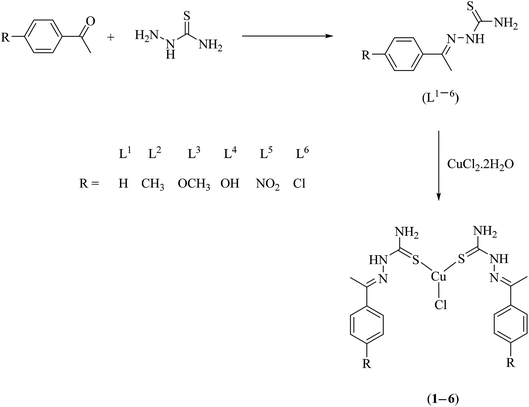
Open Access Article
Open Access ArticleCopper complexes as prospective anticancer agents: in vitro and in vivo evaluation, selective targeting of cancer cells by DNA damage and S phase arrest†
Dharmasivam Mahendirana,
Sethu Amuthakalaa,
Nattamai S. P. Bhuvaneshb,
Raju Senthil Kumarc and
Aziz Kalilur Rahiman *a
*a
aPost-Graduate and Research Department of Chemistry, The New College (Autonomous), Chennai 600 014, India. E-mail: akrahmanjkr@thenewcollege.in; akrahmanjkr@gmail.com; Fax: +91 44 2835 2883; Tel: +91 44 2835 0297
bDepartment of Chemistry, Texas A & M University, College Station, TX 77842, USA
cDepartment of Pharmaceutical Chemistry, Swamy Vivekanandha College of Pharmacy, Elayampalayam, Tiruchengodu 637 205, India
First published on 9th May 2018
Abstract
A series of six new bis(thiosemicarbazone)copper(I) complexes of the type [Cu(L1–6)2Cl] (1–6) have been synthesized and characterized. The molecular structure of the ligand L4 was determined by the single crystal XRD method. All the complexes adopted trigonal planar (Y-shaped) geometry. All the complexes strongly bind with CT-DNA via intercalative mode, which was further supported by molecular docking studies. Further, the complexes were effectively bind with BSA as observed by UV-Vis and fluorescence spectra. All the complexes effectively cleave pBR322 DNA through hydrolytic pathway as evidenced from T4 ligase experiments. All the complexes interact with the anticancer receptor focal adhesion kinase (FAK) via electrostatic, van der Waals, hydrogen bonding, σ–π and π–π interactions. In vitro cytotoxicity of the complexes were assessed by MTT assay against four cancer cell lines such as human breast adenocarcinoma (MCF-7), cervical (HeLa), epithelioma (Hep-2) and Ehrlich ascites carcinoma (EAC), and two normal cell lines namely normal human dermal fibroblasts (NHDF) and L6 myotubes with respect to the commercially used anticancer drug cisplatin. All the complexes induce apoptosis in EAC cells, which was confirmed by AO/EB, Hoechst 33258 and PI staining methods. The complexes block cell cycle progression of EAC cells in S phase (DNA synthesis). The cellular uptake studies confirmed the ability of the complexes to go into the cytoplasm and accumulation in the cell nuclei. In the in vivo anticancer studies, the complexes significantly reduce the tumour volume in female Swiss albino mice. Overall, our results ensure the role of thiosemicarbazone-based copper(I) complexes as prospective anticancer agents, induction of apoptosis and S phase arrest with the mitochondrial controlled pathway.
