Open Access Article
Open Access ArticleIdentification of potential diagnostic biomarkers of cerebral infarction using gas chromatography-mass spectrometry and chemometrics†
Ming-jiao Li‡
a,
Hong Xiao‡a,
Yi-xing Qiua,
Jian-hua Huanga,
Rong-yong Man*c,
Yan Qina,
Guang-hua Xiongb,
Qing-hua Pengb,
Yu-qing Jiana,
Cai-yun Penga,
Wei-ning Zhangd and
Wei Wang *a
*a
aTCM and Ethnomedicine Innovation & Development Laboratory, Sino-Pakistan TCM and Ethnomedicine Research Center, School of Pharmacy, Hunan University of Chinese Medicine, Changsha, China. E-mail: wangwei402@hotmail.com
bHunan Provincial Key Laboratory of Diagnostic and Therapeutical Research in Chinese Medicine, Hunan University of Chinese Medicine, Changsha, China
cDepartment of Neurology, The First People's Hospital of Huaihua, Huaihua, China
dDepartment of Cardiovascularology, Shenzhen Yantian People's Hospital, Shenzhen, China
First published on 21st June 2018
Abstract
Cerebral infarction (CI) is one of the most common cerebrovascular diseases and remains a major health problem worldwide. In this study, we evaluated the potential diagnostic biomarkers and important relevant metabolic pathways associated with CI. Metabolomics based on gas chromatography-mass spectrometry coupled with the multivariate pattern recognition technique were used to characterize the potential serum metabolic profiles of CI. Forty healthy controls and thirty-three cerebral infarction patients were recruited for the nontargeted global metabolites' study and subsequent targeted fatty acid analysis. Overall, thirty-four endogenous metabolites were found in serum from the untargeted global study, four of which were detected to be significantly different between the CI group and healthy controls, including L-lysine, octadecanoic acid (fatty acid), L-tyrosine and lactic acid. Additionally, fourteen free fatty acids were identified by the subsequent targeted fatty acid analysis, and seven of them were detected to be significantly different between the CI group and healthy controls, which were mainly associated with arachidonic acid metabolism and fatty acid metabolism. Our results suggest several potential diagnostic biomarkers, and serum metabolism research is demonstrated as a powerful tool to explore the pathogenesis of CI.
1. Introduction
Cerebral infarction (CI) refers to ischemic necrosis or the softening of localized brain tissue, which is caused by ischemia, hypoxia and blood supply disorder in the brain.1 It is one of the most common cerebrovascular diseases with characteristics of high morbidity, disability and mortality, and is one of the leading causes of adult disability and death.2–4 The main clinical manifestation of CI includes sudden collapse, unconscious, hemiplegia, speech disorder and intellectual disability.5,6 CI not only threatens human health and life, but also causes great pain and heavy burden to patients, families and the society. Many survivors suffer from severe disability, which makes CI a serious threat to the survival of patients. Medical evidence indicates that early intervention in risk factors can effectively reduce the incidence and recurrence rate of this disease.7 However, the diagnosis of stroke is currently based on clinical history, physical examination, neuroimaging and laboratory tests.8 Additionally, its early diagnosis is intricate and imperfect, and its pathogenesis is still unclear.Biomarkers are recommended to play the following roles in the stroke research field: diagnosing the disease, predicting its outcome, providing information about the risk of future stroke, giving possible stroke mechanism for biomarker-guided treatment or drug response and surrogate endpoints in clinical trials. Although various types of biomarkers have been explored, extensively available, rapid and sensitive diagnostic tests have not been achieved. Thus, comprehensive analysis of the different types of biomarkers and suitable metabolomics studies are highly needed.9
Metabolomics is a powerful and safe approach that directly represents the molecular phenotype of organisms, which has been increasingly utilized in clinical research for uncovering sensitive biomarkers for disease diagnosis.10 Gas chromatography-mass spectrometry (GC-MS)-based metabolomics is ideal for identifying and quantitating small molecular metabolites, such as small acids, alcohols, hydroxyl acids, amino acids, sugars, fatty acids, sterols, catecholamines, drugs, and toxins, where chemical derivatization is usually employed to make these compounds volatile enough for gas chromatography.11 Mass spectra from GC/MS provide reference libraries such as the National Institute of Standards and Technology (NIST) for metabolite identification and tracking, which is a strong point in metabolomic profiling analysis.12 Some previous related studies have been reported. Sun et al. proposed the identification of serum biomarkers and exploration of the metabolic alterations of ischemic stroke using UPLC/Q-TOF MS/MS. They found twelve metabolites with significant alterations, which were closely related to ischemic stroke, and three metabolites (uric acid, sphinganine, and adrenoyl ethanolamide) were identified as potential biomarkers.13 Jung et al. investigated the metabolic profiles of plasma and urine samples from patients with cerebral infarctions using 1H-NMR spectroscopy coupled with multivariate statistical analysis. The data displayed that the excretion of lactate, pyruvate, glycolate, and formate increased, and excretion of glutamine and methanol decreased in the plasma of stroke patients, and the levels of citrate, hippurate, and glycine decreased in the urine of stroke patients. These results demonstrated that a metabolomics approach may be useful for the understanding of stroke pathogenesis and the effective diagnosis of cerebral infarction.14 Wang et al. developed a metabolomics method using GC-MS to characterize the metabolite features of acute ischemic stroke. Their results showed that amino acids are the most important affected metabolites, and tyrosine, lactate, and tryptophan may be considered as potential biomarkers of AIS.15 In addition, Wan et al. studied the correlation between the metabolism of fatty acids and ischemia stroke in rat brains based on metabolomics using GC-MS. Eleven fatty acids were obtained, and nine of them were confirmed as potential biomarkers.16
The aim of this study was to comprehensively analyze the discovered potential diagnostic biomarkers in metabolism research and evaluate the important metabolites in the relevant metabolic pathways associated with cerebral infarction.
