Open Access Article
Open Access ArticleCyclization of secondarily structured oligonucleotides to single-stranded rings by using Taq DNA ligase at high temperatures†
Yixiao Cui‡
 a,
Xutiange Han‡a,
Ran An
a,
Xutiange Han‡a,
Ran An *ab,
Guangqing Zhoua,
Makoto Komiyamaac and
Xingguo Liang
*ab,
Guangqing Zhoua,
Makoto Komiyamaac and
Xingguo Liang *ab
*ab
aCollege of Food Science and Engineering, Ocean University of China, Qingdao 266003, China. E-mail: ar@ouc.edu.cn; liangxg@ouc.edu.cn
bLaboratory for Marine Drugs and Bioproducts, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266003, China
cNational Institute for Materials Science (NIMS), Namiki, Tsukuba 305-0044, Japan
First published on 23rd May 2018
Abstract
Single-stranded DNA rings play important roles in nanoarchitectures, molecular machines, DNA detection, etc. Although T4 DNA ligase has been widely employed to cyclize single-stranded oligonucleotides into rings, the cyclization efficiency is very low when the oligonucleotides (l-DNAs) take complicated secondary structures at ambient temperatures. In the present study, this problem has been solved by using Thermus aquaticus DNA ligase (Taq DNA ligase) at higher temperatures (65 and 70 °C) where the secondary structures are less stable or completely destroyed. This method is based on our new finding that this ligase successfully functions even when the splint strand is short and forms no stable duplex with l-DNA (at least in the absence of the enzyme). In order to increase the efficiency of cyclization, various operation factors (lengths and sequences of splint, as well as the size of the DNA ring) have been investigated. Based on these results, DNA rings have been successfully synthesized from secondarily structured oligonucleotides in high yields and high selectivity. The present methodology is applicable to the preparation of versatile DNA rings involving complicated secondary structures, which should show novel properties and greatly widen the scope of DNA-based nanotechnology.
Introduction
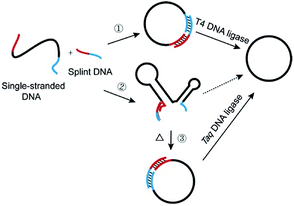
Recently, much attention has been focused on DNA-based nanoarchitectures which are applicable to nanomachines, drug delivery systems, etc.1–4 Among the many nanostructures fabricated from DNA, single-stranded DNA rings are characterized by dynamic and mobile features which cannot be otherwise fulfilled, and they are one of the most important and promising parts in nanoarchitectonics.5–7 For example, catenanes, in which two or more DNA rings are mechanically interlocked, have been elegantly employed in molecular switches, molecular motors, and switchable catalysts.7–10 Versatile applications of DNA rings to molecular biology, medicine, and biotechnology have also been reported.11–19 In most cases, DNA rings were prepared by cyclizing linear single-stranded DNA (l-DNA) with the use of T4 DNA ligase at room temperature.20–23 The cyclization has been greatly facilitated by adding an oligonucleotide (splint strand), which is complementary with the 3′- and 5′-terminal portions of l-DNA (see pathway 1 in Scheme 1). There are two critical problems which must be solved for more efficient production of DNA rings and also for their more sophisticated applications. The first problem is the formation of enormous amounts of polymeric byproducts in their production processes, which has been well recognized for a long time. The targeted intramolecular cyclization is inevitably accompanied by intermolecular polymerization of l-DNA, and thus the yield and selectivity for desired DNA rings are limited. The most popular solution is high dilution method, in which the intermolecular reactions are suppressed by decreasing the concentration of l-DNA.24,25 By this strategy, however, the amounts of DNA rings obtainable are intrinsically small. Recently, this problem has been solved, at least for T4 DNA ligase-mediated cyclization, by employing abnormally diluted buffers as well as a step-by-step operation, since intermolecular polymerization was minimized under these conditions.26Another critical problem is the formation of secondary structures of DNAs in solutions at ligation temperatures, which hampers the formation of DNA rings and should also govern the scope of applications of DNA rings. In spite of its essential roles, this factor has never been attracting much attention to date. An eminent solution to this problem is the primary concern of this paper. In contrast with rigid helix-structure of double-stranded DNA, single-stranded DNA is flexible and easily folded to complicated secondary structures through intramolecular base-pairing and/or stacking interactions.27 In fact, many of naturally occurring single-stranded DNAs have been reported to form secondary structures, which strongly affect their biological functions.28 Various functional DNAs artificially developed (e.g. aptamers, G quadruplex and DNAzymes) are also complicatedly folded.4 Besides, these secondary structures were often designed on DNA rings to construct molecular machines.29 In these structures, the two ends of oligonucleotide are often oriented unfavorably to be intramolecularly cyclized into rings. The secondary structures should also suppress the binding of splint strand to l-DNA at lower temperatures, since additional energy is required to deform the structures. Accordingly, the versatility of l-DNAs available (thus of DNA rings) is significantly limited in conventional methods (pathway 2 in Scheme 1). Furthermore, the secondary structures in DNA rings, if any, should strongly affect the dynamic, biological, and other properties of nanostructures, and thus unique rings of unprecedented features and potentialities should be obtainable. Accordingly, enzymatic cyclization of secondarily structured oligonucleotide is an urgent topic in the relevant science.