Introduction
Thiosemicarbazones and its derivatives have attracted scholarly attention because of their structure and prominent role in cancer chemotherapy.1,2 For example, 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (triapine) is a potential inhibitor of ribonucleotide reductase, which exhibits promising anticancer activity in several clinical trials on patients with hematological diseases. Similarly, di-2-pyridylketone-4,4-dimethyl-3-thiosemicarbazone (Dp44mt) and di-2-pyridylketone-4-cyclohexyl-4-methyl-3-thiosemicarbazone (DpC) are examined as drug candidates and entered into the phase I clinical trials.3 In addition, the di-2-pyridylketone thiosemicarbazones (DpT) have showed significant in vitro and in vivo antitumor activity against various cancer cell lines.1 On the other hand, CuII-diacetyl-bis(N4-methylthiosemicarbazone) radiolabeled with copper radioactive isotopes is entered in human clinical trials as an imaging agent of hypoxia in head and neck cancer.4 The bis(thiosemicarbazonato)copper(II) complexes have also been investigated as an effective therapeutic agent for the treatment of neurodegenerative disorders,5 and further, such complexes robustly inhibit the oxidative phosphorylation and DNA synthesis.6,7 In vitro and in vivo studies of glyoxal-bis[N(4)-methylthiosemicarbazonato]copper(II) complexes have demonstrated the strong inhibition of the growth of brain tumor and prostate cancer cells,8 delivers Cu into neurons, activating PI3K and inhibiting GSK3, resulting in the reduction of neurotoxic Aβ trimers and τ-phosphorylation and improving cognition in APP/PS1 mice.5 In addition, the significant therapeutic potential of thiosemicarbazone-based copper complexes for Parkinson's and Alzheimer's diseases in animal models are also reported.5,9 The thiosemicarbazone-based copper(II) complexes significantly inhibit topoisomerase-IIα when compared to the free thiosemicarbazones. Such complexes were also inhibit the proliferation of breast cancer cells (MCF-7).10 Further, the thiosemicarbazone-based copper(II) complexes containing anthracene moiety strongly interact with DNA/BSA and exhibit remarkable anticancer activity against HeLa cancer cell line.11 It was recently reported that the iminodiacetate thiosemicarbazones-based copper(II) complexes functions as ribonucleotide reductase R2 inhibitors.12As a whole, the thiosemicarbazones and bis(thiosemicarbazones)-based copper(II) complexes are broadly interrogated. However, the reports of bis(thiosemicarbazones)-based copper(I) complexes are very limited.13,14 Notably, three coordinate copper(I) complexes have been investigated for DNA interaction, antibacterial and anticancer activity on human colon and breast cancer cells.15–18 The copper(I) chelator MT-3 (metallothionein-3) was significantly protecting cells from Cu-Aβ toxicity compared to copper(II) chelator HSA (Human Serum Albumin). For this reason, +I state of Cu is as much biologically relevant as the +II state.19 Many Cu(I) complexes are found to strongly inhibit the ribo nucleoside diphosphate reductase.20 The redox cycling applications between Cu2+ and Cu+ can begin the production of highly reactive hydroxyl radicals, which can subsequently damage biomolecular species such as lipids, proteins and DNA.21 The N,N′-disubstituted thioureas-based copper(I) complexes showed moderate cytotoxicity against A498, EVSA-T, H226, IGROV, M19, MCF-7 and WIDR cell lines.22 The Cu(I) ions are famous for their high cytotoxicity as an impact of oxidative damage (particularly against eukaryotic and prokaryotic cells), while as Cu(I) helicates might be accurately directed to molecular targets into the shilpit cells.23 Numerous organisms encode a small cytoplasmic high affinity Cu(I) fastening protein, termed a copper chaperone that assumes a role in constraining the collateral damage of copper toxicity.24 Recently, the copper(I) phosphine complexes were reported, which shows significant cytotoxic and anti-angiogenic property and induced a marked reduction of tumor mass in in vivo experiments without a significant body weight loss with smaller adverse side effects than cisplatin.25 To the best of our knowledge, DFT calculations, DNA/BSA interaction, in vitro and in vivo anti-proliferative, cell cycle arrest, generation of ROS, Western blot and molecular modeling studies of bis(thiosemicarbazones)-based copper(I) complexes and their derivatives are scare in literature.17
Based on the above information, we are prompted to target selected cancer cells by both in vitro and in vivo, and study their mechanism of action. To achieve this, we have synthesized new thiosemicarbazone based-copper(I) complexes. In vitro cytotoxicity of the complexes was tested against four human breast adenocarcinoma (MCF-7), cervical (HeLa), epithelioma (Hep-2) and Ehrlich ascites carcinoma (EAC) cancerous and two normal (normal human dermal fibroblasts (NHDF) and L6 myotubes) cell lines. In vivo anticancer activity of the complexes was tested against EAC tumor model using female Swiss albino mice. We have also explored the DFT calculations to elucidate the details of the geometry optimization, electronic structures and frontier molecular orbitals (FMOs). The DNA and focal adhesion kinase (FAK) receptor docking studies were performed by using AutoDock 4.2 tools.
