Open Access Article
Open Access ArticleYunnanterpene G, a spiro-triterpene from the roots of Cimicifuga foetida, downregulates the expression of CD147 and MMPs in PMA differentiated THP-1 cells†
Ni-Hong Lua,
Zi-Wei Zhangb,
Rui-Wei Guob,
Li-Xia Yang *b,
Ya-Xian Songa,
Jin-Shan Yeb and
Yan-Kun Shib
*b,
Ya-Xian Songa,
Jin-Shan Yeb and
Yan-Kun Shib
aDepartment of Postgraduate, Kunming Medical University, Yunnan 650500, P. R. China
bDepartment of Cardiology, Kunming General Hospital of Chengdu Military Area, Yunnan 650032, P. R. China. E-mail: doctorlxyang@126.com
First published on 20th April 2018
Abstract
A new cycloartane triterpene, yunnanterpene G (1), containing an oxaspiro[5.4]decane moiety, was purified from the roots of Cimicifuga foetida. The new structure was determined from spectroscopic data and the X-ray diffraction method. Biological evaluations revealed that compound 1 significantly inhibited the mRNA expression of the atherosclerosis-related adhesion molecule CD147 (extracellular matrix metalloproteinase inducer, EMMPRIN), and proteolytic enzymes matrix metalloproteinase 2 (MMP-2), MMP-9 and MMP-14, in a dose-dependent manner in phorbol-12-myristate-13-acetate-induced human monocytic THP-1 cells by quantitative real-time PCR method. At the same time, the migration ability of the induced THP-1 cells was potently inhibited. Furthermore, western blot experiments showed that compound 1 at 25 μM strongly suppressed phosphorylation of NF-κB p65 and p38 MAPK in the differentiated THP-1 cells.
1. Introduction
Atherosclerosis is a type of chronic inflammatory disease.1–3 Thus, proper regulation of the key inflammatory factors involved in this disease is crucial for therapeutic strategies. CD147, extracellular matrix metalloproteinase inducer (EMMPRIN) or basigin, is a member of the immunoglobulin super family.4 Ample evidence has highlighted the potential effect of CD147 in the pathological process of atherosclerosis.1,5–10 Especially, it has been proven that overexpressed CD147 in activated macrophages stimulated production of several matrix metalloproteinases (MMPs), including MMP-1, MMP-2, MMP-9, and MMP-14, in advanced atherosclerotic plaque, which is critical for plaque rupture and myocardial infarction.11–14 Moreover, anti-atherogenic drugs, such as fluvastatin, besides LDL (low-density lipoprotein) reducing effect, decreasing the MMPs and EMMPRIN levels also contribute to their clinical outcomes.15–17 Thus, substances that suppress EMMPRIN and MMP expression may be potential candidates for ameliorating the development of atherosclerosis.Cimicifuga spp. (now included in the genus of Actaea), such as C. racemosa, C. foetida, C. dahurica, C. heracleifolia, C. simplex, and C. japonica, are famous herb medicines in Europe, the United States, and East Asia.18–20 9,19-Cycloartane triterpenes (CTs) are the main active chemical profile of Cimicifuga genus18–20 and these CTs show diverse activities, such as cytotoxicity,21,22 anti-angiogenic,23 anti-inflammatory,24,25 neuro-protective,26 soluble epoxide hydrolase inhibitory,27 anti-antiosteoporotic,28 anti-AIDS,29 and anti-Alzheimer30 activities. However, knowledge of the regulation of EMMPRIN and MMPs by CTs from Cimicifuga spp. is still unknown. In the present study, a spiro-triterpene, yunnanterpene G (1), was isolated from the roots of C. foetida (Fig. 1). Although containing the same 16,23:23,26-diepoxy moiety, the spiro-atom C-23 in 1 was assigned as “R” with the help of X-ray crystallography, rather than “S” in the analogues yunnanterpene A–F.20 Significantly, quantitative real-time PCR (q-PCR) experiments showed that compound 1 noticeably suppressed the mRNA expression of CD147, and MMP-2, MMP-9 and MMP-14, in a dose-dependent manner (10 to 50 μM) in phorbol-12-myristate-13-acetate (PMA)-induced THP-1 cells (PMA differentiated THP-1 cell line is a representative macrophage model, in vitro31). Consequently, motility of the differentiated THP-1 cells was potently inhibited by 1 at the same concentration range. Furthermore, 1 markedly reduced phosphorylation of NF-κB p65 and p38 MAPK at a concentration of 25 μM. Described herein are the isolation, structure elucidation, and biological activities of compound 1.