2. Materials and methods
2.1. Equipment, chemicals and reagents
Data were obtained on a GCMS-QP2010 (Shimadzu Co., Tokyo, Shimadzu). Chromatographic grade chemicals including pyridine, methoxyamine hydrochloride, N,O-bis(trimethylsilyl) trifluoroacetamide (BSTFA) containing 1% trimethylchlorosilane (TMCS), 2-isopropylmalic acid, heptadecanoic acid (internal standard), n-hexane and methanol were purchased from Sigma-Aldrich (St. Louis, MO, USA). Sodium sulfate anhydrous was obtained from ChinaSun Specialty Products Co., Ltd. 20 reference standards, namely, methyl hexanoate (C6:0), methyl octanoate (C8:0), methyl decanoate (C10:0), methyl undecanoate (C11:0), methyl dodecanoate (C12:0), methyl tridecanoate (C13:0), methyl myristate (C14:0), methyl myristoleate (C14:1), methyl pentadecanoate (C15:0), methyl cis-10-pentadecenoate (C15:1), methyl palmitate (C16:0), methyl palmitoleate (C16:1), methyl heptadecanoate (C17:0), methyl cis-10-heptadecenoate (C17:1), methyl stearate (C18:0), methyl linolelaidate (C18:2n6t), methyl linoleate (C18:2n6c), methyl γ-linolenate (C18:3n3), methyl linolenate (C18:3n6) and cis-4,7,10,13,16,19-docosahexaenoic acid methyl ester (C22:6ns) were purchased from Nanjing SenBeiJia Biological Technology Co., Ltd., China, and were used to validate the analysis method and identify the metabolites. Drying was carried out using an HGC-24 nitrogen blowing instrument (Tianjin Hen'ao Company, China).2.2. Samples collection
This study included seventy-three Chinese individuals (forty healthy participants and thirty-three cerebral infarction patients) who were treated at the First People's Hospital of Huaihua (Huaihua, Hunan, China). All participants were informed of the research procedure and signed informed consent documents before inclusion in our study. All protocols involving the use of human subjects were reviewed and approved by the Ethics Committee of the People's Hospital of Huaihua (Huaihua, Hunan, China), and all experiments were performed in accordance with the guidelines of the National Institutes of Health (NIH). The detailed information for these patients and healthy participants are displayed in Table S1 in the ESI.†2.3. Serum collection and preparation
All subjects fasted for at least 12 h before their blood was drawn and excluded the effect of alcohol, cigarettes, greasy food and excess physical activity. Serum was collected before breakfast in the morning and placed in 1.5 mL Eppendorf tubes without anticoagulant and stored at −80 °C in a refrigerator until GC-MS analysis.In the nontargeted global metabolites' study, aliquots (150 μL) of serum were thawed at 4 °C and spiked with internal standard (75 μL, 1.06 mg mL−1 2-isopropylmalic acid). For protein removal, 450 μL methanol was added, and the sample was vortex-mixed for 15 s and centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 rpm for 15 min. Subsequently, the supernatant was transferred to a derivatization reaction glass tube and dried under nitrogen. The dry residue was dissolved in 75 μL of methoxyamine solution (20 mg mL−1 in pyridine), vortex-mixed for 15 s and placed in a 70 °C water bath for 1 h to perform the oximation reaction for protecting the carbonyl. Subsequently, silylation reaction to increase the volatility of the metabolites was performed by adding a mixture of 150 μL BSTFA and 1% TMCS to the sample and incubating it for 1 h in a 70 °C water bath. After centrifugation at 15
800 rpm for 15 min. Subsequently, the supernatant was transferred to a derivatization reaction glass tube and dried under nitrogen. The dry residue was dissolved in 75 μL of methoxyamine solution (20 mg mL−1 in pyridine), vortex-mixed for 15 s and placed in a 70 °C water bath for 1 h to perform the oximation reaction for protecting the carbonyl. Subsequently, silylation reaction to increase the volatility of the metabolites was performed by adding a mixture of 150 μL BSTFA and 1% TMCS to the sample and incubating it for 1 h in a 70 °C water bath. After centrifugation at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 rpm for 8 min, the supernatant was transferred to a conical insert (200 μL) of a 1.5 mL glass vial for subsequent GC/MS analysis. Quality control (QC) samples,17 which were used to assess the performance of the method and promote the credibility of the data analysis, were prepared by combining 20 μL aliquots from each serum sample of two groups and vortex-mixing them for 1 min. The sample preparation for the QC samples was performed as described above, and the samples were divided into 150 μL aliquots equally. All samples were analyzed randomly, and the QC samples were injected once every 5 samples to monitor the stability of the analytical system.
800 rpm for 8 min, the supernatant was transferred to a conical insert (200 μL) of a 1.5 mL glass vial for subsequent GC/MS analysis. Quality control (QC) samples,17 which were used to assess the performance of the method and promote the credibility of the data analysis, were prepared by combining 20 μL aliquots from each serum sample of two groups and vortex-mixing them for 1 min. The sample preparation for the QC samples was performed as described above, and the samples were divided into 150 μL aliquots equally. All samples were analyzed randomly, and the QC samples were injected once every 5 samples to monitor the stability of the analytical system.
In the targeted fatty acid analysis, the preparation of the serum samples was performed according to previously described procedures.18 The serum samples (each 100 μL) were thawed at 4 °C and mixed with 15 μL internal standard of heptadecanoic acid (dissolved in methanol at a concentration of 0.68 mg mL−1), and a small amount of sodium sulfate anhydrous was added to remove the water in the serum. Then, 1 mL of 5% sulfuric acid solution in methanol was used to dissolve the serum, and it was placed in a 70 °C water bath for 30 min for derivatization, and then extracted twice with n-hexane (each time 1 mL). The supernatant was transferred to a glass tube and dried under nitrogen, then diluted with 100 μL n-hexane and transferred to gas vials for GC-MS analysis. Similar to the nontargeted global metabolite study, to monitor the data quality and process variations, quality control (QC) samples17 were prepared by combining 10 μL aliquots from each serum sample of all seventy three serum samples and vortex-mixing them for 1 min and equally dividing them into 100 μL aliquots.