In this study, l-DNAs having complicated secondary structures are efficiently and selectively cyclized to DNA rings at high temperatures where these structures are unstable (pathway 3 in Scheme 1). In place of heat-sensitive T4 DNA ligase, thermostable Taq DNA ligase from aquatic thermophilic bacteria30–32 is successfully used on the basis of our unexpected finding of new aspect of this enzyme (see the Results section). Various reaction conditions (the length of splint strand, GC content in the splint, size of DNA ring, and the concentration of Taq DNA ligase buffer) are optimized to suppress the formation of polymeric byproducts. As the results, single-stranded DNA rings of various sizes are synthesized in high yield and selectivity, even when the corresponding l-DNA is folded to complicated secondary structure in solutions at room temperature. Without complicated purification procedures, DNA rings of high purity (>90%) have been easily obtained in a preparative scale.
Materials and methods
Materials
Single-stranded oligonucleotides (in Tables 1 and S1†) were purchased from GENEWIZ (Suzhou, China). The sequence of l-DNA59 was a part of l-DNA74, which was from phage M13, and their secondary structures were determined by Mfold calculation.33 Prior to the cyclization experiments, a phosphate residue was enzymatically introduced to the 5′-position of these linear DNAs (l-DNAs) by using T4 polynucleotide kinase (Thermo Scientific, Pittsburgh, PA. USA). The products were purified by PAGE. Thermus aquaticus (Taq) DNA ligase was obtained from NEW ENGLAND BioLabs (Beijing, China), together with 10× Taq DNA ligase buffer. T4 DNA ligase was purchased from Thermo Scientific (Pittsburgh, PA. USA), together with 10× T4 DNA ligase buffer. Exonuclease I was from Thermo Scientific (Pittsburgh, PA. USA). All other chemicals were from Sigma-Aldrich (St. Louis, MO. USA).| Name | Sequences (5′ → 3′) | Length (nt) |
|---|---|---|
| l-DNA59 | GACGTTGGAGTCCACGTTCTTTAATAGTGGACTCTTGTTCCAAACTGGAACAACACTCA | 59 |
| splint59(6+6) | AACGTCTGAGTG | 12 |
| splint59(13+12) | GGACTCCAACGTCTGAGTGTTGTTC | 25 |
| l-DNA74 | CCTTTGACGTTGGAGTCCACGTTCTTTAATAGTGGACTCTTGTTCCAAACTGGAACAACACTCAACCCTATCTC | 74 |
| splint74(6+6) | CAAAGGGAGATA | 12 |
| splint74(13+12) | CCAACGTCAAAGGGAGATAGGGTTG | 25 |
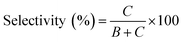 | (1) |
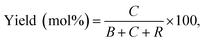 | (2) |
Results
Successful cyclization of l-DNA by Taq DNA ligase at high temperatures where the splint hardly forms stable duplex with l-DNA
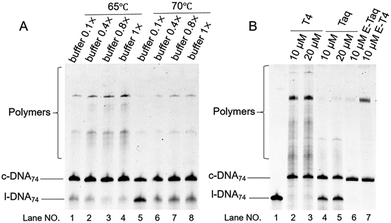
The secondarily structured DNAs we used here have apparent Tm of around 65 °C (the values of l-DNA59 and l-DNA74, measured by high resolution melting method,34,35 are 65.1 and 66.4 °C respectively; Fig. S1†). Thus, the enzymatic cyclization of these DNAs into rings must be achieved at 65 °C or higher where the secondary structures are largely deformed. However, this requirement is not easily compatible with the reaction conditions conventionally used for Taq DNA ligase, especially for a relatively short splint. As far as we know, most of the previous ligations of nicked DNA were carried out under the conditions that the duplexes in both sides of nick site are sufficiently long and stably formed at the reaction temperature. For example, a splint of 40 nt or longer was often employed for the ligation at 60 °C or lower.36–38 However, such long splints cannot be used for