2. Results and discussion
2.1. Structural elucidation of yunnanterpene G
Yunnanterpene G (1), colorless crystals, has the molecular formula C30H44O6 as determined by HRESIMS at m/z 523.3029 [M + Na]+ (calcd 523.3036), consistent with 9 sites of unsaturation. The IR spectrum showed absorptions ascribed to OH (3411 cm−1), carbonyl (1705 cm−1), and olefinic (1631 cm−1) groups. In the 1H NMR spectrum (Table 1), downfield shifted cyclopropane methylene signals at δH 0.86 and 1.29 (each 1H, brs), an olefinic proton resonance at δH 5.17 (dd, J = 6.1, 2.6 Hz), a secondary methyl signal at δH 1.47 (d, J = 6.6 Hz) and signals of five tertiary methyl groups at δH 1.01–1.91 were observed. In addition, the 13C NMR (Table 1) and HMQC spectra revealed the existence of a carbonyl group at δC 214.8 (C-3, s), a ketal carbon at 106.6 (C-23, s) and five oxygenated carbon atoms at δC 71.7 (C-12, d), 75.5 (C-16, d), 85.8 (C-24, d), 77.8 (C-25, s), and 78.1 (C-26, t). The aforementioned data suggested that 1 was a highly oxygenated CT containing a six-ring skeleton.| Position | 1Hb | 13Cc | Position | 1Hb | 13Cc |
|---|---|---|---|---|---|
| a Signals overlapped.b Recorded at 500 MHz in pyridine-d5.c Recorded at 150 MHz in pyridine-d5. | |||||
| 1 | 1.80 m, 1.51 brd (6.5) | 31.8 t | 16 | 4.84 dd (14.0, 7.4) | 75.5 d |
| 2 | 2.70 td (13.9, 6.3), 2.24 brd (11.7) | 36.8 t | 17 | 1.91a | 57.1 d |
| 3 | 214.8 s | 18 | 1.69 s | 14.3 q | |
| 4 | 50.1 s | 19 | 1.29 brs, 0.86 brs | 28.3 t | |
| 5 | 1.70a | 44.1 d | 20 | 2.62 m | 24.5 d |
| 6 | 1.70 m, 1.52 m | 21.9 t | 21 | 1.47 d (6.6) | 22.5 q |
| 7 | 5.17 dd (6.1, 2.6) | 113.1 d | 22 | 2.10 m, 2.04 m | 40.7 t |
| 8 | 149.1 s | 23 | 106.6 s | ||
| 9 | 22.5 s | 24 | 4.45 brd (5.1) | 85.8 d | |
| 10 | 27.7 s | 25 | 77.8 s | ||
| 11 | 2.82 dd (15.1, 8.9), 1.49 m | 39.8 t | 26 | 4.32 d (8.7), 4.24 d (8.7) | 78.1 t |
| 12 | 4.23a | 71.7 d | 27 | 1.91 s | 22.9 q |
| 13 | 49.5 s | 28 | 1.12 s | 22.5 q | |
| 14 | 48.8 s | 29 | 1.01 s | 19.9 q | |
| 15 | 2.14 m, 1.52 m | 43.8 t | 30 | 1.04 s | 26.8 q |
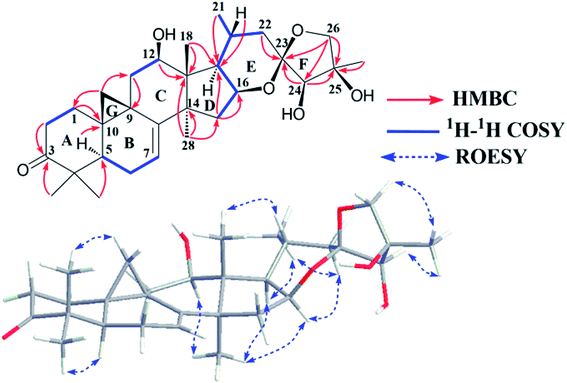
Study of the 1H–1H COSY (Fig. 1) and HSQC spectra of 1 revealed the existence of partial structures C-1/C-2 (–CH2CH2–), C-5/C-6/C-7 (–CHCH2CH–), C-11/C-12 (–CH2CH–) and a pair of geminal signals for CH2-19 at δH 0.86 and 1.29 (each 1H, brs), consistent with typical rings A, B, C, and G of CTs with a carbonyl group at C-3, a hydroxy group at C-12 and a pair of double bonds at C-7 and C-8.21–23 The HMBC correlations of H-2 (δH 2.70 and 2.24) and Me-29 (δH 1.01) to the carbonyl carbon at δC 214.8 (C-3), and H-11 (δH 2.82 and 1.49) to the hydroxymethine at δC 71.7 (C-12) and downfield shifted cyclopropane methylene (δH 0.86 and 1.29) further supported the aforementioned deduction.
