri amurensis cortex by ultra-performance liquid chromatography-electrospray ionization/quadrupole-time-of-flight mass spectrometry coupled with pattern recognition approach, Analyst, 2013, 138, 3303–3312 RSC.
- X. W. Liu, F. Zhang, S. H. Gao, B. Jiang and W. S. Chen, Metabolite profiling of Zi-Shen pill in rat boilogical specimens by UPLC-Q-TOF/MS, Chin. J. Nat. Med., 2015, 13, 145–160 Search PubMed.
- K. He, S. M. Kou, Z. Y. Zou, Y. R. Hu, M. Feng, B. Han, X. G. Li and X. L. Ye, Hypolipidemic effects of alkaloids from rhizoma coptidis in diet-induced hyperlipidemic hamsters, Planta Med., 2016, 82, 690–697 CrossRef PubMed.
- M. N. Triba, L. L. Moyec, R. Amathieu, C. Goossens, N. Bouchemal, P. Nahon, D. N. Rutledge and P. Savarin, PLS/OPLS models in metabolomics: the impact of permutation of dataset rows on the K-fold cross-validation quality parameters, Mol. BioSyst., 2015, 11, 13–19 RSC.
- L. N. Zhang, L. Wang, Z. Q. Shi, P. Li and H. J. Li, A metabolomic strategy based on integrating headspace gas chromatography-mass spectrometry and liquid chromatography-mass spectrometry to differentiate the five cultivars of Chrysanthemum flower, RSC Adv., 2018, 8, 9074–9082 RSC.
- W. K. Jia, D. Zhao, T. Shen, S. F. Ding, Y. Y. Zhao and C. L. Hu, An optimized classification algorithm by BP neural network based on PLS and HCA, Applied Intelligence, 2015, 43, 176–191 CrossRef.
- P. Geng, J. M. Harnly, J. H. Sun, M. L. Zhang and P. Chen, Feruloyl dopamine-O-hexosides are efficient marker compounds as orthogonal validation for authentication of black cohosh (Actaea racemosa)-an UHPLC-HRAM-MS chemometrics study, Anal. Bioanal. Chem., 2017, 409, 2591–2600 CrossRef PubMed.
- B. Yang, X. Y. Zhang and R. S. Zhang, A comparative study on analytical method of total alkaloids from cortex Phellodendri amurens by reversed phase high performance liquid chromatography (RP-HPLC) and pressurized capillary electrochromatography (pCEC), Rev. Bras. Farmacogn., 2010, 21, 16–23 CrossRef.
- Y. Han, L. Xu, Q. Wang, Y. Huang and W. Meng, Chemical Composition, Antioxidant and Antimicrobial Activity of the Essential Oil of Phellodendron amurense (Rupr.) from China, Asian J. Chem., 2015, 27, 841–884 CrossRef.
- Y. Zhang, L. Xu, J. Qiu, M. Sun, C. Xia, Z. Zhou and T. Liu, Provenance variations in berberine content of Phellodendron amurense, a rare and endangered medicinal plant grown in Northeast China, Scand J For Res, 2014, 29, 725–733 CrossRef.
- H. Q. Zhao, S. D. Zhou, M. M. Zhang, J. H. Feng, S. S. Wang, D. J. Wang, Y. L. Geng and X. Wang, An in vitro AChE inhibition assay combined with UF-HPLC–ESI-Q-TOF/MS approach for screening and characterizing of AChE inhibitors from roots of Coptis chinensis Franch, J. Pharm. Biomed. Anal., 2016, 120, 235–240 CrossRef PubMed.
- F. C. Meng, Z. F. Wu, Z. Q. Yin, L. G. Lin, R. Wang and Q. W. Zhang, Coptidis rhizoma and its main bioactive components: recent advances in chemical investigation, quality evaluation and pharmacological activity, Chin. Med., 2018, 13, 13 CrossRef PubMed.
- B. J. Xue, Y. Y. Zhao, Q. Miao, P. P. Miao, N. Chen, Y. J. Zhang and S. C. Ma, Vitro Intestinal Absorption and Metabolism of Magnoflorine and its Potential Interaction in Coptidis Rhizoma Decoction in Rat, Eur. J. Drug Metab. Pharmacokinet., 2017, 42, 281–293 CrossRef PubMed.
- Y. F. Xian, Z. X. Lin, S. P. Ip, Z. R. Su, J. N. Chen and X. P. Lai, Comparison the neuropreotective effect of Cortex Phellodendri chinensis and Cortex Phellodendri amurensis against beta-amyloid-induced neurotoxicity in PC12 cells, Phytomedicine, 2013, 20, 187–193 CrossRef PubMed.
- M. L. Chen, Y. F. Xian, S. P. Ip, S. H. Tsai, J. Y. Yang and C. T. Che, Chemical and Biological Differentiation of Cortex Phellodendri Chinensis and Cortex Phellodendri Amurensis, Planta Med., 2010, 76, 1530–1535 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra03369b |
| This journal is © The Royal Society of Chemistry 2018 |