selective DNA ring production, since they cover a large portion of l-DNAs and the cyclic intermediates are hard to be formed due to higher rigidity, especially for a short l-DNA. Very recently, however, we have unexpectedly found that much shorter splint is sufficiently effective for this enzymatic ligation at 65 °C (Fig. S2†). The ligation satisfactorily proceeded even with the use of an 18 nt splint, which formed 9 bp duplexes on both sides of the nick. Both duplexes have low Tm values (50 °C or lower), and are never stably formed at 65 °C, at least in the absence of the enzyme. Apparently, temporarily formed duplexes are responsible for this enzymatic ligation. The binding between l-DNA and splint could be promoted by the assistance of the enzyme. These remarkable findings have paved the way to the production of single-stranded DNA rings with the use of Taq DNA ligase and short splint (e.g. 25 nt) at 65 °C, where the secondary structures of l-DNAs are unstable. Certainly, similar cyclization can be also successful for l-DNAs without obvious secondary structures (date not shown).Superiority of Taq DNA ligase over T4 DNA ligase to cyclize secondarily structured DNA into DNA ring
In Fig. 1, two linear DNAs (l-DNAs) were treated with Taq DNA ligase and T4 DNA ligase in the presence of two splints of different lengths (the sequences are shown in Table 1). The reactions were achieved in 1× Taq DNA ligase buffer and 1× T4 DNA ligase buffer from commercial sources. According to Mfold calculation ([Mg2+] = 10 mM at 25 °C), both of l-DNA59 and l-DNA74 (59 and 74 nt) take complicated secondary structures at ambient temperatures, which are unfavorable for the cyclization (see A and B in Fig. 1). Accordingly, l-DNA59 was hardly cyclized by T4 DNA ligase at 25 °C, when 12 nt oligonucleotide (splint59(6+6)) was used as helper strand (lane 2). Only intermolecular ligation of several l-DNA59 molecules occurred to produce the polymeric byproducts (the bands of low mobility in the top of gel). In order to deform the secondary structure involving two loops and allow the cyclization, a longer splint is necessary. Thus, 25 nt splint (splint59(13+12)), which is complementary with 13 nt in the 5′-end of l-DNA59 and 12 nt in the 3′-end, was used for cyclization by T4 DNA ligase in lane 3. Some amount of DNA ring (c-DNA59) was formed (the band at the bottom of the gel). However, the polymers were still the major products, and the selectivity for the DNA ring was only 38%.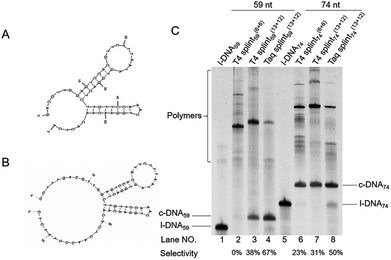 | ||
| Fig. 1 Cyclization of l-DNAs by Taq DNA ligase and T4 DNA ligase. Both l-DNA59 (A) and l-DNA74 (B) take secondary structures, as determined by Mfold calculation ([Mg2+] = 10 mM at 25 °C). In (C), the cyclization was carried out under the following conditions: [l-DNA]0 = 5 μM and [12 or 25 nt splint] = 10 μM. Lane 1, l-DNA59 only; lane 2, DNA59 with splint59(6+6) at 25 °C by T4 DNA ligase; lane 3, l-DNA59 with splint59(13+12) at 25 °C by T4 DNA ligase; lane 4, l-DNA59 with splint59(13+12) at 65 °C by Taq DNA ligase; lane 5, l-DNA74 only; lane 6, l-DNA74 with splint74(6+6) at 25 °C by T4 DNA ligase; lane 7, l-DNA74 with splint74(13+12) at 25 °C by T4 DNA ligase; lane 8, l-DNA74 with splint74(13+12) at 65 °C by Taq DNA ligase. Prior to the reactions, l-DNAs were phosphorylated at the 5′-position by using T4 polynucleotide kinase. The sequences of l-DNAs and splints are listed in Table 1 and detailed reaction conditions are shown in Experimental section. The Tm of l-DNA59 and l-DNA74 was 65.1 °C and 66.4 °C (Fig. S1†). | ||