The spin system –CH2CHCHCH(CH3)CH2– due to C-15, C-16, C-17, C-20, Me-21 and C-22 was further deduced from 1H–1H COSY correlations (Fig. 1). In the HMBC spectrum, associations from M-18 (δH 1.69) to C-13 (δC 49.5) and C-17 (δC 57.1), Me-28 (δH 1.12) to C-13 (δC 49.5), C-14 (δC 48.8), and C-15 (δC 43.8), and H-15 (δH 2.14 and 1.52) to C-17 (δC 57.1) established ring D as a cyclopentane ring. In addition, the linkages of the ketal carbon C-23 (δC 106.6) to C-22 (δC 40.7) and C-24 (δC 85.5) were deduced from the HMBC correlations of the methylene (H-22) protons (δH 2.10 and 2.04) and hydroxymethine (H-24) proton (δH 4.45) to C-23, respectively. Similarly, the connections of C-24, C-26 and Me-27 to C-25 were established by HMBC couplings of H-24 at δH 4.45, geminal signals of the oxygenated methylene protons (H-26) at δH 4.32 and 4.24 (each 1H, d, J = 8.7 Hz), and CH3-27 at δH 1.91 to C-25 (δC 77.8), respectively. Thus, to fulfill the unsaturation requirement, the molecular weight, and the chemical shift of C-16, C-23, C-25 and C-26, a 16,23:23,26-diepoxy moiety was established on the basis of rings E and F and the final hydroxy group was attached to C-25. Finally, the planar structure of 1 was constructed as shown.
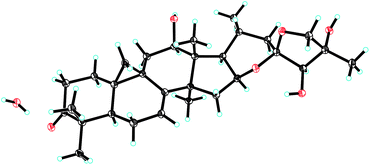
The relative configuration of 1 was established by the ROESY spectrum (Fig. 1). The cross-peaks of H-12, H-16 and H-17 with CH3-28 (biogenetically α-oriented) in the ROESY spectrum assigned their orientations as α. The α-orientation of CH3-21 was deduced from ROESY correlation of H-20 and CH3-18 (biogenetically β-oriented). In addition, CH3-27 (δH 1.91) showed correlations with H-24 (δH 4.45) and H-26b (δH 4.24), indicating same orientations of those protons and methyl in ring F. However, it was not possible to identify the absolute stereochemistry of C-23, C-24, and C-25 by ROESY experiment. Fortunately, 1 was crystallized from MeOH and analyzed by X-ray crystallography, which allowed unambiguous assignment of the absolute configuration by a Flack parameter of 0.07(11). Accordingly, the absolute configurations at C-23, C-24 and C-25 were assigned as R, R, and R, respectively (Fig. 2). Therefore, the structure of 1, named yunnanterpene G, was determined as shown.
2.2. Alteration of morphology and phenotype of PMA-induced THP-1 cells
Normal THP-1 cells are round in shape and do not adhere to the plastic surfaces of culture plates.32 In the presence of 100 nM PMA for 24 h, the cells became flat and amoeboid in shape, and adhered to the dish bottom (Fig. S11A and S11B†). Flow cytometry analysis revealed that 98% of the PMA-induced THP-1 cells were CD68 positive and 32% of these cells were CD11b positive, which confirmed the differentiation of monocyte to macrophage (Fig. S11C and S11D†).2.3. Effects of compound 1 on viability of PMA-induced THP-1 cells
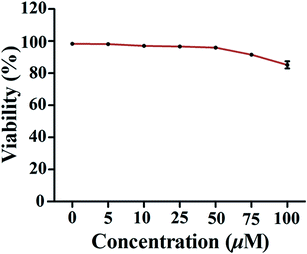
The MTT assay was employed to evaluate the effect of 1 on the viability of PMA-induced macrophages. As shown in Fig. 3, compound 1 only caused weak reduction (about 10–15%) in cell viability when the concentration was over 75 μM. Therefore, doses ranging from 10 to 50 μM used in subsequent experiments were considered to be non-cytotoxic.2.4. The effect of compound 1 on expression of CD147 and MMP mRNA of PMA-induced THP-1 cells
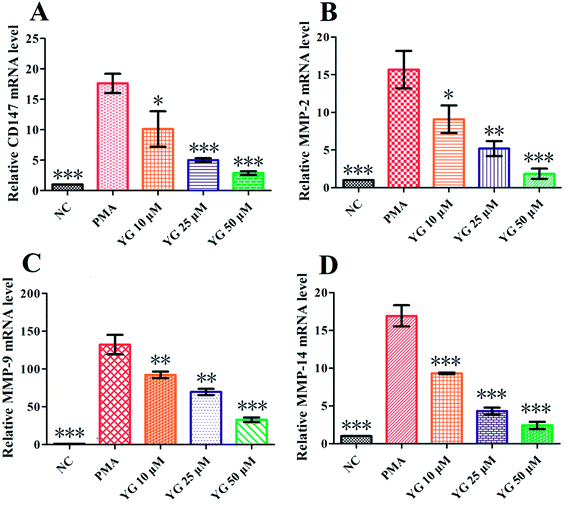
CD147 and MMP mRNA levels in PMA-differentiated THP-1 cells were tested by q-PCR using the 2−ΔΔCt method. As shown in Fig. 4A, the expression of CD147 was significantly higher in PMA-induced THP-1 cells than the NC group (p < 0.001). After 24 hours treatment with 1 on the differentiated THP-1 cells, CD147 mRNA expression was notably downregulated in the 10, 25, and 50 μM treatment groups (p < 0.05, p < 0.001, and p < 0.001, respectively; Fig. 4A). Since CD147 could stimulate production of MMP-2, MMP-9, and MMP-14 in the activated macrophages,11–14 the effects of compound 1 on mRNA expressions of these metalloproteinases were further studied. As shown in Fig. 4B–D, the levels of MMP-2, MMP-9, and MMP-14 mRNA were markedly and dose-dependently suppressed by 1.2.5. The effect of compound 1 on the migration of PMA-induced THP-1 cells