Experimental
Materials
Thiosemicarbazide, acetophenone, 4-chloroacetophenone, 4-hydroxyacetophenone, 4-methylacetophenone, 4-methoxyacetophenone and 4-nitroacetophenone used for ligand synthesis were purchased from Sigma-Aldrich (USA). Solvents of analytical grade were purchased from E. Merck, and used as received without further purification. Agarose (molecular biology grade) and ethidium bromide were procured from Sigma-Aldrich (USA). Calf-thymus DNA (CT-DNA) and supercoiled pBR322 DNA were purchased from Bangalore Genei (India). Tris(hydroxymethyl)aminomethane-hydrochloride (Tris–HCl) buffer (pH, 7.3) was used for all DNA binding and cleavage studies.The ligands 2-(1-phenylethylidene)hydrazinecarbothioamide (L1), 2-(1-(4-tolyl) ethylidene)hydrazinecarbothioamide (L2), 2-(1-(4-methoxyphenyl)ethylidene)hydrazinecarbothioamide (L3), 2-(1-(4-hydroxyphenyl)ethylidene)hydrazinecarbothioamide (L4), 2-(1-(4-nitrophenyl)ethylidene)hydrazinecarbothioamide (L5) and 2-(1-(4-chlorophenyl)ethylidene)hydrazinecarbothioamide (L6) were synthesized by following the procedure as described in the literature.26
Physical measurements
The elemental analysis (CHN) of the compounds was carried out with a Carlo Erba model-1106 elemental analyzer. IR spectra were recorded on a Perkin-Elmer FT IR 8300 model spectrophotometer using KBr disc technique in the range 4000–400 cm−1. Electronic absorption spectra were recorded using Perkin-Elmer Lambda-35 spectrophotometer in the range 200–800 nm. 1H NMR spectral data were collected on Varian-VNMRS-400 in CDCl3 and DMSO(d6) solution with tetramethylsilane (TMS) as an internal standard at ambient temperature. Fluorescence spectra were recorded on a Horiba Jobin Yvon FluoroLog SPEX-F311 spectrofluorometer. Circular dichroic spectra were recorded in the spectral region 200–300 nm with a Jasco J-815 spectropolarimeter at 25 °C using 0.1 cm path quartz cell. Electrospray ionization (ESI) mass spectra were recorded on Q-Tof mass spectrometer using acetonitrile as a carrier solvent.General procedure for synthesis of bis(thiosemicarbazone)copper(I) complexes (1–6)
A methanolic solution (20 mL) of 4-substituted-thiosemicarbazone (L1–6, 1 mmol) was added slowly with constant stirring to a methanolic solution (20 mL) of CuCl2·2H2O (0.09 g, 0.5 mmol), and the resulting solution was refluxed for 4 h. After cooling the reaction mixture to room temperature, the solid product obtained was collected by filtration, washed with diethyl ether, dried under vacuum, and recrystallized from methanol.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1587 ν(N–H)bending, 832 ν(C
N), 1587 ν(N–H)bending, 832 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). UV-Vis [λmax (nm) (ε, M−1 cm−1)] in MeOH: 228 (36
S). UV-Vis [λmax (nm) (ε, M−1 cm−1)] in MeOH: 228 (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 760; π–π*), 346 (3370; n–π*). 1H NMR (400 MHz, CDCl3) (d ppm): 9.69 (s, 1H, NH), 8.99 (s, 1H, NH2), 7.45–7.56 (m, 5H, aromatic), 2.36 (s, 3H, Me). ESI-MS (m/z): 485.54 [Cu(L1)2Cl]+.
