Open Access Article
Open Access ArticleNontargeted SWATH acquisition mode for metabolites identification of osthole in rats using ultra-high-performance liquid chromatography coupled to quadrupole time-of-flight mass spectrometry†
Man Liao,
Xinpeng Diao,
Xiaoye Cheng,
Yupeng Sun and
Lantong Zhang *
*
Department of Pharmaceutical Analysis, School of Pharmacy, Hebei Medical University, 361 East Zhongshan Road, Shijiazhuang, Hebei 050017, P. R. China. E-mail: zhanglantong@263.net; Fax: +86-311-86266419; Tel: +86-311-86266419
First published on 19th April 2018
Abstract
Osthole (OST), 7-methoxy-8-isopentenoxycoumarin, is the characteristic constituent found in Cnidium monnieri (L.) Cuss. and possesses excellent pharmacological activities, including anticancer, anti-apoptosis and neuroprotection. In this study, a rapid and reliable method based on ultra-high-performance liquid chromatography coupled to quadrupole time-of-flight mass spectrometry (UHPLC-Q-TOF-MS) and MetabolitePilot2.0™ software with principal component variable grouping (PCVG) filtering was developed to observe probable metabolites of OST firstly. The high resolution mass data were acquired by data-independent acquisition mode (DIA), i.e., sequential window acquisition of all theoretical fragmentation spectra (SWATH), which could significantly improved the hit rate of low-level and trace metabolites. A novel data processing method ‘key product ions (KPIs)’ were employed for metabolites rapid hunting and identification as an assistant tool. A total of 72 metabolites of OST were detected in vitro and in vivo, including 39 metabolites in rat liver microsomes (RLMs), 20 metabolites in plasma, 32 metabolites in bile, 32 metabolites in urine and 37 metabolites in feces. The results showed that mono-oxidation, demethylation, dehydrogenation, sulfate conjugation and glucuronide conjugation were major metabolic reactions of OST. More significant, oxydrolysis, 3,4-epoxide-aldehylation, phosphorylation, S-cysteine conjugation and N-acetylcysteine conjugation were considered as unique metabolic pathways of OST, and phosphorylation, S-cysteine conjugation and N-acetylcysteine conjugation reactions were characterized in rat biological samples for the first time. Preparation of active metabolites will be greatly helpful in elucidating the potential biological mechanism of OST, and the proposed metabolic pathways of it might provide further understanding of the safety and efficacy of simple coumarins.
1. Introduction
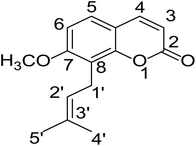
Osthole (OST, Fig. 1), a natural simple coumarin, is the active component in many popular medical plants and fruits, including Cnidium monnieri (L.) Cuss.1 Peucedanum ostruthium,2 Citrus aurantium3 and C. sinensis cv. Doppio Sanguigno.4 In recent years, pharmacological studies have revealed that OST had diverse effects as anticancer,5 anti-apoptosis6,7 and neuroprotection.8,9 Currently, OST has attracted tremendous attention due to its insect control application on crop planting and it has become a novel plant-based pesticides at home and abroad.3,10,11Prototype drugs undergoes rapid metabolism after administration and the metabolites might exert higher biological activity or reduce the toxicity.12 Tsai T. H. et al.,13 Liu M. et al.14 and Yun F. et al.15 all developed the determination of OST in rat plasma and indicated that OST appeared to be absorbed fast but eliminated slowly. So, it is vital to investigate biotransformation of OST in vitro and in vivo to ensure further safety and efficacy.
To our knowledge, only eighteen metabolites of OST were tentatively identified in rat urine by UPLC-QTOF/MS involving 7-demethylation, 8-dehydrogenation, hydroxylation, 3,4-epoxide, glucuronide conjugation and sulfate conjugation metabolic pathways.16,17 Lv X. et al.18 observed thirteen metabolites in rat urine after oral administration of OST by HPLC-DAD-MS/MS analysis and 2D-NMR techniques. Besides, five demethylation or multiple isomers of dehydrogenation metabolites were observed in rat liver microsomes using HPLC-Qtrap-MS/MS.19 Liver is considered as the most important organ in the biotransformation of drugs with a variety of required enzymes especially for cytochrome P450 (CYP450) enzymes.20 Zhang L. F. et al.21 investigated that OST presented characteristics of enzymatic kinetics metabolism in rat hepatocytes and CYP3A4 was involved in the metabolism of OST in vitro. Xiao H. et al.22 showed that ketoconazole, sulfaphenazole and quinidine could inhibit the disappearance of OST and CYP3A4 was involved in the metabolism of OST at all the tested concentrations of OST. However, CYP2C9 and CYP2D6 mediated the metabolism at high concentration of OST.
As a predominant analytical technique for unknown compounds analysis, high-resolution mass spectrometry (HRMS) especially TOF analyzer23 plays an important role in qualitative studies of metabolites with higher accuracy and sensitivity. ‘Data-independent acquisition’ (DIA) strategies, i.e., sequential window acquisition of all theoretical fragment spectra (SWATH), have been developed in the study of metabolite identification24,25 and proteomics26 recently, which could acquire whole precursor ions at any time in the chromatographic separation by parsing the mass range into small specific mass windows. In every scan cycle, the analyzer rapidly and sequentially acquires high hit rates of MS/MS spectra of all mass windows across the specific mass range.24 A practical analysis strategy, which combined Principal Component Variable Grouping (PCVG) filtering in the MetabolitePilot™2.0 software and key product ions (KPIs) in Masterview™1.1 post-processing software, holds the advantage of eliminating hybrid spectra and rapidly hunting possible metabolites. The assistant data processing techniques possessed in MetabolitePilot™2.0 software, including extracted ion chromatography (EIC), mass defect filtering (MDF), production filtering (PIF), neutral loss filtering (NLF) and isotope pattern filtering (IPF) could also be used to analyze the structures of metabolites.25 Therefore, it is essential to explore metabolism of OST in vitro in the hepatic model and in vivo metabolism in rats by a powerful and novel analytical technique for metabolite identification in complex matrices with its high resolution and accurate mass measurements.