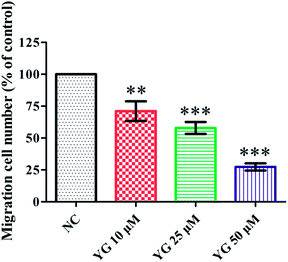
In the pathological process of atherosclerosis, the enhanced migration or invasion of peripheral macrophages is always observed.13,14 Thus, the effect of compound 1 on the migration of PMA-induced THP-1 cells was evaluated by scratch wound assay. Compound 1 (after 24 h incubation) reduced the motility of PMA-induced THP-1 cells in a dose-dependent manner. Fig. 5 shows that the number of migrated cells was significantly fewer than those of vehicle-treated wells in the 10, 25, and 50 μM treatment groups (p < 0.01, p < 0.001, and p < 0.001).2.6. The NF-κB and MAPK pathways were involved in the inhibitory effect of compound 1 on CD147 and MMP mRNA expression in PMA-induced THP-1 cells
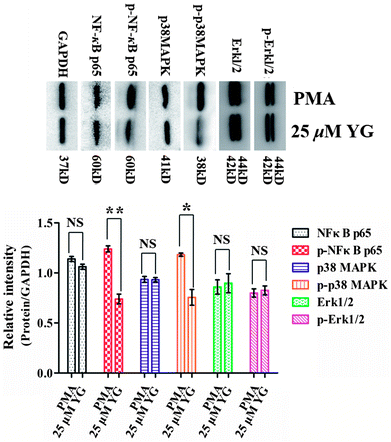
In atherosclerosis, CD147 is involved in a variety of signaling pathways, including NF-κB, MAPK, and JAK/STAT, to regulate pathological processes in different ways.33–36 Therefore, it was of interest to examine whether these pathways were also affected by compound 1 in PMA-induced macrophages. In the present study, after differentiated THP-1 cells had been treated with 1 (25 μM) for 4 h, the phosphorylations of NF-κB p65 and p38 MAPK were significantly inhibited (p < 0.05 and p < 0.01, respectively; Fig. 6). Therefore, these data suggest that NF-κB and p38 MAPK signaling pathways are involved in the regulation of the effect of 1 on EMMPRIN and MMP expression in PMA-induced THP-1 cells.The stability of atherosclerotic plaques is the key point of the transformation for stable angina pectoris or a life-threatening acute coronary syndrome, including acute myocardial infarction and unstable angina pectoris.13,14,37 In that process, MMPs, from macrophages and macrophage-derived foam cells of arterial wall, are responsible for extracellular matrix degradation and remodeling, which ultimately weaken the atherosclerotic lesion and promote plaque rupture in the arterial wall.11–14 CD147, an adhesion molecule, is an upstream regulator of MMPs in atherosclerotic plaque.7–10 Importantly, the clinical outcomes of statins have been shown to attribute to both their LDL reducing and CD147 and MMP lowering effects.15–17 In addition, CD147-blocking antibodies, antagonistic peptides, and siRNA have been used in animal models to reduce tissue inflammation.38 CD147+/− mice and mAb anti-CD147-treated WT-mice are largely protected from myocardial I/R injury.39 Thus, preventing the rupture of atherosclerotic plaques by inhibition of CD147 and MMPs may have benefit in atherosclerosis therapy.
Although prominent biological activities of CTs from Cimicifuga spp. have been reported, to date, however, knowledge of the inhibition of those chemical constituents on CD147 and MMPs is mainly not yet clear. For the first time, yunnanterpene G, a CT from C. foetida, showed notable suppression of mRNA expression of CD147, MMP-2, MMP-9 and MMP-14 in PMA-induced THP-1 cells. Moreover, the migration of differentiated THP-1 cells was also inhibited by yunnanterpene G. Finally, a preliminary mechanistic study suggested that the aforementioned bioactivities of yunnanterpene G were related to its inhibition on NF-κB and p38 MAPK pathways. Yunnanterpene G represents a new type of active structural prototype, and, given the important roles of CD147 and MMP expression in the development of atherosclerosis, this compound and its modified analogues deserve further studies for their health-related properties in stabilizing atherosclerotic plaque, and thus ameliorating the development of atherosclerosis in future.