760; π–π*), 346 (3370; n–π*). 1H NMR (400 MHz, CDCl3) (d ppm): 9.69 (s, 1H, NH), 8.99 (s, 1H, NH2), 7.45–7.56 (m, 5H, aromatic), 2.36 (s, 3H, Me). ESI-MS (m/z): 485.54 [Cu(L1)2Cl]+.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1567 ν(N–H)bending, 828 ν(C
N), 1567 ν(N–H)bending, 828 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). UV-Vis [λmax (nm) (ε, M−1 cm−1)] in MeOH: 227 (35
S). UV-Vis [λmax (nm) (ε, M−1 cm−1)] in MeOH: 227 (35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 290; π–π*), 351 (3560; n–π*). 1H NMR (400 MHz, CDCl3) (d ppm): 9.74 (s, 1H, NH), 8.94 (s, 1H, NH2), 7.17–7.86 (m, 5H, aromatic), 2.31 (s, 3H, Me). ESI-MS (m/z): 513.59 [Cu(L2)2Cl]+.
290; π–π*), 351 (3560; n–π*). 1H NMR (400 MHz, CDCl3) (d ppm): 9.74 (s, 1H, NH), 8.94 (s, 1H, NH2), 7.17–7.86 (m, 5H, aromatic), 2.31 (s, 3H, Me). ESI-MS (m/z): 513.59 [Cu(L2)2Cl]+.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1596 ν(N–H)bending, 836 ν(C
N), 1596 ν(N–H)bending, 836 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). UV-Vis [λmax (nm) (ε, M−1 cm−1)] in MeOH: 234 (39
S). UV-Vis [λmax (nm) (ε, M−1 cm−1)] in MeOH: 234 (39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 580; π–π*), 348 (3480; n–π*). ESI-MS (m/z): 545.60 [Cu(L3)2Cl]+.
580; π–π*), 348 (3480; n–π*). ESI-MS (m/z): 545.60 [Cu(L3)2Cl]+.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1573 ν(N–H)bending, 831 ν(C
N), 1573 ν(N–H)bending, 831 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). UV-Vis [λmax (nm) (ε, M−1 cm−1)] in MeOH: 231 (39
S). UV-Vis [λmax (nm) (ε, M−1 cm−1)] in MeOH: 231 (39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100; π–π*), 353 (3610; n–π*). ESI–MS (m/z): 517.54 [Cu(L4)2Cl]+.
100; π–π*), 353 (3610; n–π*). ESI–MS (m/z): 517.54 [Cu(L4)2Cl]+.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1579 ν(N–H)bending, 1521 ν(NO2)asym, 1372 ν(NO2)sym, 840 ν(C
N), 1579 ν(N–H)bending, 1521 ν(NO2)asym, 1372 ν(NO2)sym, 840 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). UV-Vis [λmax (nm) (ε, M−1 cm−1)] in MeOH: 236 (39
S). UV-Vis [λmax (nm) (ε, M−1 cm−1)] in MeOH: 236 (39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 840; π–π*), 349 (3510; n–π*). ESI-MS (m/z): 575.52 [Cu(L5)2Cl]+.
840; π–π*), 349 (3510; n–π*). ESI-MS (m/z): 575.52 [Cu(L5)2Cl]+.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1598 ν(N–H)bending, 842 ν(C
N), 1598 ν(N–H)bending, 842 ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). UV-Vis [λmax (nm) (ε, M−1 cm−1)] in MeOH: 229 (38
S). UV-Vis [λmax (nm) (ε, M−1 cm−1)] in MeOH: 229 (38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 210; π–π*), 354 (3680; n–π*). ESI-MS (m/z): 554.44 [Cu(L6)2Cl]+.