2.4. Chromatographic and mass spectrometric conditions
A DB-5MS capillary column (30 m × 0.25 mm × 0.25 μm, Agilent, USA) was used for the first nontargeted global metabolites' study in the GC system. The temperature program was optimized as follows: initial temperature was held at 70 °C for 4 min, programmed to 110 °C at a heating rate of 20 °C min−1, and programmed to 270 °C at a rate of 8 °C min−1 and then held for 5 min. The total program time was 31 min with a 6.50 min solvent cut time. Helium (99.999%) was used as the carrier gas with a flow rate of 1.0 mL min−1. The injection volume was 1 μL with a split ratio of 10:1. The temperature of the injector, ion source and interface was 280 °C, 200 °C and 275 °C, respectively. The mass spectrometer was operated under electron impact (EI) mode at an energy of 70 eV and detector voltage of 0.94 kV in full-scan mode at 0.2 s per scan (m/z 35–800).For the subsequent targeted fatty acid analysis, 1 μL aliquot of the derivatized sample was injected into a VF-23ms capillary column (30 m × 0.25 mm × 0.25 μm, Agilent, EU) using the split mode at a ratio of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The oven temperature was initially set to 70 °C, then increased to 170 °C at a heating rate of 30 °C min−1 and increased to 180 °C at a rate of 8 °C min−1, then held for 2 min, programmed to 200 °C at a rate of 4 °C min−1, increased to 215 °C at a rate of 20 °C min−1, then maintained for 1.5 min, and finally programmed to 225 °C at a rate of 40 °C min−1 for 5 min before cooling. Helium (99.999%) was used as the carrier gas with a flow rate of 0.98 mL min−1, and the temperature of the injector, ion source and interface was set to 250 °C, 200 °C and 250 °C, respectively. The mass spectrometer was operated in electron impact (EI) mode at energy of 70 eV and detector voltage of 0.90 kV in full-scan mode at 0.2 s per scan (m/z 30–450).
1. The oven temperature was initially set to 70 °C, then increased to 170 °C at a heating rate of 30 °C min−1 and increased to 180 °C at a rate of 8 °C min−1, then held for 2 min, programmed to 200 °C at a rate of 4 °C min−1, increased to 215 °C at a rate of 20 °C min−1, then maintained for 1.5 min, and finally programmed to 225 °C at a rate of 40 °C min−1 for 5 min before cooling. Helium (99.999%) was used as the carrier gas with a flow rate of 0.98 mL min−1, and the temperature of the injector, ion source and interface was set to 250 °C, 200 °C and 250 °C, respectively. The mass spectrometer was operated in electron impact (EI) mode at energy of 70 eV and detector voltage of 0.90 kV in full-scan mode at 0.2 s per scan (m/z 30–450).
2.5. Qualitative and quantitative analysis
Based on a total ion chromatogram (TIC), the NIST (National Institute of Standards and Technology) MS database was used for identifying the structures of the common peaks. The full scan mass spectra of these metabolites were searched and analyzed using biochemical databases including the Human Metabolome Database (HMDB) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.The GC-MS Postrun analysis software (Shimadzu) was directly used for the quantitative analysis. The internal standards 2-isopropylmalic acid and heptadecanoic acid were used for normalization in the relative quantitative analysis for the nontargeted global metabolites' study and subsequent targeted fatty acid analysis, respectively.
2.6. Multivariate statistical analysis
The raw data acquired by the analytical instruments were processed for peak detection and alignment using the profiling solution software Minitab 17 to exclude the abnormal value. The multivariate pattern recognition technique called random forest (RF) was carried out to cluster analyze the metabolites results. Its programs were written in Matlab by our own group.19 Also, a Mann–Whitney U test with a false discovery rate (FDR) adjustment was used to compare the differences between the cerebral infarction patients (CIP) group and healthy group (HP), which were considered statistically significant when the q-value was less than 0.05. MetaboAnalyst 3.0 was used for the pathway analysis based on the metabolites information.3. Results
3.1. GC-MS-based metabolomics method
Before analysis, the reliability of the GC-MS method was evaluated using all the standard parameters including precision, stability and reproducibility. Instrumental precision was evaluated by analyzing mixed reference standards six times within a single day. For the evaluation of sample stability, a QC sample was selected randomly and repeatedly analyzed six times at 2 h intervals. Reproducibility was determined by analyzing six independent QC samples using the above described procedure within one day. Relative standard deviation (RSD) was used to express variations in the retention time and contents of all common metabolites (Table S2 and 3†). Precision, stability and reproducibility of the retention time were less than 0.0059 min, 0.0098 min, and 0.0120 min, respectively. Also, their RSD ranged from 0.58–5.66%, 3.50–8.96%, and 0.70–6.14% respectively. It is worth mentioning that a downward trend was observed because of material degradation in these six analyses except for docosahexaenoic acid during the stability measurement. The increase in concentration of docosahexaenoic acid may be due to solvent evaporation. The results indicate good instrumental precision, good stability of the samples at least for a 10 h period at room temperature and good reproducibility of the sample processing method.The typical total ion chromatograms (TICs) from the serum metabolic profiling of the CI patients group (CIP), healthy participants' group (HP) and QC group from the nontargeted global metabolites' study are shown in Fig. S1.† Thirty-four endogenous metabolites were identified using reference standards and NIST 08, which were double-checked via HMDB and KEGG and further quantitatively analyzed using the internal standard 2-isopropylmalic acid (shown in Table 1).