3. Conclusions
In summary, yunnanterpene G (1), a new spiro-triterpene from the roots of C. foetida, downregulates expression of CD147 and MMPs in PMA-induced THP-1 cells through suppressing NF-κB and p38 MAPK signaling pathways' activation. Since suppression of CD147 and MMPs could stabilize atherosclerotic plaque, these results highlight a promising role of yunnanterpene G in retarding the development of the vulnerability of such plaque. More sophisticated animal studies are needed to clarify yunnanterpene G's in vivo anti-atherosclerosis effect.4. Experimental section
4.1. General experimental procedures
Column chromatography (CC) was run on silica gel (200–300 mesh, Qingdao Marine Chemical, Inc.) and Lichroprep RP-18 (40–63 μm, Merck). Semipreparative HPLC was carried out on an Agilent 1100 liquid chromatography system using a YMC-Pack 10 mm × 250 mm column (Pro C18 RS). Precoated TLC plates (200–250 μm thickness, silica gel 60 F254, Qingdao Marine Chemical, Inc.) were used for thin-layer chromatography. 1D and 2D NMR spectra were obtained using Bruker DRX-500 and Avance III-600 MHz spectrometers (Bruker, Zűrich, Switzerland) with solvent signal as internal reference. ESIMS and HRESIMS were run with a Shimadzu LCMS-IT-TOF mass spectrometer (Shimadzu, Kyoto, Japan) or an Agilent G6230 TOF MS (Agilent Technologies, Palo Alto, USA). Infrared spectra were obtained using a Shimadzu IR-450 instrument with KBr pellets. A JASCO P-1020 digital polarimeter was applied to record optical rotations, using MeOH as solvent. X-ray diffraction was realized with a Bruker SMART APEX CCD crystallography system. Flow cytometry analysis was conducted with a PARTEC brand flow cytometer (Germany). Quantitative PCR (q-PCR) was conducted with a ProFlex™ PCR system (Thermo Fisher, Shanghai, China). SDS-PAGE was carried out with a Mini-Protean Tetra Electrophoresis System (Bio-Rad, Shanghai, China).4.2. Materials
4.3. Extraction and isolation
MeOH (10 L, 3 times, 7 days each) was used to extract the dried and milled fruits of Cimicifuga foetida (3.0 kg) at room temperature. MeOH was evaporated under vacuum at 50 °C to afford the extract (406.8 g). The extract was divided to fractions A (46.7 g), B (78.2 g), C (67.5 g), D (58.1 g) and E (40.5 g) by silica gel CC (5.0 kg, 20 × 200 cm) eluted with CHCl3–MeOH [100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 (10 L), 50
0 (10 L), 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (20 L), 10
1 (20 L), 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (20 L), 5
1 (20 L), 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (15 L), 0
1 (15 L), 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (7 L)]. Subsequently, five sub-fractions (B.1–B.7) were obtained through further silica CC (3 kg, 10 × 150 cm), eluting with CHCl3–Me2CO from 40
100 (7 L)]. Subsequently, five sub-fractions (B.1–B.7) were obtained through further silica CC (3 kg, 10 × 150 cm), eluting with CHCl3–Me2CO from 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 gradient to 5
1 gradient to 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Fractions (B.5.1–B.5.5) were obtained by RP-18 CC (eluted with MeOH–H2O, gradient from 50
1. Fractions (B.5.1–B.5.5) were obtained by RP-18 CC (eluted with MeOH–H2O, gradient from 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 to 100
50 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) of fraction B.5. Compound 1 (6.5 mg) was purified from fraction B.5.3 (6.7 g) by silica gel CC (80 g, 5 × 40 cm) eluted with CHCl3–Me2CO (gradient from 20
0) of fraction B.5. Compound 1 (6.5 mg) was purified from fraction B.5.3 (6.7 g) by silica gel CC (80 g, 5 × 40 cm) eluted with CHCl3–Me2CO (gradient from 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 5
1 to 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and then repeated semipreparative HPLC (eluted with CH3CN–H2O, gradient from 50
1), and then repeated semipreparative HPLC (eluted with CH3CN–H2O, gradient from 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 to 75
50 to 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25).
25).
4.4. X-ray crystal structure analysis
Colorless crystals of 1 were obtained from CH3OH. Intensity data were collected at 100 K using a Bruker APEX DUO diffractometer equipped with an APEX II CCD, using Cu Kα radiation. Cell refinement and data reduction were performed with Bruker SAINT. The structures were solved by direct methods using SHELXS-97.22 Refinements were performed with SHELXL-97, using full-matrix least-squares, with anisotropic displacement parameters for all the non-hydrogen atoms. The H atoms were placed in calculated positions and refined using a riding model. Molecular graphics were computed with PLATON. Crystallographic data (excluding structure factor tables) for the structures reported have been deposited with the Cambridge Crystallographic Data Centre as supplementary publication no. CCDC 1813083.Yunnanterpene G (1): C30H44O6·H2O, M = 518.67, a = 6.1629(2) Å, b = 18.1237(6) Å, c = 12.4853(4) Å, α = 90°, β = 102.545(2)°, γ = 90°, V = 1361.25(8) Å3, T = 100(2) K, space group P21, Z = 2, μ(CuKα) = 0.713 mm−1, 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 631 reflections measured, 4459 independent reflections (Rint = 0.0451). The final R1 values were 0.0456 (I > 2σ(I)). The final wR(F2) values were 0.1194 (I > 2σ(I)). The final R1 values were 0.0472 (all data). The final wR(F2) values were 0.1209 (all data). The goodness of fit on F2 was 1.047. Flack parameter = 0.07(11). The Hooft parameter is 0.08(11) for 1885 Bijvoet pairs.