In order to destroy the secondary structure of l-DNA59 and promote its intramolecular cyclization, the reaction was carried out at 65 °C by Taq DNA ligase (lane 4). It is worth mentioning that the apparent Tm of l-DNA59 is 65.1 °C (the corresponding value of l-DNA74 is 66.4 °C; Fig. S1†). Exactly as designed, the formation of polymeric by-products were greatly suppressed, and the DNA ring was now obtained as the dominant product (67% selectivity). This method was also applicable to the cyclization of l-DNA74, a longer oligonucleotide taking similar structure to l-DNA59 (see Fig. 1B). By using Taq DNA ligase and 25 nt splint (splint74(13+12)) at 65 °C, the DNA ring was prepared in 50% selectivity. This selectivity is much higher than the corresponding value (31%) for the cyclization using T4 DNA ligase at 25 °C (compare lane 8 with lane 7). The ring structures of the products obtained by the present method have been confirmed in terms of complete resistance against Exonuclease I (Fig. S3†). It is concluded that thermostable Taq DNA ligase is a highly powerful and advantageous tool, compared with conventionally employed T4 DNA ligase, to prepare DNA rings from secondarily structured oligonucleotides. Note that a relatively higher DNA concentration (5 μM l-DNA) was used here. This advantage of Taq DNA ligase can be further amplified in diluted Taq DNA ligase buffers, and the selectivity for the formation of DNA rings is further increased up to almost 100% (vide infra).
Optimization of reaction conditions for efficient formation of DNA ring by Taq DNA ligase
 | ||
| Fig. 2 Effects of the length of splint on the cyclization of l-DNA74 by Taq DNA ligase at (A) 65 °C and (B) 70 °C. The reaction conditions: [l-DNA74]0 = 5 μM; [splint] = 10 μM; 40 U Taq DNA ligase (in 20 μL). In (C) and (D), the yields and the selectivity at 65 °C (circles) and 70 °C (triangles) are plotted as a function of splint length. The results in lanes 2 and 3 in (A) and lanes 2–4 in (B) were not incorporated to the plots in (D), since the band intensities of the products were too small to determine their ratios precisely. The sequences of splints are listed in Table S1.† | ||
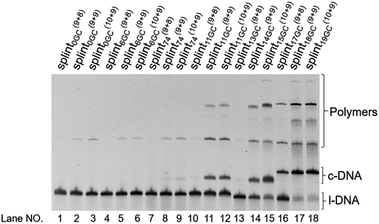 | ||
| Fig. 3 Cyclization of l-DNA (74 nt) by Taq DNA ligase using the splints of various GC contents. Reaction conditions: [l-DNA]0 = 5 μM, [splint] = 10 μM, and 40 U Taq DNA ligase (in 20 μL) at 65 °C for 12 h. The sequences of all the l-DNAs and the splints are shown in Table S1.† | ||
In the lanes adjacent to the lanes described above, the splints are 1 nt shorter (in the 3′-side) or 1 nt longer (in the 5′-side) than the parent splints.
These truncated and grafted splints (9 + 8 and 10 + 9, respectively) provided virtually the same results with respect to GC contents as the parent 18 nt splint. Necessity of sufficient number of G-C pairs for the cyclization has been further confirmed. The absence of ring products in lanes 10 and 13 (with 9 + 8 splints) indicates that the splint of 18 nt (9 + 9) or longer is desirable for efficient catalysis by Taq DNA ligase under the conditions employed here, although some circular DNA was obtained when 17 nt splint with 100% GC content was used (lane 16). Consistently, no products of intermolecular polymerization are observed in lanes 10 and 13.