2. Material and methods
2.1. Chemicals and reagents
OST (purity > 98%, 110822-200305) which was extracted from Cnidii fructus was applied by National Institutes for Food and Drug Control. Isoscopoletin (purity > 98%, 111741-200501) was also purchased from National Institutes for Food and Drug Control. Daphnetin (purity > 98%, MUST-13071104) was purchased from Chengdu Mansite Bio-Technology Co., Ltd. Isopsoralen (purity > 98%, YJ-131203) and 7-hydroxycoumarin (purity > 98%, J0222AS) were purchased from Jiangsu Yongjian Pharmaceutical Co., Ltd. and Dalian Meilun Biotech Co., Ltd., respectively. RLMs were laboratory-made27 and the protein concentration of them was determined by the Lowry method in Department of Pharmaceutical Analysis in School of Pharmacy of Hebei Medical University. They were stored at −80 °C until being used. Phosphate buffer saline (PBS, 10×), β-nicotine-amide adenine dinucleotide phosphate (NADPH, purity > 98%) in reduced form, UDP-D-glucuronide trisodium salt (UDPGA, purity > 98%) and alamethicin (purity > 98%) were purchased from Solarbio Science Co., Ltd. (Beijing, China). Magnesium chloride hexa-hydrate was obtained from Yong Da Chemical Co., Ltd. (Tianjin, China). Sodium carboxymethyl cellulose (CMC-Na) was purchased from Tianjin Chemical Co., Ltd. (Tianjin, China). These chemicals were all of analytical grade. Acetonitrile, methanol and formic acid were HPLC grade, which were purchased from Fisher Scientific (Waltham, MA, USA). De-ionized water (18.2 MΩ cm−1) was prepared by Milli-Q water purification system (Millipore, Bedford, MA, USA).2.2. Standard solutions preparation
OST was dissolved in 90% methanol at the exact concentrations of 1.13 mg mL−1 prepared for in vitro incubation in RLMs. Isoscopoletin, daphnetin, 7-hydroxycoumarin and isopsoralen were weighed appropriately and dissolved in 90% methanol to reach their limit of detections.2.3. Apparatus
UHPLC was performed on a Nexera-X2 UHPLC system (Shimadzu Corp., Kyoto, Japan), which consisted of two LC-30AD pumps, a SIL-30AC autosampler, a CTO-30A column oven, a CBM-20A system controller and a DGU-20A on-line degassing unit. Chromatographic separations were achieved on a Poroshell 120 EC-C18 column (100 mm × 2.1 mm, i.d. 2.6 μm) with a Poroshell 120 EC-C18 UHPLC guard column (Agilent Corp, Santa Clara, CA, USA). Mass spectrometry detection was performed on a hybrid quadrupole time-of-flight tandem mass spectrometer equipped with Turbo V sources and a Turbo ion spray interface (AB Sciex Triple-TOF 5600+, Concord, Ontario, Canada).2.4. UHPLC-Q-TOF-MS conditions
The mobile phases consisted of eluent A (0.1% formic acid in water, v/v) and eluent B (acetonitrile). The gradient elution program was described as follows: 0–2 min, 20% B; 2–25 min, 20–60% B, 25–26 min, 60–95% B, 26–30 min, 95% B. Before the next injection, the program would maintain for 3 min for the column equilibration. The injection volume was 3 μL and the flow rate was 0.3 mL min−1. The column was maintained at 25 °C and the autosampler tray temperature was set at 4 °C.The optimized parameters of MS analysis were set as follows: nebulizing gas (GAS1): 55 psi, TIS gas (GAS2): 55 psi, turbo spray temperature: 550 °C and ion spray voltage (IS): 5500 V with 35 psi curtain gas. Intact protonated molecular ions [M + H]+ were detected via TOF MS scan (60 V declustering potential (DP)), 30 eV collision energy (CE), 100–800 Da TOF MS scan range, and 50 ms accumulation time. SWATH MS/MS experiments acquired spectra in high sensitivity mode (60 V DP, 35 eV CE with ±15 eV collision energy spread, 50–600 Da TOF MS scan range, and 160 ms accumulation time). Twelve SWATH quadrupole isolation windows were 35 Da wide ranging from 100 to 500 Da. The total cycle time for the TOF MS and twelve SWATH MS/MS scans was 2000 ms.
2.5. In vitro metabolism of OST by RLMs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 10 min. The organic phase was collected and evaporated under nitrogen gas. The residues were reconstituted in 90% acetonitrile (200 μL) and the supernatant was filtered through a 0.22 μm polytetrafluoroethylene (PTFE) membrane filter and stored at −20 °C until analyzed. The control sample was incubated without NADPH generating system followed by the same treatment while the blank sample was incubated without parent drug.
000 rpm at 4 °C for 10 min. The organic phase was collected and evaporated under nitrogen gas. The residues were reconstituted in 90% acetonitrile (200 μL) and the supernatant was filtered through a 0.22 μm polytetrafluoroethylene (PTFE) membrane filter and stored at −20 °C until analyzed. The control sample was incubated without NADPH generating system followed by the same treatment while the blank sample was incubated without parent drug.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 10 min, the supernatant was filtered through a 0.22 μm PTFE membrane filter and stored at −20 °C until analyzed. The control sample was incubated without UDPGA followed the same treatment while the blank sample was incubated without OST.
000 rpm at 4 °C for 10 min, the supernatant was filtered through a 0.22 μm PTFE membrane filter and stored at −20 °C until analyzed. The control sample was incubated without UDPGA followed the same treatment while the blank sample was incubated without OST.2.6. Animal and drug administration
Male Sprague Dawley rats (Certificate no. SCXK 2013-1-003, 12–14 weeks of age), weighing 240 ± 20 g, were applied by Experimental Animal Research Center, Hebei Medical University, China. The rats were maintained at ambient temperature (22–24 °C) and 60% relative humidity with a 12 h light/dark cycle. All animals were kept in an environmentally controlled breeding room for at least 1 week with water and standard chow ad libitum, and fasted for 12 h with free access to water before the formal tests. Then the rats were randomly divided into six groups, including three treated groups (group 2, 4 & 6) for plasma, bile, urine and feces drug samples (n = 6) and three control groups (group 1, 3 & 5) for blank samples (n = 3), respectively. Each group was maintained in metabolic cages for collection of samples separately. The suspension of OST powder was suspended in 0.4% CMC-Na water solution, and was given to rats at a dose of 40 mg kg−1 by oral gavage. The same dose of 0.4% CMC-Na water solution was administered to rats of the control groups. All protocols in our laboratory were approved by the Guide for the Care and Use of Laboratory Animals (National Institutes of Health), and also approved by the Animal Ethics Committee of Hebei Medical University.2.7. Sample collection and preparation
Blood sample was collected from posterior orbital venous of group 2 in heparinized tubes before (0 h) and at 0.17, 0.5, 1, 2, 3, 6, 8, 10, 12 h after administration.13–15 Then, the samples were centrifuged at 4500 rpm for 5 min to obtain the plasma. The plasma samples from different time points were combined into one sample.