631 reflections measured, 4459 independent reflections (Rint = 0.0451). The final R1 values were 0.0456 (I > 2σ(I)). The final wR(F2) values were 0.1194 (I > 2σ(I)). The final R1 values were 0.0472 (all data). The final wR(F2) values were 0.1209 (all data). The goodness of fit on F2 was 1.047. Flack parameter = 0.07(11). The Hooft parameter is 0.08(11) for 1885 Bijvoet pairs.
4.5. Cell culture and differentiation
Human monocytic THP-1 cells were cultured in DMEM/F12 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in a humidified atmosphere of 5% CO2. For the induction of cell differentiation, cells (5 × 105 to 106 per ml) were seeded in DMEM/F12 serum medium with 100 nM PMA for 24 h. After incubation, nonattached cells were removed by aspiration, and the adherent cells were washed with DMEM/F12.4.6. Flow cytometry analysis
Expression of CD68 and CD11b on the surface of the differentiated THP-1 cells was determined by flow cytometry. Cells (5 × 105) were washed 3 times with phosphate-buffered saline (PBS) and then were treated successively with fluorescein isothiocyanate (FITC)-conjugated anti-CD68 antibody, and FITC-conjugated anti-CD11b antibody, for 20 minutes in a dark condition. Cells were washed with PBS and then analyzed with a PARTEC CyFlow® Cube flow cytometer. Data were processed using the CytExpert software.4.7. MTT assay
The differentiated THP-1 cells were suspended in DMEM/F12 medium with 10% FBS, plated onto gelatinized 96-well culture plates (5 × 105 cells per ml, 0.1 ml per well), and incubated at 37 °C with 5% CO2 for 24 h. Then, the medium was replaced with 0.1 ml of DMEM/F12, containing different concentrations of yunnanterpene G (0, 5, 10, 25, 50, 75, and 100 μM), and incubated for 48 h at 37 °C with 5% CO2. Cell viability was determined by MTT assay. In the MTT assay, 20 μl of MTT was added to each well (final MTT concentration was 0.5 g L−1) for 4 h before the addition of 150 μl DMSO to solubilize the reactive dye. The absorbance value of each well was recorded at 570 nm using a Bio-Rad microplate reader. All the experiments were performed in triplicate.4.8. Isolation of total RNA and RT-PCR
Total RNA was extracted from differentiated THP-1 cells treated with yunnanterpene G (10, 25, and 50 μM) for 24 h (5 × 105 cells per ml) using RevertAid™ First Strand cDNA Synthesis Kit according to the manufacturer's instructions. Synthesis of cDNA from the isolated total RNA was conducted using the PrimeScript RT reagent kit. In brief, 5× PrimeScript™ Buffer (2 μl), PrimeScript RT Enzyme Mix 1 (0.5 μl), oligo dt Primer (0.5 μl) and Random 6 mers (0.5 μl) were added to 1 μg RNA from each sample. The reaction volume was brought to 10 μl with RNase-free water, mixed gently, and incubated at 37 °C for 15 minutes to activate the reverse transcriptase enzyme and 85 °C for 5 seconds to inactivate the reaction.After reverse transcription, cDNA was used for real-time quantitative RT-PCR with a ProFlex™ PCR system using SYBR Premix Ex Taq (Takara). The RT-PCR was performed in a final volume of 25 μl containing 12.5 μl SYBR green master mix, 1 μl cDNA, 0.5 μl each forward and reverse primer, and 10.5 μl nuclease-free water. Primer information is in Table S1.† Thermal cycling conditions for all genes were as follows: template pre-denaturation (10 min at 95 °C), denaturation (15 seconds at 95 °C), annealing and extension (30 seconds at 60 °C) for 40 cycles. Experiments were performed in triplicate for each data point. GAPDH mRNA was amplified as a housekeeping gene, and fold changes in each target mRNA expression relative to GAPDH were calculated by the 2−ΔΔCt method. Expression of mRNA is defined as the change in mRNA copy numbers relative to negative control cells (undifferentiated THP-1 cells). All the experiments were performed in triplicate.