| Peak no. | Retention time | Metabolite | Concentration (mg mL−1) | KEGG | HMDB | |
|---|---|---|---|---|---|---|
| HP (n = 40) | CIP (n = 33) | |||||
| 1 | 6.725 | Oxalic acid | 0.022 ± 0.008 | 0.027 ± 0.012 | C00209 | HMDB02329 |
| 2 | 7.24 | Lactic acid | 0.325 ± 0.134 | 0.376 ± 0.230 | C00186 | HMDB00190 |
| 3 | 7.733 | L-Valine | 0.006 ± 0.003 | 0.006 ± 0.003 | C00183 | HMDB00883 |
| 4 | 8.301 | Butyric acid | 0.003 ± 0.001 | 0.007 ± 0.005 | C00246 | HMDB00039 |
| 5 | 8.541 | Oxalic acid | 0.120 ± 0.078 | 0.172 ± 0.088 | C00209 | HMDB02329 |
| 6 | 8.852 | (R)-3-Hydroxybutyric acid | 0.006 ± 0.003 | 0.010 ± 0.007 | C01089 | HMDB00011 |
| 7 | 10.245 | Urea | 0.238 ± 0.140 | 0.250 ± 0.148 | C00086 | HMDB00294 |
| 8 | 10.683 | Phosphoric acid | 0.082 ± 0.026 | 0.104 ± 0.042 | C00009 | HMDB02142 |
| 9 | 11.248 | Glycine | 0.007 ± 0.007 | 0.005 ± 0.005 | C00037 | HMDB00123 |
| 10 | 12.203 | L-Serine | 0.001 ± 0.001 | 0.004 ± 0.005 | C00065 | HMDB00187 |
| 11 | 12.512 | L-Threonine | 0.003 ± 0.002 | 0.008 ± 0.007 | C00188 | HMDB00167 |
| 12 | 14.728 | L-Proline | 0.044 ± 0.022 | 0.047 ± 0.016 | C00148 | HMDB00162 |
| 13 | 15.245 | Erythronic acid | 0.008 ± 0.003 | 0.007 ± 0.002 | — | HMDB00613 |
| 14 | 16.213 | L-Glutamine | 0.005 ± 0.003 | 0.007 ± 0.005 | C00064 | HMDB00641 |
| 15 | 16.358 | L-Phenylalanine | 0.006 ± 0.004 | 0.007 ± 0.005 | C00079 | HMDB00159 |
| 16 | 19.076 | Ornithine | 0.015 ± 0.013 | 0.019 ± 0.016 | C00077 | HMDB00214 |
| 17 | 19.554 | N-Acetyl-D-glucosamine | 0.033 ± 0.016 | 0.037 ± 0.026 | C00140 | HMDB00215 |
| 18 | 19.972 | D-Galactose | 0.010 ± 0.004 | 0.019 ± 0.010 | C00124 | HMDB00143 |
| 19 | 20.145 | D-Glucose | 1.087 ± 0.371 | 1.197 ± 0.543 | C00031 | HMDB00122 |
| 20 | 20.396 | Mannitol | 0.236 ± 0.099 | 0.245 ± 0.118 | C00392 | HMDB00765 |
| 21 | 20.523 | L-Lysine | 0.003 ± 0.001 | 0.014 ± 0.013 | C00047 | HMDB00182 |
| 22 | 20.647 | L-Tyrosine | 0.005 ± 0.004 | 0.006 ± 0.004 | C00082 | HMDB00158 |
| 23 | 20.935 | D-Turanose | 0.008 ± 0.004 | 0.007 ± 0.003 | C19636 | HMDB11740 |
| 24 | 21.163 | D-Arabinose | 0.011 ± 0.009 | 0.009 ± 0.008 | C00216 | HMDB29942 |
| 25 | 21.812 | Stearic acid | 0.002 ± 0.001 | 0.013 ± 0.016 | C01530 | HMDB00827 |
| 26 | 22.165 | Hexadecanoic acid | 0.196 ± 0.148 | 0.235 ± 0.168 | C00249 | HMDB00220 |
| 27 | 22.644 | Myoinositol | 0.011 ± 0.004 | 0.012 ± 0.005 | C00137 | HMDB00211 |
| 28 | 24.08 | L-Tryptophan | 0.009 ± 0.007 | 0.011 ± 0.007 | C00078 | HMDB00929 |
| 29 | 24.174 | Linoleic acid | 0.072 ± 0.024 | 0.084 ± 0.032 | C01595 | HMDB00673 |
| 30 | 24.24 | Oleic acid | 0.068 ± 0.028 | 0.090 ± 0.039 | C00712 | HMDB00207 |
| 31 | 24.534 | Stearic acid | 0.119 ± 0.096 | 0.144 ± 0.120 | C01530 | HMDB00827 |
| 32 | 25.948 | Arachidonic acid | 0.018 ± 0.009 | 0.023 ± 0.013 | C00219 | HMDB01043 |
| 33 | 26.662 | Cholesterol | 0.168 ± 0.081 | 0.186 ± 0.102 | C00187 | HMDB00067 |
| 34 | 28.629 | Glycerol 1-hexadecanoate | 0.074 ± 0.092 | 0.086 ± 0.102 | — | HMDB31074 |
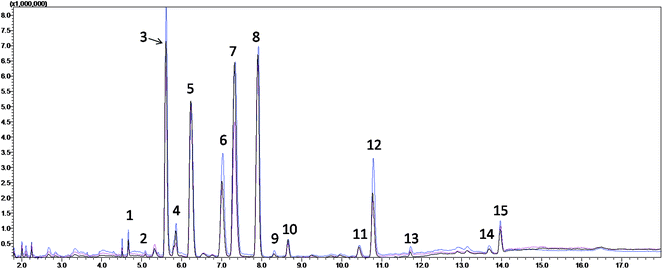
The TICs from the serum metabolite profiling of the CIP, HP and QC from the subsequent fatty acid analysis are shown in Fig. 1, which demonstrate that the serum FFAs of the three groups were similar, but the concentrations of some metabolites were different. After the qualitative and quantitative analyses, fourteen fatty acids were collected, and the detailed information is shown in Table 2. Seven of them were found to be significantly different between the CIP and HP with P values from the Mann–Whitney U test less than 0.05. Among them, myristic acid, palmitic acid, 9-octadecenoic acid, 6,9,12-octadecatrienoic acid, arachidonic acid and docosapentaenoic acid remained significant after FDR correction for multiple comparisons (q < 0.05) (Table 3).