Six rats of group 4 were immediately anesthetized with urethane (1.0 g kg−1, ip) by intraperitoneal injection and fixed on a wooden plate after administered OST by oral gavage. An abdominal incision was made and the bile duct was cannulated with PE-10 tubing (ID = 0.07 cm) for continuous collection of bile samples from 0–24 h.
Urine and feces samples were collected 48 h from group 6 after oral administration and free accessing to purified water. The urine and feces samples were combined as one sample, respectively. The blank group was administered purified water with the same treatment as the administration group. All samples were stored at −80 °C until further treatment.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 10 min. The organic phase was removed under nitrogen gas. The blank biological samples were treated as the other samples. All samples were stored at −80 °C. Before analysis, the residues were dissolved in 90% acetonitrile (600 μL) completely and the supernatant was filtered through a 0.22 μm PTFE membrane filter.
000 rpm at 4 °C for 10 min. The organic phase was removed under nitrogen gas. The blank biological samples were treated as the other samples. All samples were stored at −80 °C. Before analysis, the residues were dissolved in 90% acetonitrile (600 μL) completely and the supernatant was filtered through a 0.22 μm PTFE membrane filter.2.8. Data processing
The analytic strategies were described in detail as follows: (1) the total ion current chromatograms (TIC) of samples and chemical structure of parent drug were imported into MetabolitePilot™2.0 software. (2) Primary metabolite identification was conducted by MetabolitePilot™2.0 software based on accurate measurements of m/z values and processing of the post-data obtained from PCVG, EIC, MDF, PIF, NLF and IPF screening of possible metabolites. The elemental compositions and chemical formulae of them could be calculated. Potential metabolites of target groups can be observed according to fragmentation rules of parent drug, polarity, retention times and the predicted metabolic pathways entered in the software. (3) Then, proposed metabolites inferred by current metabolic pathways could be validated after MS/MS spectra analysis or authentic standards comparison. Then the repeatable constituents in blank samples and test samples were removed by PeakView™ 2.2 software and MasterView™ 1.1 software. The mass error of quasi-molecular was limited in ±5 ppm, and the MS/MS error of the product ions was less than 20 ppm. Product ions m/z 189.0554 and m/z 91.0567 were set as the KPIs. KPIs technique, which was an assistant tool in MasterView™ 1.1 software could also observe probable isomers metabolites or metabolites with similar structures extracted by the same product ions rapidly. (4) Finally, the Clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values and the minimize energy values provided by the program ChemBioDraw Ultra 14.0 were used for distinguishing isomers and accounting for the stability of the molecule, respectively.
P values and the minimize energy values provided by the program ChemBioDraw Ultra 14.0 were used for distinguishing isomers and accounting for the stability of the molecule, respectively.
3. Results and discussion
3.1. Fragmentation studies of OST
The chemical structure was shown in Fig. 1. The MS/MS fragmentation behavior of OST was investigated in positive ion mode (collision energy, 35 ± 15 eV) (Fig. S1†). After microsomal incubation in vitro and intragastric gavage (i.g.) administration to rats, OST was eluted at 19.68 min and yielded [M + H]+ at m/z 245.1175 (1.2 ppm, elemental composition C15H16O3) in RLMs, plasma, bile, urine and feces samples under the present chromatographic and mass spectrometry. OST gave abundant product ions at m/z 230.0937 and m/z 202.0625 via side-chain cleavage of the consecutive loss of CH2. The major MS/MS characteristic product ions was at m/z 189.0554 conducted by the loss of the side-chain C4H7. Then a series of characteristic ions produced by m/z 189.0554 were showed at m/z 161.0625 [M + H–C4H7–CO]+, m/z 159.0441 [M + H–C4H7–CH2O]+, m/z 131.0498 [M + H–C4H7–CO–CH2O]+, m/z 103.0554 [M + H–C4H7–2CO–CH2O]+, m/z 91.0567 [M + H–C5H9–2CO–CH2O]+, m/z 77.0404 [M + H–C7H11–2CO–CH2O]+ and m/z 65.0453 [C5H5]+. The proposed fragmentation pathways of OST were exhibited in Fig. S1.†3.2. Phase I metabolites identification
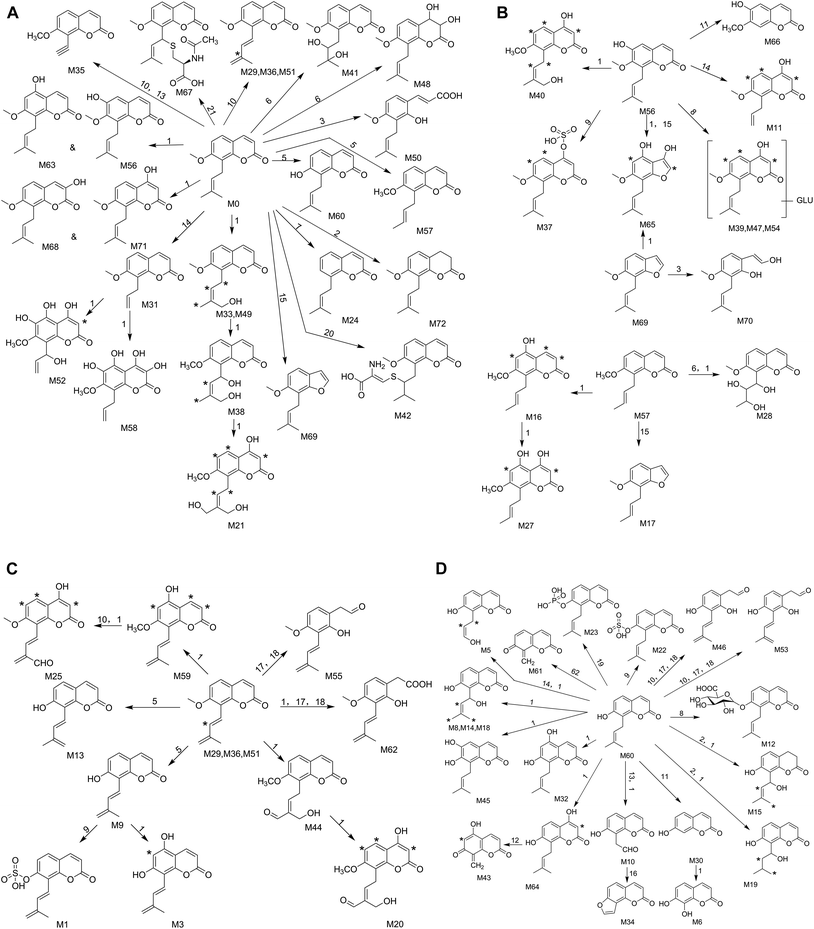