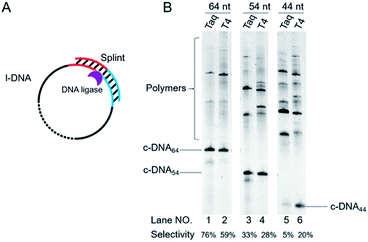 | ||
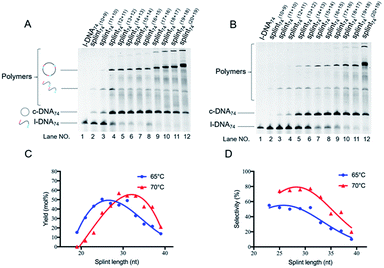
| Fig. 4 Schematic of view of cyclization of l-DNA of different lengths by Taq DNA ligase (A). The reaction conditions in (B): [l-DNA64, l-DNA54, or l-DNA44]0 = 5 μM, [splint74(15+14)] = 10 μM, and 40 U Taq DNA ligase (in 20 μL) at 65 °C for 12 h. For the purpose of comparison, [splint74(6+6)] = 10 μM and the results with T4 DNA ligase at 25 °C are also presented. The sequences of l-DNAs and splints are listed in Table S1.† | ||
| Enzyme | [l-DNA74]0 (μM) | Product analysis | Purity (%) of DNA ring after Exonuclease I digestionc | |
|---|---|---|---|---|
| Yield (mol%) | Selectivity (%) | |||
| a [l-DNA74]0/[splint74(15+14)] = 1/2, Taq DNA ligase = 40 U (in 20 μL), 0.1× Taq DNA ligase buffer, 70 °C, 12 h.b [l-DNA74]0/[splint74(15+14)] = 1/2, T4 DNA ligase = 10 U (in 20 μL), 0.1× T4 DNA ligase buffer, 25 °C, 12 h.c The purity of the DNA ring obtained after simple treatment of the enzymatic products with Exonuclease I (without any separation procedures). | ||||
| Taq ligasea | 10 | 58 | 91 | 94 |
| 20 | 46 | 75 | 83 | |
| T4 ligaseb | 10 | 40 | 40 | 61 |
| 20 | 29 | 29 | 46 | |
With the use of T4 DNA ligase in 0.1× T4 DNA ligase buffer at 25 °C, however, notable amounts of polymeric byproducts were formed when [l-DNA74]0 = 10 μM (lane 2 in Fig. 5B). The selectivity was 40%. At [l-DNA74]0 = 20 μM, the selectivity was only 29% (lane 3 in Fig. 5B and Table 2). Apparently, this method is not very useful to cyclize secondarily structured l-DNA, although it is powerful for the cyclization of l-DNA without secondary structures.26 Furthermore, it is noteworthy that the major byproduct was a cyclic oligomer, which is resistant against Exonuclease I (the band near the top of gel). Thus, even after the digestion of the product in lane 2 by Exonuclease I, the desired “monomeric” ring (c-DNA74) was contaminated with notable amount of this cyclic oligomer (lane 7 in Fig. 5B), and its purity was only 61% (see Table 2). Superiority of Taq DNA ligase for the production of DNA rings is completely evidenced.
Discussion
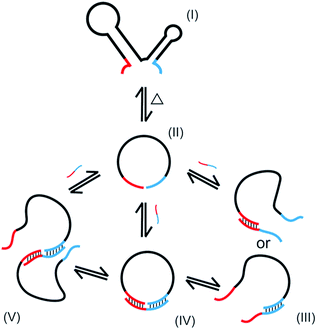
In the present study, oligonucleotides having complicated secondary structure in solutions (l-DNA) are for the first time intramolecularly cyclized in high yield and selectivity to single-stranded DNA rings. By using thermostable Taq DNA ligase at 65–70 °C, the problems of conventional method using T4 DNA ligase (dominant production of polymeric byproducts and the resultant low selectivity for the desired DNA rings) have been solved. The essential points of the present preparation of DNA rings are (i) to achieve the cyclization at high temperatures in which the secondary structures are unstable, (ii) to use splint strand of appropriate length and GC content which binds with l-DNA with reasonable stability at high temperatures, and (iii) to decrease the concentration of Taq DNA ligase buffer to minimize the production of polymeric byproducts.The NAD+-dependent Taq DNA ligase, like ATP-dependent T4 DNA ligase, also involves the lysine-mediated adenylation of the 5′-phosphate at the end of one DNA strand.30 In the following step, the resultant pyrophosphate is attacked by the 3′-OH of an adjacent DNA strand, joining two strands by a stable phosphodiester linkage. In the mixtures for the present cyclization, multiple equilibria hold as schematically depicted in Scheme 2 (only the minimum set of species, some of which are only temporarily formed, is shown here). The oligonucleotide is folded to complicated secondary structure I in solutions, but deformed to less structured form II at higher temperature. When a splint strand is added to the solution, several composite species are formed in equilibria. In III, either 3′-end of l-DNA or its 5′-end binds to the splint, whereas both ends simultaneously bind to the splint in IV. The composite IV is the major active species for the intramolecular cyclization. The binding of the splint to l-DNA could be promoted by the assistance of Taq DNA ligase. When two molecules of II are bound each other by one splint, the composite V is formed, which is responsible for the formation of polymeric byproducts. Note that the yield of DNA ring in the present enzymatic cyclization is primarily proportional to the concentration of the composite IV (with some contribution of III). On the other hand, the selectivity for the formation of DNA ring is governed by the ratio of the concentration of IV to that of V.