4.9. Wound-healing migration assay
The differentiated THP-1 cells were seeded and grown into full confluence in 6-well plates. Cells were starved with 2% FBS DMEM/F12 media for 12 h to inactivate cell proliferation and then wounded by pipette tips. Fresh DMEM/F12 medium with or without 10, 25, and 50 μM yunnanterpene G was added to the scratched monolayers. Images were taken after 24 hours using an inverted microscope (magnification, 10×; Nikon). The migration cell number of PMA-induced THP-1 cell group was defined as control. All the experiments were performed in triplicate.4.10. Western blot assay
The differentiated THP-1 cells were serum-starved overnight, then pretreated with 25 μM yunnanterpene G for 4 h. Cells were lysed with buffer containing 50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 2 mM sodium pyrophosphate, 25 mM β-glycerophosphate, 40 mmol L−1 NaF, 10 mmol L−1 Na4P2O7, proteinase inhibitor cocktail and 1 mmol L−1 phenylmethylsulfonyl fluoride. Protein concentrations were determined by Bradford assay and equalized before loading. About 80 μg cellular proteins were separated using gradient SDS-PAGE gels and probed with specific antibodies (abcam) including NF-κB p65, phospho-NF-κB (phospho S536) p65, p38 MAPK, phospho-p38 MAPK (phospho T180/Y182), Erk1/2, phosphor-Erk1/2 (phospho Thr202/Tyr204), HRP, and GAPDH. Blots were visualized with enhanced chemiluminescence reagent.4.11. Statistical analysis
Data were presented as mean ± SD. Statistical analysis of data was performed with Student's t-test for unpaired data in two trails. (*) p < 0.05, (**) p < 0.01, and (***) p < 0.001 were considered significant.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We greatly acknowledge the help from Doctor Haiyuan Li for the experiment design and language polishing.Notes and references
- C. P. Wang, R. Jin, X. L. Zhu, J. C. Yan and G. H. Li, J. Cardiovasc. Transl. Res., 2015, 8, 59–66 CrossRef PubMed.
- R. Ross, N. Engl. J. Med., 1999, 340, 115–126 CrossRef CAS PubMed.
- R. P. Choudhury and J. M. Lee, Nat. Clin. Pract. Cardiovasc. Med., 2005, 2, 309–315 CrossRef CAS PubMed.
- K. T. Iacono, A. L. Brown, M. I. Greene and S. J. Saouaf, Exp. Mol. Pathol., 2007, 83, 283–295 CrossRef CAS PubMed.
- E. E. Gabison, T. Hoang-Xuan, A. Mauviel and S. Menashi, Biochimie, 2005, 87, 361–368 CrossRef CAS PubMed.
- K. D. Curtin, I. A. Meinertzhagen and R. J. Wyman, J. Cell Sci., 2005, 118, 2649–2660 CrossRef CAS PubMed.
- Y. W. Yoon, H. M. Kwon, K. C. Hwang, E. Y. Choi, B. K. Hong, D. Kim, H. S. Kim, S. H. Cho, K. S. Song and G. Sangiorgid, Atherosclerosis, 2005, 180, 37–44 CrossRef CAS PubMed.
- D. A. Siwik, G. M. Kuster, J. V. Brahmbhatt, Z. Zaidi, J. Malik, H. Ooi and G. Ghorayeb, J. Mol. Cell. Cardiol., 2008, 44, 210–217 CrossRef CAS PubMed.
- F. G. Spinale, M. L. Coker, L. J. Heung, B. R. Bond, H. R. Gunasinghe, T. Etoh, A. T. Goldberg, J. L. Zellner and A. J. Crumbley, Circulation, 2000, 102, 1944–1949 CrossRef CAS PubMed.
- E. Y. Choi, D. Kim, B. K. Hong, H. M. Kwon, Y. G. Song, K. H. Byun, K. C. Whang and H. S. Kim, Exp. Mol. Med., 2002, 34, 391–400 CrossRef CAS PubMed.
- R. Li, L. Huang, H. Guo and B. P. Toole, J. Cell. Physiol., 2001, 186, 371–379 CrossRef CAS PubMed.
- T. C. Major, L. Liang, X. Lu, W. Rosebury and T. M. Bocan, Arterioscler., Thromb., Vasc. Biol., 2002, 22, 1200–1207 CrossRef CAS.
- R. Schmidt, A. Bultmann, M. Ungerer, N. Joghetaei, O. Bülbül, S. Thieme, T. Chavakis, B. P. Toole, M. Gawaz, A. Schömig and A. E. May, Circulation, 2006, 113, 834–841 CrossRef CAS PubMed.
- D. L. Brown, M. S. Hibbs, M. Kearney, C. Loushin and J. M. Isner, Circulation, 1995, 91, 2125–2131 CrossRef CAS PubMed.
- N. Abe, T. Osanai, T. Fujiwara, K. Kameda, T. Matsunaga and K. Okumura, Life Sci., 2006, 78, 1021–1028 CrossRef CAS PubMed.
- S. Bellosta, D. Via, M. Canavesi, P. Pfister, R. Fumagalli, R. Paoletti and F. Bernini, Arterioscler., Thromb., Vasc. Biol., 1998, 18, 1671–1678 CrossRef CAS.
- N. Ferri, G. Colombo, C. Ferrandi, E. W. Raines, B. Levkau and A. Corsini, Arterioscler., Thromb., Vasc. Biol., 2007, 27, 1043–1049 CrossRef CAS PubMed.
- J. X. Li and Z. Y. Yu, Curr. Med. Chem., 2006, 13, 2927–2951 CrossRef CAS PubMed.