A total of 62 phase I metabolites were identified including 39 metabolites in RLMs, 12 metabolites in plasma, 22 metabolites in bile, 22 metabolites in urine and 36 metabolites in feces samples. M6, M30, M34 and M66 were confirmed in comparison with authentic standards daphnetin, 7-hydroxycoumarin, isopsoralen and isoscopoletin, respectively. Among them, most were produced via mono-oxidation, demethylation and dehydrogenation. Oxydrolysis, demethylation, 3,4-epoxide and aldehylation could be considered as the characteristic metabolic pathways of OST. Other metabolic pathways, such as loss of side-chain alkyl, loss of CO, di-oxidation, tri-oxidation, qur-oxidation, hydrolysis and hydrogenation, were also observed in the samples. Characteristic biotransformation pathways and the most abundant phase I metabolites were summarized as follows. The detail information about retention times and product ions of the other metabolites were listed in Table 1.| Compound ID | Retention time (min) | Formula | Calculated m/z [M + H] | Experimental m/z [M + H] | Error (ppm) | Product ions | Potential pathways | Source | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RLMs | P | B | U | F | ||||||||
| a *: Confirmation in comparison with authentic standards; key product ions (KPIs) were underlined and indicated in bold face; RLMs: rat liver microsomes; P: plasma; B: bile; U: urine; F: feces; “+”, detected. | ||||||||||||
| M0 | 19.68 | C15H16O3 | 245.1172 | 245.1175 | 1.2 | 230.0937, 202.0625, 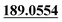 , 161.0625, 159.0441, 131.0498, 103.0554, , 161.0625, 159.0441, 131.0498, 103.0554, 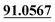 |
— | + | + | + | + | + |
| M1 | 2.19 | C14H12O6S | 309.0427 | 309.0422 | −1.6 | 229.0860, 175.0398, 131.0494, 91.0559 | Demethylation, dehydrogenation, sulfate conjugation | + | + | + | ||
| M2 | 2.80 | C15H14O4 | 259.0965 | 259.0960 | −1.9 | 243.1018, 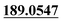 , 131.0490 , 131.0490 |
Dehydrogenation, mono-oxidation | + | ||||
| M3 | 3.51 | C14H12O4 | 245.0808 | 245.0820 | 4.9 | 191.0325, 107.0493 | Dehydrogenation, mono-oxidation, demethylation | + | + | |||
| M4 | 3.60 | C15H14O4 | 259.0965 | 259.0962 | −1.2 | 243.1014, 189.0550 | Dehydrogenation, mono-oxidation | + | + | |||
| M5 | 3.71 | C12H10O4 | 219.0652 | 219.0654 | 0.9 | 175.0388, 147.0444 | Demethylation, loss of C2H4, mono-oxidation | + | + | + | + | |
| M6* | 4.65 | C9H6O4 | 179.0339 | 179.0335 | −2.2 | 151.0387, 77.0402 | Demethylation, mono-oxidation, loss of C5H8 | + | ||||
| M7 | 4.84 | C15H14O4 | 259.0965 | 259.0962 | −1.2 | 243.1014, 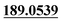 |
Dehydrogenation, mono-oxidation | + | + | + | ||
| M8 | 5.18 | C14H14O4 | 247.0965 | 247.0959 | −2.4 | 175.0393, 131.0495, 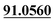 |
Demethylation, mono-oxidation | + | + | |||
| M9 | 5.21 | C14H12O3 | 229.0859 | 229.0856 | −1.3 | 175.0387, 103.0552, 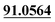 |
Demethylation, dehydrogenation | + | + | + | ||
| M10 | 5.30 | C11H8O4 | 205.0495 | 205.0501 | 2.9 | 175.0381, 131.0499, 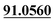 |
Demethylation, loss of C3H6, oxidation | + | + | + | ||
| M11 | 5.40 | C13H12O4 | 233.0808 | 233.0800 | −3.4 | 205.0500, 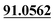 |
Loss of C2H4, mono-oxidation | + | ||||
| M12 | 5.65 | C20H22O9 | 407.1337 | 407.1337 | 0.0 | 231.0997, 175.0385, 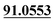 |
Demethylation, glucuronide conjugation | + | + | |||
| M13 | 5.78 | C14H12O3 | 229.0859 | 229.0856 | −1.3 | 175.0389, 103.0550, 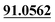 |
Demethylation, dehydrogenation | + | + | + | ||
| M14 | 5.79 | C14H14O4 | 247.0965 | 247.0969 | 1.6 | 175.0395, 131.0494, 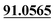 |
Demethylation, mono-oxidation | + | + | |||
| M15 | 5.80 | C14H16O4 | 249.1121 | 249.1123 | 0.8 | 231.1017, 177.0554, 147.0446, 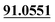 |
Demethylation, hydrogenation, mono-oxidation | + | ||||
| M16 | 6.34 | C14H14O4 | 247.0965 | 247.0969 | 1.6 | 205.0556, 175.0390 | Demethylation, mono-oxidation | + | + | |||
| M17 | 6.74 | C13H14O2 | 203.1067 | 203.1065 | −1.0 | 161.0624, 131.0493, 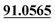 |
Demethylation, loss of CO | + | + | |||
| M18 | 6.90 | C14H14O4 | 247.0965 | 247.0973 | 3.2 | 175.0397, 131.0493 | Demethylation, mono-oxidation | + | ||||
| M19 | 6.95 | C14H16O4 | 249.1121 | 249.1118 | −1.2 | 175.0390, 131.0498, 103.0553 | Demethylation, hydrogenation, mono-oxidation | + | + | |||
| M20 | 7.41 | C15H14O6 | 291.0863 | 291.0853 | −3.4 | 205.0564, 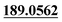 |