| Peak no. | Retention time | Metabolite | Concentration (mg mL−1) | KEGG | HMDB | |
|---|---|---|---|---|---|---|
| HP (n = 40) | CIP (n = 33) | |||||
| 1 | 4.665 | Myristic acid | 0.0217 ± 0.0064 | 0.0159 ± 0.0047 | C06424 | HMDB00806 |
| 2 | 5.091 | Pentadecanoic acid | 0.0036 ± 0.0010 | 0.0031 ± 0.0009 | C16537 | HMDB00826 |
| 3 | 5.605 | Palmitic acid | 0.6772 ± 0.2024 | 0.7599 ± 0.1354 | C00249 | HMDB00220 |
| 4 | 5.851 | Palmitoleic acid | 0.0777 ± 0.0222 | 0.0932 ± 0.0279 | C08362 | HMDB03229 |
| 5 | 7.01 | Stearic acid | 0.2972 ± 0.0811 | 0.3087 ± 0.0894 | C01530 | HMDB00827 |
| 6 | 7.314 | 9-Octadecenoic acid | 0.8948 ± 0.2613 | 1.0746 ± 0.2652 | C01712 | HMDB00573 |
| 7 | 7.901 | Linoleic acid | 0.8509 ± 0.1831 | 0.8926 ± 0.1901 | C01595 | HMDB00673 |
| 8 | 8.303 | 6,9,12-Octadecatrienoic acid | 0.0097 ± 0.0028 | 0.0158 ± 0.0044 | C06426 | HMDB03073 |
| 9 | 8.654 | 9,12,15-Octadecatrienoic acid | 0.0422 ± 0.0117 | 0.0491 ± 0.0147 | C06427 | HMDB01388 |
| 10 | 10.426 | 8,11,14-Eicosatrienoic acid | 0.0350 ± 0.0100 | 0.0404 ± 0.0117 | C03242 | HMDB02925 |
| 11 | 10.776 | Arachidonic acid | 0.2149 ± 0.0616 | 0.2760 ± 0.0820 | C00219 | HMDB01043 |
| 12 | 11.71 | Eicosapentaenoic acid | 0.0152 ± 0.0043 | 0.0157 ± 0.0046 | C06428 | HMDB01999 |
| 13 | 13.681 | Docosapentaenoic acid | 0.0143 ± 0.0040 | 0.0226 ± 0.0064 | C16513 | HMDB06528 |
| 14 | 13.963 | Docosahexaenoic acid | 0.0838 ± 0.0229 | 0.0827 ± 0.0239 | C06429 | HMDB02183 |
| Peak no. | Potential biomarker | Peak area in HP (mean ± SD) | Peak area in CIP (mean ± SD) | P value of Mann–Whitney U | FDR | Related pathway |
|---|---|---|---|---|---|---|
| a P < 0.05.b q < 0.05. | ||||||
| 1 | Myristic acid | 0.0217 ± 0.0064 | 0.0159 ± 0.0047 | 0.002a | 0.014b | Fatty acid biosynthesis |
| 3 | Palmitic acid | 0.6772 ± 0.2024 | 0.7599 ± 0.1354 | 0.011a | 0.026b | Fatty acid biosynthesis |
| Fatty acid elongation | ||||||
| Fatty acid degradation | ||||||
| 4 | Palmitoleic acid | 0.0777 ± 0.0222 | 0.0932 ± 0.0279 | 0.032a | 0.064 | Fatty acid biosynthesis |
| 6 | 9-Octadecenoic acid | 0.8948 ± 0.2613 | 1.0746 ± 0.2652 | 0.005a | 0.017b | — |
| 8 | 6,9,12-Octadecatrienoic acid | 0.0097 ± 0.0028 | 0.0158 ± 0.0044 | 0.005a | 0.017b | Linoleic acid metabolism |
| Biosynthesis of unsaturated fatty acids | ||||||
| 11 | Arachidonic acid | 0.2149 ± 0.0616 | 0.2760 ± 0.0820 | 0.009a | 0.025b | Arachidonic acid metabolism |
| Linoleic acid metabolism | ||||||
| Biosynthesis of unsaturated fatty acids | ||||||
| 13 | Docosapentaenoic acid | 0.0143 ± 0.0040 | 0.0226 ± 0.0064 | 0.001a | 0.010b | Biosynthesis of unsaturated fatty acids |
3.2. Metabolites data analysis
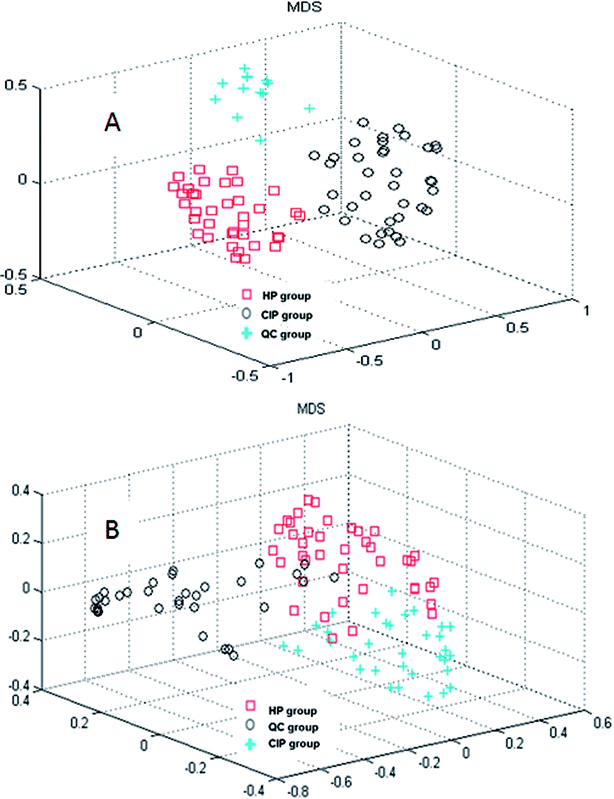
In the current study, random forest (RF) was used to analyze the metabolites information and identify the latent makers between CIP and HP. Its advantages in dealing with this metabolomics data was demonstrated in our previous studies.20–22 RF can provide two useful tools: proximity matrix and variable importance calculation. Proximity matrix coupled with MDS displayed the cluster situation of the samples from different groups (Fig. 2).3.3. Potential biomarkers discovery between CIP and HP
From the plots of the variables importance (VIP) measure, several metabolites play a great role in the classification of the CIP and HP samples in the nontargeted global metabolites' study (Fig. S2†), including L-lysine, octadecanoic acid, L-tyrosine and lactic acid. Also, the subsequent fatty acid analysis was conducted in this research. The VIP measure shows that myristic acid, 9-octadecenoic acid, linoleic acid and 6,9,12-octadecatrienoic acid play a great role in the model classification (Fig. 3).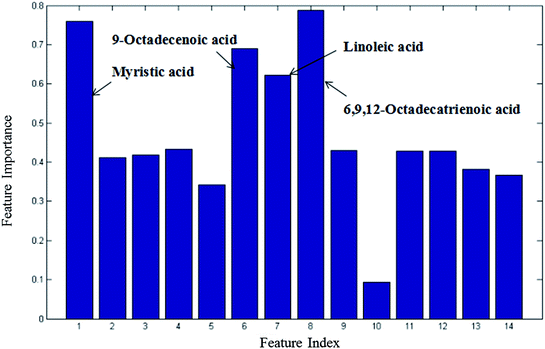 | ||
| Fig. 3 Variable importance measure between CIP and HP in the fatty acid analysis obtained by RF (all 14 metabolites). | ||
Furthermore, MetaboAnalyst 3.0 was used to identify the metabolic pathways associated with the identified combinations of metabolites in the fatty acid analysis. MetaboAnalyst 3.0 (http://www.metaboanalyst.ca/) is a comprehensive tool suite for metabolomic data analysis, which combines the results from a powerful pathway enrichment analysis concerning the conditions under study and uses the high-quality KEGG pathway database as its back end knowledge base.13 It was found that two metabolic pathways of arachidonic acid metabolism and fatty acid metabolism play important roles in the incidence of CI (Fig. 4). Their impact indices are 0.21 and 0.03. The detailed relevant metabolic pathways are shown in Table S4, and Fig. S3 and S4 in the ESI.†