The first requisite for successful cyclization is the formation of the composite IV at high temperatures. Thus, the GC content in the splint (and the number of G-C pairs in the composite IV) must be sufficiently high (Fig. 3). When the splint length is systematically increased, the yield first increases (the left-hand side of the bell-shaped curves in Fig. 2C). In the right-hand sides of the curves, however, the increase in the splint length destabilizes IV due to increased strain in the cyclic structures of the composite. The length of single-stranded portion in the composite, which mostly militates the strain in the cyclic structure, is gradually shortened and becomes ineffective (note that the double-stranded portions are too rigid for this purpose). As the result, the intramolecular cyclization becomes less efficient when the splint is too long. This argument fairly agrees with the results in Fig. 4 which shows critical importance of the size of DNA rings for the present enzymatic cyclization. As shown in Fig. 2D, the selectivity for the production of DNA ring is virtually constant irrespective of the splint length up to 31 nt, since [IV]/[V] ratio is not much changed here. The secondary structure has been almost completely destroyed, so that the binding mode of splint to l-DNA in IV (for the intramolecular cyclization) is similar to that in V (for the intermolecular polymerization). With further increase of splint length, however, IV is destabilized by the increased strain of cyclic structure, as described above, decelerating the intramolecular cyclization. On the other hand, the stability of intermolecular composite V is never affected by this factor, and thus the [IV]/[V] ratio (and thus the selectivity) is steeply decreased with increasing splint length.
Interestingly, the Tm of splint/l-DNA for the optimal length (25 nt) of splint was only 55.2 °C (date not shown), which is about 10 °C lower than the reaction temperature of 65 °C. Similarly, for the ligation at 70 °C, the optimal length of splint was 29 nt, and the Tm of splint/l-DNA (60.8 °C, date not shown) was also about 10 °C lower than the reaction temperature. Longer splints gave more polymeric byproducts, indicating that a Tm of about 10 °C lower than the reaction temperature is favorable for cyclization.
By reducing the concentration of Taq DNA ligase buffer, the selectivity for the formation of DNA rings was greatly promoted (Fig. 5). The formation of polymeric byproducts was almost completely suppressed in 0.1× Taq DNA ligase buffer, and the selectivity for the ring formation was close to 100%. This result is consistent with the previous finding in the cyclization by T4 DNA ligase that concurrent polymerization was suppressed in highly diluted T4 DNA ligase buffer.26 The remarkable effect of T4 DNA ligase buffer concentration was ascribed to dominant contribution of nonprocessive ligation process in these buffers, in which the ligase dissociates from the DNA during the catalysis.39,40 Compared with processive ligation in which all the catalysis is completed without dissociation of the enzyme, entropic advantage of the intramolecular ligation (cyclization) over the intermolecular polymerization is more significant. Whether a similar mechanism is in fact operative in the present cyclization or not cannot be concluded at present, since detailed kinetic information on Taq DNA ligase is not available.