Dehydrogenation, tri-oxidation | + | ||||
| M21 | 7.41 | C15H16O6 | 293.1020 | 293.1022 | 0.7 | 205.0500, 131.0501, 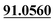 |
Tri-oxidation | + | + | |||
| M22 | 7.77 | C14H14O6S | 311.0584 | 311.0580 | −1.3 | 175.0397, 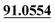 |
Demethylation, sulfate conjugation | + | + | + | ||
| M23 | 7.78 | C14H15O6P | 311.0679 | 311.0678 | −0.3 | 231.1021, 175.0396 | Demethylation, phosphorylation | + | + | + | ||
| M24 | 9.00 | C14H14O2 | 215.1067 | 215.1055 | −3.7 | 173.0597, 131.0500 | Loss of OCH2 | + | ||||
| M25 | 9.06 | C15H12O5 | 273.0758 | 273.0745 | −4.8 | 205.0505, 131.0499 | Dehydrogenation, di-oxidation | + | + | |||
| M26 | 9.17 | C15H14O4 | 259.0965 | 259.0977 | 4.6 | 243.1018, 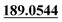 |
Dehydrogenation, mono-oxidation | + | + | + | + | + |
| M27 | 9.19 | C14H14O5 | 263.0914 | 263.0925 | 4.2 | 221.0447, 205.0493, 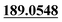 |
Demethylation, di-oxidation | + | ||||
| M28 | 9.19 | C14H16O6 | 281.1020 | 281.1018 | −0.7 | 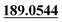 , 177.0540, 147.0437 , 177.0540, 147.0437 |
Demethylation, mono-oxidation, oxydrolysis | + | ||||
| M29 | 9.34 | C15H14O3 | 243.1016 | 243.1019 | 1.2 | 213.0910, 201.0904, 189.0552, 159.0441 | Dehydrogenation | + | ||||
| M30* | 9.54 | C9H6O3 | 163.0390 | 163.0395 | 1.2 | 135.0441, 107.0490 | Demethylation, loss of C5H8 | + | + | |||
| M31 | 9.68 | C13H12O3 | 217.0859 | 217.0874 | 3.1 | 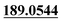 , 159.0437 , 159.0437 |
Loss of C2H4 | + | + | |||
| M32 | 9.90 | C14H14O4 | 247.0965 | 247.0969 | 2.3 | 191.0478, 163.0303, 107.0190 | Demethylation, mono-oxidation | + | ||||
| M33 | 9.93 | C15H16O4 | 261.1121 | 261.1118 | 1.6 | 243.1027, 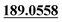 , 131.0501, 103.0556 , 131.0501, 103.0556 |
Mono-oxidation | + | + | + | ||
| M34* | 10.11 | C11H6O3 | 187.0390 | 187.0392 | −1.1 | 159.0442 | Demethylation, loss of C3H6, oxidation, loss of H2O | + | + | |||
| M35 | 10.12 | C12H8O3 | 201.0546 | 201.0540 | 1.1 | 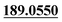 , 161.0620 , 161.0620 |
Dehydrogenation, loss of C3H8 | + | + | |||
| M36 | 10.26 | C15H14O3 | 243.1016 | 243.1010 | −3.0 | 213.0913, 201.0904, 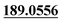 , 159.0437, 131.0503 , 159.0437, 131.0503 |
Dehydrogenation | + | + | + | + | |
| M37 | 10.40 | C15H16O7S | 341.0690 | 341.0674 | −2.5 | 261.1153, 205.0497, 177.0654, 149.0606 | Mono-oxidation, sulfate conjugation | + | + | + | ||
| M38 | 10.41 | C15H16O5 | 277.1071 | 277.1077 | −4.7 | 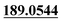 , 91.0556 , 91.0556 |
Di-oxidation | + | + | + | ||
| M39 | 10.65 | C21H24O10 | 437.1442 | 437.1439 | 2.2 | 261.1123, 205.0499 | Mono-oxidation, glucuronide conjugation | + | + | + | ||
| M40 | 10.68 | C15H16O5 | 277.1071 | 277.1073 | −0.7 | 205.0499, 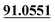 |
Di-oxidation | + | ||||
| M41 | 10.70 | C15H18O5 | 279.1227 | 279.1219 | 0.7 | 245.1169, 223.0526, 149.0239, 119.0503 | Oxydrolysis | + | + | + | ||
| M42 | 10.80 | C18H21NO5S | 364.1213 | 364.1220 | −2.9 | 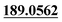 , 161.0605 , 161.0605 |
S-cysteine conjugation | + | + | + | ||
| M43 | 11.06 | C10H6O4 | 191.0339 | 191.0334 | 1.9 | 107.0506, 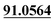 |
Demethylation, mono-oxidation, loss of C4H8 | + | ||||
| M44 | 11.07 | C15H14O5 | 275.0914 | 275.0904 | −2.6 | 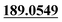 , 159.0441, 131.0496 , 159.0441, 131.0496 |
Dehydrogenation, di-oxidation | + | + | + | + | + |
| M45 | 11.13 | C14H14O4 | 247.0965 | 247.0966 | −3.6 | 191.0476, 163.0305, 107.0193 | Demethylation, mono-oxidation | + | + | + | ||
| M46 | 11.29 | C13H14O3 | 219.1016 | 219.1022 | 0.4 | 165.0541, 135.0440, 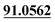 |
3,4-Epoxide, demethylation, dehydrogenation, aldehylation | + | + | |||
| M47 | 11.38 | C21H24O10 | 437.1442 | 437.1448 | 2.7 | 261.1127, 205.0503 | Mono-oxidation, glucuronide conjugation | + | + | + | ||
| M48 | 11.51 | C15H18O5 | 279.1227 | 279.1219 | 1.4 | 245.1169, 189.0555, 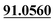 |
Oxydrolysis | + | + | |||
| M49 | 11.81 | C15H16O4 | 261.1121 | 261.1120 | −2.9 | 243.1025, 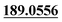 |
Mono-oxidation | + | + | + | ||
| M50 | 11.83 | C15H18O4 | 263.1278 | 263.1265 | −0.4 | 245.1179, 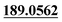 |
Ester hydrolysis | + | + | |||