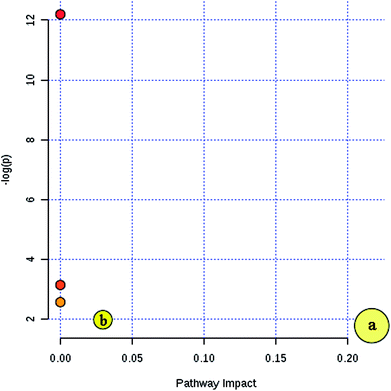 | ||
| Fig. 4 Summary of the key metabolic pathway analysis in the fatty acid analysis with MetaboAnalyst 3.0. (a) Arachidonic acid metabolism and (b) fatty acid metabolism. | ||
4. Discussion
CI is caused by a sudden blockage in an artery that provides blood to the brain, which causes the brain cells at the blockage site to die rapidly due to the lack of oxygen or nutrients in the brain. Additionally, the surrounding brain tissue is damaged owing to the release of toxic substances. These changes in metabolome levels in the biofluids are considered to be associated with the metabolic alterations in the brain.5In the current study, the nontargeted serum metabolomics analysis and targeted fatty acid analysis based on GC-MS coupled with the multivariate pattern recognition technique in CI were profiled. The results show the reliability of the GC-MS method, which has high precision, stability and reproducibility. Multivariate analysis was used to identify the potential biomarkers of CI by establishing a classification model to distinguish CIP from HP. The random forest (RF) method was applied to analyze these metabolites. The RF model is a classifier that contains multiple decision trees, and each tree casts a vote to classify the samples; then, RF selects the majority vote to decide the final classification result.22 The samples from the three groups were totally separated, as observed in Fig. 2A. Fig. 2B shows the good separation trend of the samples among the three groups. These results indicate that the metabolic pattern samples are different in all three groups. The variables importance (VIP) measure of RF can reveal the potential biomarkers with the highest importance values, which can be very useful in disease diagnosis and further exploration of pathogeneses. In the nontargeted serum metabolomics analysis, the VIP (Fig. S2†) results indicate that L-lysine, octadecanoic acid, L-tyrosine and lactic acid play a great role in the classification of CI and healthy samples. In the targeted fatty acid analysis, the results indicate that the contributions of myristic acid, 9-octadecenoic acid, linoleic acid and 6,9,12-octadecatrienoic acid play a great role in the model classification. Moreover, myristic acid, palmitic acid, 9-octadecenoic acid, 6,9,12-octadecatrienoic acid, arachidonic acid and docosapentaenoic acid have values of less than 0.05 (Table 3), and myristic acid, 9-octadecenoic acid and 6,9,12-octadecatrienoic acid are also found in the VIP measure of RF, which indicates that they might participate in the global metabolic alteration of CI.
In the nontargeted serum metabolomics analysis, L-lysine, octadecanoic acid (fatty acid), L-tyrosine and lactic acid were detected to be significantly different between the CI group and healthy controls. Among them, L-tyrosine and lactic acid were considered as potential biomarkers for acute ischemic stroke (AIS) in serum according to the study reported by Wang et al.15 Octadecanoic acid was confirmed by Wan et al., who investigated the correlation between the metabolism of fatty acids and ischemia stroke in rat brains based on metabolomics using GC-MS.16 In addition, they also performed a study to determine the potential biomarkers after analyzing the relationship between the metabolism of amino acids and ischemic cerebral stroke in rats by methods involving GC-MS and principal component analysis (PCA). Thirteen potential biomarkers of ischemic stroke were determined, which contain L-lysine and L-tyrosine.23 However, Wang et al. focused on AIS and the studies by Wan et al. were based on rats.15 In our subsequent targeted fatty acid analysis, myristic acid, palmitic acid, 9-octadecenoic acid, linoleic acid, 6,9,12-octadecatrienoic acid, arachidonic acid and docosapentaenoic acid are the special fatty acids that distinguished the CI and healthy group.