By the present method, DNA rings of various of sizes and sequences can be prepared, which should be significant for further developments of nanoarchitectonics. The selectivity of these reactions is satisfactorily high. Unfortunately, however, Taq DNA ligase, which is currently available from commercial sources may be not completely stable at higher temperatures. Accordingly, the cyclization does not easily attain 100% completion (in most cases, the conversion is 50–80%, Fig. S5†). Further increase of the thermostability of this enzyme through mutation experiments and/or discovery of new thermostable DNA ligases should be one of the solutions, and promote the practical usefulness of the present methodology still more.
Conclusion
Even when oligonucleotides are folded to complicated secondary structures in solutions and otherwise hardly cyclized, DNA rings can be successfully prepared by using Taq DNA ligase at high temperatures. The use of Taq DNA ligase is absolutely necessary, since conventionally employed T4 DNA ligase overwhelmingly produces polymeric byproducts and the amount of ring products is only limited. To guarantee both yield and selectivity of DNA rings using Taq DNA ligase, G-C rich splints of sufficient length (e.g. 25–29 nt) must be used. Furthermore, diluted Taq DNA ligase buffer significantly suppresses the formation of polymeric byproducts. Accordingly, desired DNA rings can be easily obtained in sufficiently high purity, simply by (i) cyclizing the corresponding oligonucleotide by Taq DNA ligase at 70 °C in 0.1× Taq DNA ligase buffer, and (ii) treating the reaction product with Exonuclease I to remove non-cyclic DNAs. No complicated purification procedures are necessary. Many single-stranded DNAs take secondary structures so that the scope of the DNA rings available (and of the relevant nanoarchitectonics) should be greatly extended by this finding. Furthermore, DNA rings involving complicated secondary structures can be easily obtained by the present method using Taq DNA ligase. They should show novel properties in chemical, physical, and biological functions, and are highly useful components for future nanoarchitectonics.Conflicts of interest
None declared.Acknowledgements
This work was supported by National Natural Science Foundation of China [31571937 to X. L.]; and the Fundamental Research Funds for the Central Universities [201713050 to R. A.]. Funding for open access charge: National Natural Science Foundation of China.References
- K. Wang, M. Q. He, F. H. Zhai, J. Wang, R. H. He and Y. L. Yu, Biosens. Bioelectron., 2018, 105, 159–165 CrossRef PubMed.
- H. Xu, Y. Jiang, D. Liu, K. Liu, Y. Zhang, S. Yu, Z. Shen and Z. S. Wu, Anal. Chim. Acta, 2018, 1011, 86–93 CrossRef PubMed.
- C. Teller and I. Willner, Curr. Opin. Biotechnol., 2010, 21, 376–391 CrossRef PubMed.
- M. Komiyama, K. Yoshimoto, M. Sisido and K. Ariga, Bull. Chem. Soc. Jpn., 2017, 90, 967–1004 CrossRef.
- C. Gaillard and F. Strauss, PLoS One, 2015, 10, e0119368 Search PubMed.
- L. Finn, A. Damian and F. Michael, J. Am. Chem. Soc., 2012, 134, 11884–11887 CrossRef PubMed.
- C. H. Lu, A. Cecconello, X. J. Qi, N. Wu, S. S. Jester, M. Famulok, M. Matthies, T. L. Schmidt and I. Willner, Nano Lett., 2015, 15, 7133–7137 CrossRef PubMed.
- X. J. Qi, C. H. Lu, A. Cecconello, H. H. Yang and I. Willner, Chem. Commun., 2014, 50, 4717–4720 RSC.
- C. H. Lu, A. Cecconello and I. Willner, J. Am. Chem. Soc., 2016, 138, 5172–5185 CrossRef PubMed.
- L. Hu, C. H. Lu and I. Willner, Nano Lett., 2015, 15, 2099–2103 CrossRef PubMed.
- X. Liang, H. Kuhn and M. D. Frank-Kamenetskii, Biophys. J., 2006, 90, 2877–2889 CrossRef PubMed.
- E. T. Kool, Annu. Rev. Biophys. Biomol. Struct., 1996, 25, 1–28 CrossRef PubMed.
- X. Wang, C. Li, X. Gao, J. Wang and X. Liang, Mol. Ther.--Nucleic Acids, 2015, 4, e215 CrossRef PubMed.
- Y. Liu, A. Kuzuya, R. Sha, J. Guillaume, R. Wang, J. W. Canary and N. C. Seeman, J. Am. Chem. Soc., 2008, 130, 10882–10883 CrossRef PubMed.