| M51 | 11.83 | C15H14O3 | 243.1016 | 243.1011 | −4.9 | 213.0910, 201.0900, 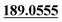 , 159.0433, 131.0501 , 159.0433, 131.0501 |
Dehydrogenation | + | + | |||
| M52 | 11.93 | C13H12O7 | 281.0656 | 281.0649 | −2.1 | 253.0707, 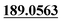 |
Qur-oxidation, loss of C2H4 | + | + | |||
| M53 | 11.93 | C13H14O3 | 219.1016 | 219.1012 | −2.5 | 165.0541, 135.0440, 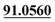 |
3,4-Epoxide, demethylation, dehydrogenation, aldehylation | + | + | |||
| M54 | 12.12 | C21H24O10 | 437.1442 | 437.1436 | −1.8 | 261.1119, 205.0497 | Mono-oxidation, glucuronide conjugation | + | + | + | ||
| M55 | 12.18 | C14H16O3 | 233.1172 | 233.1171 | −1.4 | 149.0599, 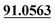 |
3,4-Epoxide, dehydrogenation, aldehylation | + | + | |||
| M56 | 12.79 | C15H16O4 | 261.1121 | 261.1130 | −0.4 | 205.0500, 175.0393, 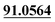 |
Mono-oxidation | + | ||||
| M57 | 12.94 | C14H14O3 | 231.1016 | 231.1014 | 3.4 | 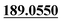 , , 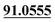 |
Demethylation | + | ||||
| M58 | 13.04 | C13H12O7 | 281.0656 | 281.0666 | −0.9 | 151.0317 | Qur-oxidation, loss of C2H4 | + | + | |||
| M59 | 13.05 | C15H14O4 | 259.0965 | 259.0958 | 3.6 | 243.1016, 205.0505, 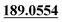 , 131.0502 , 131.0502 |
Dehydrogenation, mono-oxidation | + | + | + | ||
| M60 | 13.60 | C14H14O3 | 231.1016 | 231.1016 | −2.7 | 175.0396, 147.0445, 119.0497, 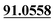 |
Demethylation | + | + | |||
| M61 | 13.61 | C10H6O3 | 175.0390 | 175.0385 | 0.0 | 160.0152, 147.0446 | Demethylation, loss of C4H8 | + | + | + | ||
| M62 | 14.39 | C14H16O4 | 249.1121 | 249.1130 | −2.9 | 231.1040, 177.0539, 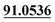 |
3,4-Epoxide, dehydrogenation, aldehylation, oxidation | + | ||||
| M63 | 14.50 | C15H16O4 | 261.1121 | 261.1118 | 3.6 | 205.0504, 175.0390, 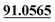 |
Mono-oxidation | + | + | + | ||
| M64 | 14.56 | C14H14O4 | 247.0965 | 247.0965 | −1.1 | 191.0472, 163.0307 | Demethylation, mono-oxidation | + | ||||
| M65 | 15.14 | C14H16O4 | 249.1121 | 249.1118 | 0.0 | 193.0495, 165.0535 | Di-oxidation, loss of CO | + | ||||
| M66* | 15.36 | C10H8O4 | 193.0495 | 193.0491 | −1.2 | 165.0548 | Mono-oxidation, loss of C5H8 | + | + | |||
| M67 | 16.28 | C20H23NO6S | 406.1319 | 406.1330 | −2.1 | 245.1173, 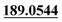 |
N-Acetylcysteine conjugation | + | + | + | ||
| M68 | 16.81 | C15H16O4 | 261.1121 | 261.1130 | 2.7 | 205.0500, 175.0391, 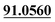 |
Mono-oxidation | + | + | + | ||
| M69 | 17.77 | C14H16O2 | 217.1223 | 217.1224 | 3.4 | 202.0985, 161.0604, 133.0656 | Loss of CO | + | + | |||
| M70 | 17.80 | C14H18O3 | 235.1329 | 235.1320 | 0.5 | 217.1223, 161.0788 | Loss of CO, hydrolysis | + | + | |||
| M71 | 18.88 | C15H16O4 | 261.1121 | 261.1122 | −3.8 | 205.0503, 175.0388, 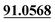 |
Mono-oxidation | + | + | + | + | |
| M72 | 20.65 | C15H18O3 | 247.1329 | 247.1326 | 0.4 | 191.0689, 161.0582, 133.0639, 103.0545, 77.0398 | Hydrogenation | + | + | |||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P among them, M56, M63, M68 and M71 were tentatively identified as 6-hydroxylosthole, 5-hydroxylosthole, 3-hydroxylosthole and 4-hydroxylosthole, respectively.17,28
P among them, M56, M63, M68 and M71 were tentatively identified as 6-hydroxylosthole, 5-hydroxylosthole, 3-hydroxylosthole and 4-hydroxylosthole, respectively.17,28M31 was detected at 9.68 min and showed the molecular formula of C13H12O3 ([M + H]+ m/z 217.0864), which was 28 Da (C2H4) lower than that of M0. The characteristic product ions at m/z 189.0544 [M + H–C2H4]+ and m/z 159.0437 [M + H–C2H4–CH2O]+ indicated that M31 might be the cleavage of side-chain alkyl C2H4.28
M69 was eluted at 17.77 min and exhibited the molecular formula of C14H16O2 ([M + H]+ m/z 217.1224), which was 28 Da (CO) lower than that of OST. The key product ions were at m/z 202.0985 [M + H–CH3], m/z 161.0604 [M + H–C4H8]+ and m/z 133.0656 [M + H–C4H8–CO]+. According to its cleavage rule of OST, decarbonylation might occur at C-2 from M0.
M35 (tR = 10.12 min) gave an [M + H]+ at m/z 201.0540 with the molecular formula of C12H8O3, which was 44 Da (C3H8) lower than that of OST. In the MS/MS spectrum, product ions at m/z 189.0550 and m/z 161.0620 were observed. Therefore, we deduced that M35 was loss of a group of C3H8 from side chain of parent drug.28
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P.