210; π–π*), 354 (3680; n–π*). ESI-MS (m/z): 554.44 [Cu(L6)2Cl]+.X-ray crystallography studies
X-ray diffraction intensity data of the ligand L4 was collected at room temperature (293 K) on a Bruker APEX 2 single crystal X-ray diffractometer equipped with graphite monochromatic Mo Kα (λ = 0.71073 Å) radiation and CCD detector. The crystal dimension of 0.25 × 0.19 × 0.04 mm3 was mounted on a glass fiber using cyanoacrylate adhesive. The unit cell parameters were determined from 36 frames measured (0.5° phi-scan) from three different crystallographic zones using the method of difference vectors. The intensity data were collected with an average four-fold redundancy per reflection and optimum resolution (0.75 Å). The intensity data collection, frames integration, Lorentz and polarization corrections and decay correction were carried out using SAINT-NT (version 7.06a) software.27 An empirical absorption correction (multi-scan) was performed using the SADABS program.27 The crystal structure was solved by direct methods using SHELXS-97 (ref. 28) and SHELXS-2014,29 and refined by full-matrix least-squares using SHELXL-2014, SHELXL-2016 (ref. 29) and Olex2.30 Molecular geometry was calculated using PARST programme.31 All non-hydrogen atoms were refined using anisotropic thermal parameters. The hydrogen atoms were included in the structure factor calculation at idealized positions using a riding model, but not refined. Images were created with the ORTEP-PLATON program.32,33DNA/BSA binding, DNA cleavage and in silico studies
DNA/BSA binding, DNA cleavage and in silico studies of DNA and focal adhesion kinase (FAK) receptor were carried out by following the procedure reported in our earlier publications.34,35In vitro and in vivo anti-proliferative studies
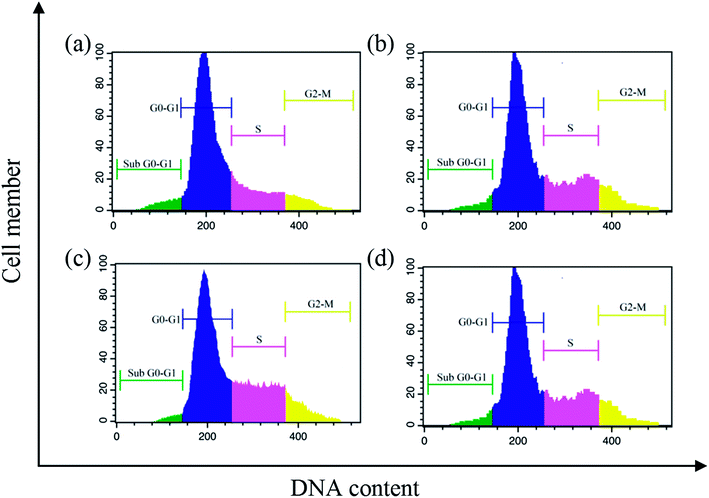
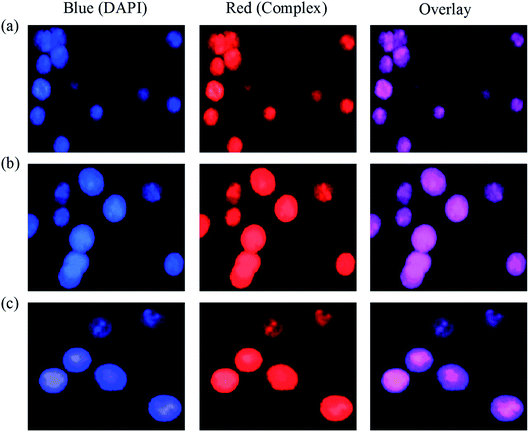
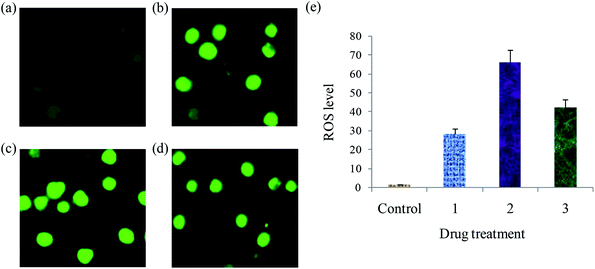
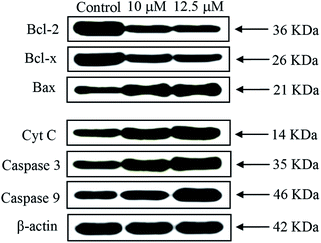
Lipophilicity, MTT assay, apoptosis, cell cycle, cellular uptake, ROS generation, Western blot and in vivo anticancer analyses were carried out by following the procedure reported in our earlier publications.36,37 The study protocol of the in vivo experiments was approved by the Institutional Animal Ethical Committee (IAEC) (No. SVCP/016/2015-16 dated 18-02-2016) and all the animal experiments were carried out in accordance with the guidelines of the Committee for the Purpose of Control and Supervision of Experiments in Animals (CPCSEA), India. For the determination of LD50, OECD guideline AOT 423 was followed.38Results and discussion
Synthesis and characterization of bis(thiosemicarbazone)copper(I) complexes