Fatty acids exist in the blood in the form of free fatty acids (FFAs) and esterified fatty acids (EFAs). In human blood, there is a dynamic transformation process between FFAs and EFAs. EFAs can be rapidly decomposed into FFAs in the presence of lipase. FFAs are the products of triglycerides in adipose tissue and the primary source of human organ energy, including that of the heart, liver, and skeletal muscle. The concentration of free fatty acids in the blood is very low and easily affected by lipid metabolism, glucose metabolism, endocrine function and other factors.24–26 In addition, FFAs are very important substances for the human body and play a role of signal molecules in a variety of life activities. Thus, their content in the serum can reflect a certain clinical significance.27
Atherosclerosis is the basic incentive factor of CI, which leads to the formation of cerebral vascular stenosis and cerebral thrombosis. The formation of atherosclerosis is mainly associated with lipid metabolism disorder and chronic inflammatory reaction after the damage of blood vessels. FFAs can be involved in inflammatory responses in various ways. First, FFA metabolites can stimulate the expression of various inflammatory factors such as tumor necrosis factor-α (TNF-α), interleukin (IL-1 and IL-8) and plasminogen activator inhibitor-1 (PAI-1), which can accelerate the inflammatory response of atherosclerosis. Second, metabolites can regulate the expression of the relevant genes involved in the formation of atheromatous plaques such as NF-kb transcription factor. Moreover, FFA metabolites can induce vascular smooth muscle cells, releasing reactive nitrogen, reactive oxygen and oxidative stress by a cascade effect to accelerate the formation of CI.28,29
The level of FFAs is extremely small in normal brain tissue. However, the occurrence of cerebral infarction causes hypoxia in brain cells. Sequentially, this leads to an increase in glycolysis and the incomplete oxidation of fatty acids, resulting in acidosis in the cytochylema due to an increase in ketone concentration. Phospholipases can lead to the degradation of various membranous structures that are rich in phospholipids in neurons. Phospholipases are activated when the intracellular pH value decreases, which weaken the cell membrane and damage the brain tissue, therefore causing the release of a large amount of FFAs. Hence, the contents of palmitic acid, 9-octadecenoic acid, linoleic acid, 6,9,12-octadecatrienoic acid, arachidonic acid, and docosapentaenoic acid increased in the CI group. Moreover, they may further promote the occurrence of CI.30–33 The membrane phospholipid in the biofilm contains arachidonic acid, which is released after the hydrolysis of phospholipase A2, and a high concentration of arachidonic acid can cause cell apoptosis. Arachidonic acid can produce a thrombus A2 after a series of enzymatic reactions, which can promote platelet aggregation, vasoconstriction and the promotion of coagulation and thrombosis. Furthermore, oxygen free radicals are produced in the metabolic process of arachidonic acid, which can affect the structure and function of red blood cells in the blood and other various of pathological processes such as damaging the vascular endothelial cells, affecting the capillaries and destroying the blood–brain barrier, thus leading to further brain damage.34–38
According to the metabolic pathway analysis based on MetaboAnalyst 3.0, arachidonic acid metabolism and fatty acid metabolism are highly correlated with CI. In previous research, it was also found that arachidonic acid metabolic pathway-related gene interactions increase the incidences of stroke and vulnerable plaques.39 Also, by searching the KEGG database, these important fatty acids are associated with relevant fatty acid pathways such as fatty acid biosynthesis, as shown in Table 3, unsaturated fatty acids biosynthesis, fatty acid elongation, fatty acid degradation, arachidonic acid metabolism and linoleic acid metabolism. Furthermore, it was found that myristic acid is connected with fatty acid biosynthesis pathways, which might be involved in the synthesis of other fatty acids and may be the cause for the decrease in the level of myristic acid in CIP.
The limitations of the present study are as follows: only a relatively small number of CI samples was analyzed, and the recruited patients do not rule out other recessive confounding factors such as mental state since depression is associated with CI in young adults aged 15–49 years.40 In addition, although significant differences in the contents of these serum FFAs were found in this study, which were associated with arachidonic acid metabolism and fatty acid metabolism, larger subject number and further studies are proposed to validate these findings. Also, a study with CI patients at different stages is recommended to be conducted.
5. Conclusion
In conclusion, the presented GC-MS method is a simple, quick and accurate analysis method for determining serum free fatty acid metabolic profiles. Serum metabolites play a great role in the separation of the cerebral infarction and healthy groups. L-lysine, octadecanoic acid (fatty acid), L-tyrosine and lactic acid were highly considered as important metabolites for CI. Furthermore, myristic acid, palmitic acid, 9-octadecenoic acid, 6,9,12-octadecatrienoic acid, arachidonic acid, docosapentaenoic acid and linoleic acid were screened as potential fatty acids biomarkers related to CI. To some degree, the concentration of FFAs in the serum can explain the occurrence of CI and these variations maybe highly associated with arachidonic acid metabolism and fatty acid metabolism, which may have great significance for the diagnosis and clinical treatment of CI.Author contributions
W. W. and M. R. Y. conceived the studies. W. W. and H. J. H. designed the studies. L. M. J. and X. H. conducted experiments and analyzed the results. L. M. J., X. H., Q. Y. X., Q. Y., X. G. H., P. C. Y., J. Y. Q., P. Q. H., Z. W. N. and W. W. wrote or helped to draft the manuscript. All authors have reviewed the manuscript.Conflicts of interest
The authors declare no competing financial interests.Abbreviations and acronyms
| CI | Cerebral infarction |
| CIP | CI patients |
| FFA | Free fatty acids |
| GC-MS | Gas chromatography-mass spectrometry |
| HP | Healthy participants |
| QC | Quality control |
| RF | Random forest |
| TIC | Total ion chromatogram |
Acknowledgements
This work was supported by Key Laboratory for Quality Evaluation of Bulk Herbs of Hunan Province, National Natural Science Foundation of People’s Republic of China (Grant number 81673579 and 81703819), State Administration of Traditional Chinese Medicine of the People's Republic of China (ZYBZH-Y-HUN-23), Natural Science Foundation of Hunan province (Grant number 2016JJ6118) and Administration of Traditional Chinese Medicine of Hunan Province (Grant number 201673 and 201714).Notes and references
- A. H. Ropper, R. D. Adams, R. F. Brown and M. Victor, Adams and Victor's Principles of Neurology, McGraw-Hill Medical Pub. Division, New York, 2005, ISBN 0-07-141620-X, pp. 686–704 Search PubMed.
- S. Mendis, Int. J. Stroke, 2010, 5, 86–91 CrossRef PubMed.
- P. F. Cheng, J. X. Wang and W. H. Shao, J Stroke Cerebrovasc Dis, 2016, 25, 1326–1334 CrossRef PubMed.
- S. Mendis, Int. J. Stroke, 2013, 8, 3–4 CrossRef PubMed.
- S. Ellis and M. Small, Stroke, 1997, 28, 67–71 CrossRef PubMed.
- C. S. Kase, P. A. Wolf, M. Kelly-Hayes, W. B. Kannel, A. Beiser and R. B. D'Agostino, Stroke, 1998, 29, 805–812 CrossRef PubMed.
- R. R. Heuser, Prog. Cardiovasc. Dis., 2017, 59, 549–554 CrossRef PubMed.
- N. E. Husseini and D. T. Laskowitz, Expert Rev. Neurother., 2010, 10, 189–203 CrossRef PubMed.
- S. J. Kim, G. J. Moon and O. Y. Bang, J. Stroke, 2013, 15, 27–37 CrossRef PubMed.
- X. J. Wang, A. H. Zhang, Y. Han, P. Wang, H. Sun, G. C. Song, T. W. Dong, Y. Yuan, X. X. Yuan, M. Zhang, N. Xie, H. Zhang, H. Dong and W. Dong, Mol. Cell. Proteomics, 2012, 11, 370–380 CrossRef PubMed.
- O. Fiehn, Curr Protoc Mol Biol, 2016, 114, 1–32 Search PubMed.