P.M32, M45 and M64 all showed diagnostic product ions at m/z 191.0478 and m/z 163.0303, which suggested that demethylation reaction might generate at C-7's methoxy group. M32 and M45 were both detected product ion at m/z 107.0190, which revealed that mono-oxidized could occur at C-5 or C-6 compared with that of parent drug. According to their values of Clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, M32 was identified C-5 oxidized metabolites and M45 was identified C-6 oxidized metabolites of M60. M64 was temporarily confirmed as C-3 or C-4 oxidized metabolite of M60.28
P, M32 was identified C-5 oxidized metabolites and M45 was identified C-6 oxidized metabolites of M60. M64 was temporarily confirmed as C-3 or C-4 oxidized metabolite of M60.28
M8, M14 and M18 was only detected the typical product ion at m/z 175.0393, which indicated that demethylation reaction might occur at C-7's methoxy group and mono-oxidation reaction might occur at side chain.16
3.3. Phase II metabolites identification
A total of 10 phase II metabolites were identified in rats, and metabolic pathways involved sulfate conjugation, glucuronide conjugation, phosphorylation, S-cysteine conjugation and N-acetylcysteine conjugation. Deductions of representative phase II metabolite structures were summarized as follows. The retention times and product ions of the other metabolites were listed in Table 1.3.4. Metabolic profiles of OST
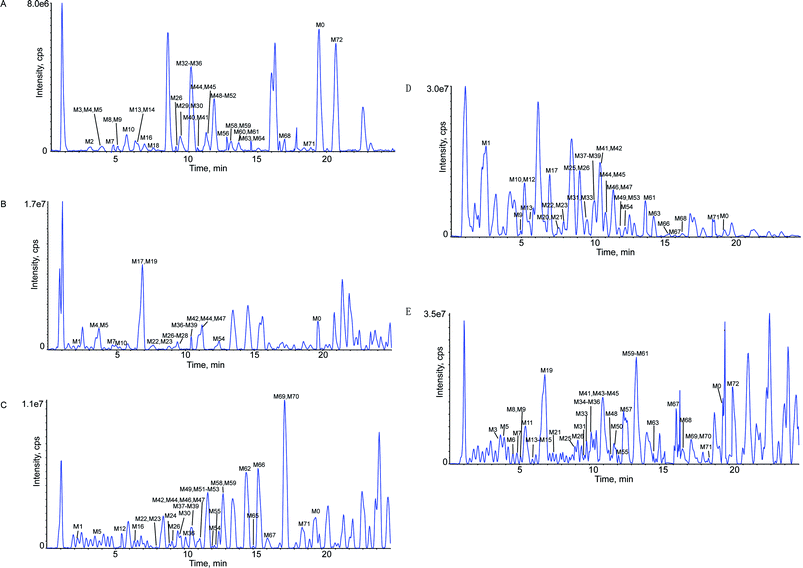
This study was conducted to elucidate the metabolism of OST in vitro (RLMs) and in vivo (plasma, bile, urine, feces of rats). The involved biotransformation and mass changes were listed in the Table S1.† TIC for the sample group in RLMs, rat plasma, bile, urine and feces were exhibited at Fig. 2. Based on the identified metabolites (Table 1), the proposed metabolic pathways for OST were shown in Fig. 3, and the representative MS/MS spectra and proposed fragmentation pathways of M12, M33, M37, M41, M46, M56, M60 and M62 were showed in Fig. S2.† The main metabolic pathways for OST were mono-oxidation, demethylation, dehydrogenation, oxydrolysis, 3,4-epoxide, aldehylation. The predominant phase II metabolite reaction mainly underwent sulfate conjugation, glucuronide conjugation, phosphorylation, S-cysteine conjugation and N-acetylcysteine conjugation.Thirty-nine metabolites were identified in RLMs, indicated that most metabolites could be excreted in RLMs samples and RLMs samples are suitable for rapid identification of metabolites of OST in vitro. It also verified that liver is considered as the most important organ in the biotransformation of drugs with a variety of required enzymes especially for cytochrome P450 (CYP) enzymes. They are the major drug metabolizing enzymes that play an important role in xenobiotic metabolism.31 Rat bile, urine and feces also possessed high activity for OST metabolism, which were observed 32, 32 and 37 metabolites, respectively. Only twenty metabolites were detected in rat plasma, implied that rat plasma might hold low biotransformation activity in plasma.
Among all the metabolites, M6, M30, M34 and M66 were confirmed by comparing with standard substances in terms of the retention times and MS/MS product ions. Comparing with the metabolism in different bio-samples, it was found that oxidation, dehydrogenation, demethylation and oxydrolysis were major biotransformations in vitro, and loss of side-chain alkyl, 3,4-epoxide, aldehylation, hydrogenation, loss of CO, a series of conjugation reactions in vivo. More significantly, the metabolites which underwent loss of side-chain alkyl, loss of CO, loss of OCH2, oxydrolysis, S-cysteine conjugation, sulfate conjugation, phosphorylation and N-acetylcysteine conjugation reactions were tentatively identified in rats or RLMs for the first time. M6 (daphnetin), M30 (7-hydroxycoumarin), M34 (isopsoralen) and M66 (isoscopoletin) have been proved that hold immunosuppressive,32 antinociceptive, anti-inflammatory,33 antitumor34 and inhibitory35 properties, respectively. These results provide some insights into understanding the metabolic profiles of OST, which could guide the pharmacokinetics investigation of OST and its major metabolites in the clinic.
4. Conclusions
In conclusion, a rapid and practical analytical method based on UHPLC-Q-TOF-MS was developed for the characterization of OST metabolites in vitro and in vivo. A novel SWATH acquisition method was used for detected most metabolites of OST with a high hit rate and quality by means of PCVG filtering function for the first time. A total of 62 phase I metabolites and 10 phase II metabolites of OST were identified in vitro and in vivo, including 39 metabolites in RLMs, 20 metabolites in rat plasma, 32 metabolites in rat bile, 32 metabolites in rat urine and 37 metabolites in rat feces samples. The characteristic metabolic bio-transformation routes of phase I were oxydrolysis, demethylation, 3,4-epoxide, and aldehylation. Glucuronide conjugation, S-cysteine conjugation, sulfate conjugation, phosphorylation and N-acetylcysteine conjugation reactions were main metabolic pathways of phase II. This proposed bio-transformation routes of OST provide scientific and reliable support for full understanding of the metabolism of coumarins. Furthermore, the novel strategy was a powerful tool for constituents and metabolites identification, and preparation of the active metabolites will be greatly helpful in elucidating the potential biological mechanism of coumarin derivatives in clinical application.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
The work received financial support from the National Natural Science Foundation of China (no. 81473180). All of the authors are grateful for the support from Department of Pharmaceutical Analysis of school of Pharmacy in Hebei Medical University.References
- Chinese Pharmacopoeia Commission, The Pharmacopoeia of the People's Republic of China, Part I, China Medical Science Press, Beijing, China, 2015, p. 315 Search PubMed.
- S. Vogl, M. Zehl, P. Picker, E. Urban, C. Wawrosch, G. Reznicek, J. Saukel and B. Kopp, J. Agric. Food Chem., 2011, 59, 4371–4377 Search PubMed.
- E. P. Siskos, B. E. Mazomenos and M. A. Konstantopoulou, J. Agric. Food Chem., 2008, 56, 5577–5581 Search PubMed.