A series of bis(thiosemicarbazone)copper(I) complexes of the type [Cu(L1–6)2Cl] (1–6) were synthesized in good yields and pure forms by the reaction of thiosemicarbazone ligands (L1–6) with CuCl2·2H2O in mole ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Scheme 1). The reaction may be promoted by the thione-thiol tautomerism of thiosemicarbazones,39,40 the thiol form possibly acting as the reductant. The possibility of redox reaction with Cu(II) salts affording the Cu(I) complexes is known for certain thiosemicarbazones.41 The synthesized complexes were characterized by elemental analysis, FT IR, UV-Vis, 1H NMR and ESI-MS spectral analysis. The structure of ligand L4 was confirmed by single crystal XRD technique.
1 (Scheme 1). The reaction may be promoted by the thione-thiol tautomerism of thiosemicarbazones,39,40 the thiol form possibly acting as the reductant. The possibility of redox reaction with Cu(II) salts affording the Cu(I) complexes is known for certain thiosemicarbazones.41 The synthesized complexes were characterized by elemental analysis, FT IR, UV-Vis, 1H NMR and ESI-MS spectral analysis. The structure of ligand L4 was confirmed by single crystal XRD technique.
FT IR spectra provide valuable information regarding the formation of thiosemicarbazone ligands and their copper(I) complexes (Fig. S1†). The ligands (L1–6) exhibit characteristic bands in the range 3338–3324, 3167–3113, 1635–1608 and 865–839 cm−1 attributed to ν(N–H), ν(C–H), ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) and ν(C
N) and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) stretching vibrations, respectively. The ligand L4 shows broad band at 3409 cm−1 corresponding to ν(O–H) stretching of phenolic group. All the complexes exhibit ν(C
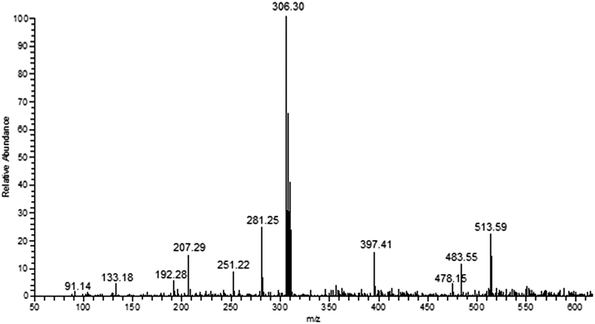
S) stretching vibrations, respectively. The ligand L4 shows broad band at 3409 cm−1 corresponding to ν(O–H) stretching of phenolic group. All the complexes exhibit ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) stretching vibration at 819–838 cm–1, which is lower than that of the free thiosemicarbazone ligands representing the coordination of the copper ion with the sulphur atoms. The formation and stoichiometric composition of the synthesized copper(I) complexes were established by ESI-MS mass spectra. The observed molecular ion peaks confirmed the proposed empirical molecular formulae. For example, complexes 1–6 show molecular ion peak at m/z 485.54, 513.59, 545.60, 517.54, 575.52 and 554.44 due to [Cu(L1−6)2Cl]+, respectively, which is corresponding to their molecular weight (Fig. 1 & S2–S4†). The base peak at m/z 292.24, 306.30, 322.30, 308.25, 337.28 and 326.72 corresponds to [Cu(L1–6)Cl]+, respectively. The observed mass spectral data confirmed the proposed molecular formulae of the synthesized copper(I) complexes. The electronic spectra, which give more information about the structural and geometrical properties of the complexes showed an intense absorption peak at 227–236 nm due to intra-ligand charge transfer transition (π–π*). The medium intense absorption band observed at 341–353 nm is attributed to LMCT or MLCT transition. The non-appearance of band over 400 nm are due to d10 electronic configuration of copper(I) complexes (Fig. S5†).