- Y. Q. Guo, T. Yin, X. M. Wang, F. Zhang, G. X. Pan, H. Lv, X. R. Wang, J. O. Orgah, Y. Zhu and H. H. Wu, J. Ethnopharmacol., 2017, 209, 264–282 CrossRef CAS PubMed.
- A. Imai, D. C. Lankin, D. Nikolić, S. Ahn, R. B. Breemen, N. R. Farnsworth, J. B. McAlpine, S. N. Chen and G. F. Pauli, J. Nat. Prod., 2016, 79, 541–554 CrossRef CAS PubMed.
- Y. Nian, H. Zhu, W. R. Tang, Y. Luo, J. Du and M. H. Qiu, J. Nat. Prod., 2013, 76, 896–902 CrossRef CAS PubMed.
- Y. Nian, H. Yan, X. N. Li, L. Zhou and M. H. Qiu, RSC Adv., 2017, 7, 38557–38564 RSC.
- Y. Nian, J. Yang, T. Y. Liu, Y. Luo, J. H. Zhang and M. H. Qiu, Sci. Rep., 2015, 5, 9026–9031 CrossRef CAS PubMed.
- Y. Su, L. Wu, Q. H. Wang, B. Y. Yang and H. X. Kuang, Bioorg. Med. Chem. Lett., 2014, 24, 5688–5691 CrossRef CAS PubMed.
- Y. Su, L. Wu, G. R. Mu, Q. H. Wang, B. Y. Yang, G. H. Cheng and H. X. Huang, Bioorg. Med. Chem., 2017, 25, 4917–4923 CrossRef CAS PubMed.
- C. N. Lv, F. Yang, R. L. Qin, Z. Y. Qi, W. R. Zhou and J. C. Lu, Bioorg. Med. Chem. Lett., 2017, 27, 3305–3309 CrossRef CAS PubMed.
- N. P. Thao, B. T. T. Luyen, J. S. Lee, J. H. Kim, N. T. Dat and Y. H. Kim, J. Nat. Prod., 2017, 80, 1867–1875 CrossRef CAS PubMed.
- J. X. Li, J. Liu, C. C. He, Z. Y. Yu, Y. S. Kadota and H. Seto, Maturitas, 2007, 58, 59–69 CrossRef CAS PubMed.
- N. Sakurai, J. H. Wu, Y. Sashida, Y. Mimaki, T. Nikaido, K. Koike, H. Itokawa and K. H. Lee, Bioorg. Med. Chem. Lett., 2004, 14, 1329–1332 CrossRef CAS PubMed.
- M. A. Findeis, F. Schroeder, T. D. McKee, D. Yager, P. C. Fraering, S. P. Creaser, W. F. Austin, J. Clardy, R. Wang, D. Selkoe and C. B. Eckman, ACS Chem. Neurosci., 2012, 3, 941–951 CrossRef CAS PubMed.
- J. Auwerx, Experientia, 1991, 47, 22–31 CrossRef CAS PubMed.
- J. Zhou, P. Zhu, J. L. Jiang, Q. Zhang, Z. B. Wu, X. Y. Yao, H. Tang, N. Lu, Y. Yang and Z. N. Chen, BMC Cell Biol., 2005, 6, 25–34 CrossRef PubMed.
- W. Yuan, H. Ge and B. He, Atherosclerosis, 2010, 213, 415–421 CrossRef CAS PubMed.
- A. J. Muslin, Clin. Sci., 2008, 115, 203–218 CrossRef CAS PubMed.
- H. Motoshima, B. J. Goldstein, M. Igata and E. Araki, J. Physiol., 2006, 574, 63–71 CrossRef CAS PubMed.
- G. Ortiz-Muñoz, J. L. Martin-Ventura, P. Hernandez-Vargas, B. Mallavia, V. Lopez-Parra, O. Lopez-Franco, B. Muñoz-Garcia, P. Fernandez-Vizarra, L. Ortega, J. Egido and C. Gomez-Guerrero, Arterioscler., Thromb., Vasc. Biol., 2009, 29, 525–531 CrossRef PubMed.
- Y. Wang, Z. Q. Huang, C. Q. Wang, L. S. Wang, S. Meng, Y. C. Zhang, T. Chen and Y. Q. Fan, Clin. Exp. Pharmacol. Physiol., 2011, 38, 11–18 CrossRef CAS PubMed.
- P. Zhu, N. Lu, Z. G. Shi, J. Zhou, Z. B. Wu, Y. Yang, J. Ding and Z. N. Chen, Arthritis Res. Ther., 2006, 8, R44 Search PubMed.
- P. Seizer, C. Ochmann, T. Schönberger, S. Zach, M. Rose, O. Borst, K. Klingel, R. Kandolf, H. R. MacDonald, R. A. Nowak, S. Engelhardt, F. Lang, M. Gawaz and A. E. May, Arterioscler., Thromb., Vasc. Biol., 2011, 31, 1377–1386 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: 1D and 2D NMR spectra, HRESIMS, UV, IR, and OR spectra of new compound 1. CCDC 1813083. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c8ra01895b |
| This journal is © The Royal Society of Chemistry 2018 |