- I. Jerković, J. Drulžić, Z. Marijanović, M. Gugić, S. Jokić and M. Roje, Nat. Prod. Commun., 2015, 10, 1315–1318 Search PubMed.
- L. Liu, J. Mao, Q. Wang, Z. Zhang, G. Wu, Q. Tang, B. Zhao, L. Li and Q. Li, Biomed. Pharmacother., 2017, 94, 1020–1027 Search PubMed.
- Y. Li, Y. Li, F. Shi, L. Wang, L. Li and D. Yang, Eur. J. Pharmacol., 2017, 818, 525–533 Search PubMed.
- X. Zhu, X. Song, K. Xie, X. Zhang, W. He and F. Liu, Int. J. Mol. Med., 2017, 40, 1143–1151 Search PubMed.
- Y. Shokoohinia, S. Khajouei, F. Ahmadi, N. Ghiasvand and L. Hosseinzadeh, Iran. J. Basic Med. Sci., 2017, 20, 438–445 Search PubMed.
- Y. Yan, L. Kong, Y. Xia, W. Liang, L. Wang, J. Song, Y. Yao, Y. Lin and J. Yang, Brain, Behav., Immun., 2018, 67, 118–129 Search PubMed.
- P. P. Song, J. Zhao, Z. L. Liu, Y. B. Duan, Y. P. Hou, C. Q. Zhao, M. Wu, M. Wei, N. H. Wang, Y. Lv and Z. J. Han, Pest Manage. Sci., 2017, 73, 94–101 Search PubMed.
- Z. Wang, J. R. Kim, M. Wang, S. Shu and Y. J. Ahn, Pest Manage. Sci., 2012, 68, 1041–1047 Search PubMed.
- X. Zhang, Y. Yao, Y. Lou, H. D. Jiang, X. W. Wang, X. J. Chai and S. Zeng, Drug Metab. Dispos., 2010, 38, 2157–2165 Search PubMed.
- T. H. Tsai, T. R. Tsai, C. C. Chen and F. C. Chen, J. Pharm. Biomed. Anal., 1996, 14, 749–753 CrossRef CAS PubMed.
- M. Liu, H. Liu, X. Lu, C. Li, Z. Xiong and F. Li, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2007, 860, 113–120 CrossRef CAS PubMed.
- F. Yun, A. Kang, J. Shan, X. Zhao, X. Bi and L. Di, Biomed. Chromatogr., 2013, 27, 676–680 CrossRef CAS PubMed.
- J. Li and W. Chan, J. Pharm. Biomed. Anal., 2013, 74, 156–161 CrossRef CAS PubMed.
- Q. Zhao, X. M. Li, H. N. Liu, F. J. Gonzalez and F. Li, Xenobiotica, 2018, 48, 285–299 CrossRef CAS PubMed.
- X. Lv, C. Y. Wang, J. Hou, B. J. Zhang, S. Deng, Y. Tian, S. S. Huang, H. L. Zhang, X. H. Shu, Y. H. Zhen, K. X. Liu, J. H. Yao and X. C. Ma, Xenobiotica, 2012, 42, 1120–1127 CrossRef CAS PubMed.
- Z. T. Yuan, H. Y. Xu, K. Wang, Z. H. Zhao and H. Ming, J. Pharm. Biomed. Anal., 2009, 49, 1226–1232 CrossRef CAS PubMed.
- Y. J. Yoon, K. B. Kim and H. Kim, Drug Metab. Dispos., 2007, 35, 1518–1524 CrossRef CAS PubMed.
- L. F. Zhang, X. Hu, P. Wang and L. Zhang, Acta Pharm. Sin. B, 2009, 44, 1131–1135 CAS.
- H. Xiao, H. Wei and Y. Yuan, Eur. J. Drug Metab. Pharmacokinet., 2015, 40, 373–377 CrossRef PubMed.
- I. Chernushevich, A. V. Loboda and B. A. Thomson, J. Mass Spectrom., 2001, 36, 849–865 CrossRef CAS PubMed.
- K. B. Scheidweiler, M. J. Jarvis and M. A. Huestis, Anal. Bioanal. Chem., 2015, 407, 883–897 CrossRef CAS PubMed.
- W. Xie, Y. Jin, L. Hou, Y. Ma, H. Xu, K. Zhang, L. Zhang and Y. Du, J. Pharm. Biomed. Anal., 2017, 145, 865–878 CrossRef CAS PubMed.
- L. Semenec, A. E. Laloo, B. L. Schulz, I. A. Vergara, P. L. Bond and A. E. Franks, Bioelectrochemistry, 2018, 119, 150–160 CrossRef CAS PubMed.
- X. Zhang, J. T. Yin, C. J. Liang, Y. P. Sun and L. T. Zhang, J. Agric. Food Chem., 2017, 65, 10959–10972 CrossRef CAS PubMed.
- G. Song, M. Jin, Y. Du, L. Cao and H. Xu, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2016, 1022, 21–29 CrossRef CAS PubMed.
- S. L. Born, A. M. Api, R. A. Ford, F. R. Lefever and D. R. Hawkins, Food Chem. Toxicol., 2003, 41, 247–258 CrossRef CAS PubMed.
- D. F. Lewis, Y. Ito and B. G. Lake, Toxicol. In Vitro, 2006, 20, 256–264 CrossRef CAS PubMed.
- F. P. Guengerich, Chem. Res. Toxicol., 2008, 21, 70–83 CrossRef CAS PubMed.
- B. Song, Z. Wang, Y. Liu, S. Xu, G. Huang, Y. Xiong, S. Zhang, L. Xu, X. Deng and S. Guan, PLoS One, 2014, 9, e96502 Search PubMed.
- T. A. Barros, L. A. de Freitas, J. M. Filho, X. P. Nunes, A. M. Giulietti, G. E. de Souza, R. R. dos Santos, M. B. Soares and C. F. Villarreal, J. Pharm. Pharmacol., 2010, 62, 205–213 CrossRef CAS PubMed.
- Y. Wang, C. Hong, C. Zhou, D. Xu and H. B. Qu, J. Evidence-Based Complementary Altern. Med., 2011, 2011, 363052 Search PubMed.
- M. Y. Ali, S. Jannat, H. A. Jung, R. J. Choi, A. Roy and J. S. Choi, Asian Pac. J. Trop. Med., 2016, 9, 103–111 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra01221k |
| This journal is © The Royal Society of Chemistry 2018 |