- H. Kuhn, V. V. Demidov and M. D. Frank-Kamenetskii, Nucleic Acids Res., 2002, 30, 574–580 CrossRef PubMed.
- J. Marciniak, A. Kummel, S. Esener, M. Heller and B. Messmer, BioTechniques, 2008, 45, 275–280 CrossRef PubMed.
- D. Y. Zhang, M. Brandwein, T. Hsuih and H. B. Li, Mol. Diagn., 2001, 6, 141–150 CrossRef PubMed.
- R. Deng, L. Tang, Q. Tian, Y. Wang, L. Lin and J. Li, Angew. Chem., Int. Ed. Engl., 2014, 53, 2389–2393 CrossRef PubMed.
- T. Murakami, J. Sumaoka and M. Komiyama, Nucleic Acids Res., 2009, 37, e19 CrossRef PubMed.
- A. E. Tomkinson, S. Vijayakumar, J. M. Pascal and T. Ellenberger, Chem. Rev., 2006, 106, 687–699 CrossRef PubMed.
- A. J. Doherty and T. R. Dafforn, J. Mol. Biol., 1999, 296, 43–56 CrossRef PubMed.
- R. Rossi, A. Montecucco, G. Ciarrocchi and G. Biamonti, Nucleic Acids Res., 1997, 25, 2106–2113 CrossRef PubMed.
- J. D. Kahn and D. M. Crothers, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 6343–6347 CrossRef.
- Z. S. Wu, Z. Shen, K. Tram and Y. Li, Nat. Commun., 2014, 5, 4279 CrossRef PubMed.
- M. Liu, Q. Zhang, Z. Li, J. Gu, J. D. Brennan and Y. Li, Nat. Commun., 2016, 7, 12074 CrossRef PubMed.
- R. An, Q. Li, Y. Fan, J. Li, X. Pan, M. Komiyama and X. Liang, Nucleic Acids Res., 2017, 45, e139 CrossRef PubMed.
- J. SantaLucia Jr and D. Hicks, Annu. Rev. Biophys. Biomol. Struct., 2004, 33, 415–440 CrossRef PubMed.
- D. Bikard, C. Loot, Z. Baharoglu and D. Mazel, Microbiol. Mol. Biol. Rev., 2010, 74, 570–588 CrossRef PubMed.
- L. Hu, C. H. Lu and I. Willner, Nano Lett., 2015, 15, 2099–2103 CrossRef PubMed.
- J. Tong, F. Barany and W. Cao, Nucleic Acids Res., 2000, 28, 1447–1454 CrossRef PubMed.
- J. Luo, D. E. Bergstrom and F. Barany, Nucleic Acids Res., 1996, 24, 3071 CrossRef PubMed.
- J. Tong, W. Cao and F. Barany, Nucleic Acids Res., 1999, 27, 788–794 CrossRef PubMed.
- M. Zuker, Nucleic Acids Res., 2003, 31, 3406–3415 CrossRef PubMed.
- J. Wang, P. Dong, W. Wu, X. Pan and X. Liang, J. Biomol. Struct. Dyn., 2018, 36, 1–13 Search PubMed.
- J. Wang, X. Pan and X. Liang, J. Anal. Methods Chem., 2016, 2016, 1–8 Search PubMed.
- J. Luo, D. E. Bergstrom and F. Barany, Nucleic Acids Res., 1996, 24, 3071–3078 CrossRef PubMed.
- G. J. S. Lohman, R. J. Bauer, N. M. Nichols, L. Mazzola, J. Bybee, D. Rivizzigno, E. Cantin and T. J. Evans, Nucleic Acids Res., 2016, 44, e14 CrossRef PubMed.
- J. Stewart, P. Kozlowski, M. Sowden, E. Messing and C. Smith, Nucleic Acids Res., 1998, 26, 961–966 CrossRef PubMed.
- A. V. Cherepanov and V. S. De, J. Biol. Chem., 2002, 277, 1695–1704 CrossRef PubMed.
- A. V. Cherepanov and V. S. De, Eur. J. Biochem., 2003, 270, 4315–4325 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra02804d |
| ‡ These authors contributed equally to this work as first authors. |
| This journal is © The Royal Society of Chemistry 2018 |