- S. H. Lin, N. Liu, Z. Yang, W. J. Song, P. Wang, H. L. Chen, M. Lucioe, P. S. Koppline, G. N. Chen and Z. W. Cai, Talanta, 2010, 83, 262–268 CrossRef PubMed.
- H. X. Sun, J. Y. Zhao, D. Zhong and G. Z. Li, PLoS One, 2017, 12, e0189009 CrossRef PubMed.
- J. Y. Jung, H. S. Lee, D. G. Kang, N. S. Kim, M. H. Cha, O. S. Bang, D. H. Ryu and G. S. Hwang, Stroke, 2011, 42, 1282–1288 CrossRef PubMed.
- D. Wang, J. Kong, J. Y. Wu, X. X. Wang and M. C. Lai, Neurosci. Lett., 2017, 642, 7–13 CrossRef PubMed.
- X. L. Wan, X. H. Xia, M. Liu and C. F. Zhang, Guangdong Med. J., 2014, 35, 3788–3791 Search PubMed.
- T. Sangster, H. Major, R. Plumb, A. J. Wilson and I. D. Wilson, Analyst, 2006, 131, 1075–1078 RSC.
- L. Z. Yi, J. He, Y. Z. Liang, D. L. Yuan, H. Y. Gao and H. H. Zhou, Chem. Phys. Lipids, 2007, 150, 204–216 CrossRef PubMed.
- J. H. Huang, R. H. He, L. Z. Yi, H. L. Xie, D. S. Cao and Y. Z. Liang, Talanta, 2013, 110, 1–7 CrossRef PubMed.
- Y. Liu, Z. B. Lin, G. G. Tan, Z. Y. Chu, Z. Y. Lou, J. P. Zhang, Z. Y. Hong and Y. F. Chai, Metabolomics, 2013, 9, 1082–1095 CrossRef.
- J. H. Huang, L. Fu, B. Li, H. L. Xie, Y. H. Qin, Y. H. Wang, S. H. Zhang, H. Y. Huang, D. F. Liao and W. Wang, RSC Adv., 2015, 5, 58952–58958 RSC.
- H. Xiao, J. H. Huang, X. W. Zhang, R. Ahmed, Q. L. Xie, B. Li, Y. M. Zhu, X. Cai, Q. H. Peng, Y. H. Qin, H. Y. Huang and W. Wang, Pancreatology, 2017, 17, 543–549 CrossRef PubMed.
- X. L. Wan, X. H. Xia, H. Y. Qi, M. Liu, C. F. Zhang and D. P. Zhang, Tradit. Chin. Drug Res. Clin. Pharmacol., 2013, 24, 588–592 Search PubMed.
- S. Wein, S. Wolffram, J. Schrezenmeir, D. Gašperiková, I. Klimeš and E. Šeböková, Diabetes/Metab. Res. Rev., 2009, 25, 185–194 CrossRef PubMed.
- R. Rabøl, P. M. Højberg, T. Almdal, R. Boushel, S. B. Haugaard, S. Madsbad and F. Dela, J. Clin. Endocrinol. Metab., 2009, 94, 1372–1378 CrossRef PubMed.
- B. Yang, F. Ding, F. L. Wang, J. Yan, X. W. Ye, W. Yu and D. Li, Sci. Rep., 2016, 6, 23446 CrossRef PubMed.
- G. Boden, Curr. Opin. Endocrinol., Diabetes Obes., 2011, 18, 139–143 CrossRef PubMed.
- P. Kishore, W. J. Li, J. Tonelli, D. E. Lee, S. Koppaka, K. H. Zhang, Y. Lin, S. Kehlenbrink, P. E. Scherer and M. Hawkins, Sci. Transl. Med., 2010, 2, 20ra15 Search PubMed.
- S. R. Kashyap, R. Belfort, E. Cersosimo, S. Lee and K. Cusi, J. CardioMetab. Syndr., 2008, 3, 141–146 CrossRef PubMed.
- D. Y. Yang, H. C. Pan, Y. J. Yen, C. C. Wang, Y. H. Chuan, S. Y. Chen, S. Y. Lin, S. L. Liao, S. L. Raung, C. W. Wu, M. C. Chou, A. N. Chiang and C. J. Chen, NeuroToxicology, 2007, 28, 1220–1229 CrossRef PubMed.
- J. Z. Chen, Y. F. Zhao, C. Huang, C. E. Yao and X. P. Zhu, J. Clin. Res., 2004, 21, 490–491 Search PubMed.
- Y. N. Zhang, Y. Xing, H. Li, S. N. Li and L. Xu, J. Brain Nerv. Dis., 2016, 24, 108–112 Search PubMed.
- I. Riballena, A. Penalba, D. Pelegrí, A. Vilar, C. I. Jarca, J. Filomena, J. Montaner and P. Delgado, Atherosclerosis, 2014, 237, 811–815 CrossRef PubMed.
- L. Makowski and G. S. Hotamisligil, Curr. Opin. Lipidol., 2005, 16, 543–548 CrossRef PubMed.
- J. S. Pankow, D. J. Couper, B. B. Duncan, R. C. Hoogeveen, M. I. Schmidt, S. H. Golden and C. M. Ballantyne, Diabetes Care, 2004, 27, 77–82 CrossRef PubMed.
- K. L. Liu, Y. C. Yang, H. T. Yao, T. W. Chia, C. Y. Lu, C. C. Li, H. J. Tsai, C. K. Lii and H. W. Chen, Mol. Nutr. Food Res., 2016, 60, 430–443 CrossRef PubMed.
- L. Ménégaut, C. Thomas, L. Lagrost and D. Masson, Curr. Opin. Lipidol., 2017, 28, 19–26 Search PubMed.
- M. S. Nielsen, E. B. Schmidt, J. Stegger, A. Gorstrasmussen, A. Tjonneland and K. Overvad, Atherosclerosis, 2013, 227, 386–390 CrossRef PubMed.
- M. J. Shao, Chin. J. Neurol., 2016, 49, 215–221 Search PubMed.
- H. Naess, H. I. Nyland, L. Thomassen, J. Aarseth and K. M. Myhr, Eur. J. Neurol., 2005, 12, 194–198 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra03132k |
| ‡ The first two authors have equal contribution to this article. |
| This journal is © The Royal Society of Chemistry 2018 |