S) stretching vibration at 819–838 cm–1, which is lower than that of the free thiosemicarbazone ligands representing the coordination of the copper ion with the sulphur atoms. The formation and stoichiometric composition of the synthesized copper(I) complexes were established by ESI-MS mass spectra. The observed molecular ion peaks confirmed the proposed empirical molecular formulae. For example, complexes 1–6 show molecular ion peak at m/z 485.54, 513.59, 545.60, 517.54, 575.52 and 554.44 due to [Cu(L1−6)2Cl]+, respectively, which is corresponding to their molecular weight (Fig. 1 & S2–S4†). The base peak at m/z 292.24, 306.30, 322.30, 308.25, 337.28 and 326.72 corresponds to [Cu(L1–6)Cl]+, respectively. The observed mass spectral data confirmed the proposed molecular formulae of the synthesized copper(I) complexes. The electronic spectra, which give more information about the structural and geometrical properties of the complexes showed an intense absorption peak at 227–236 nm due to intra-ligand charge transfer transition (π–π*). The medium intense absorption band observed at 341–353 nm is attributed to LMCT or MLCT transition. The non-appearance of band over 400 nm are due to d10 electronic configuration of copper(I) complexes (Fig. S5†).
The 1H NMR spectra of the ligands L1&2 and complexes 1 & 2 were recorded to authenticate the coordination of the thiosemicarbazone ligands to the copper(I) ion (Fig. S6 & S7†). The ligands L1&2 showed singlet signal at 2.36 and 2.31 ppm, respectively, corresponding to methyl (CH3) protons. The multiplets observed in the region 7.17–7.86 ppm for the ligands L1&2 have been assigned to the aromatic protons of thiosemicarbazone moiety. The signal positions of methyl and aromatic protons were observed at almost same positions without any appreciable shifts for the complexes. The NH2 protons of ligands L1&2 observed at 8.47 and 8.28 ppm, respectively, with a downfield shift for complexes 1 and 2 (δ = 8.99 and 8.94 ppm). The NH proton of ligands L1&2 observed at δ = 10.30 and 10.20 ppm, respectively, with upfield shift for complexes 1 and 2 (δ = 9.69 and 9.74 ppm). The observed NMR data gave additional information about the structure of the ligands as well as complexes.
Structural description of ligand L4
The ligand L4 crystallizes in the triclinic P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group with two molecules in the unit cell (a = 6.885(3), b = 7.108(3) and c = 11.230(4) Å; α = 74.336(4), β = 76.662(4), γ = 73.802(4)° and Z = 2). The crystal data and structure refinement parameters are listed in Table S1.† The three dimensional molecular structure of this ligand was determined by X-ray crystallography using SHELXS-97/SHELXS-2014 and refined by full-matrix least-squares using SHELXL-2016/Olex2 to a final R-values of 0.0382.
space group with two molecules in the unit cell (a = 6.885(3), b = 7.108(3) and c = 11.230(4) Å; α = 74.336(4), β = 76.662(4), γ = 73.802(4)° and Z = 2). The crystal data and structure refinement parameters are listed in Table S1.† The three dimensional molecular structure of this ligand was determined by X-ray crystallography using SHELXS-97/SHELXS-2014 and refined by full-matrix least-squares using SHELXL-2016/Olex2 to a final R-values of 0.0382.
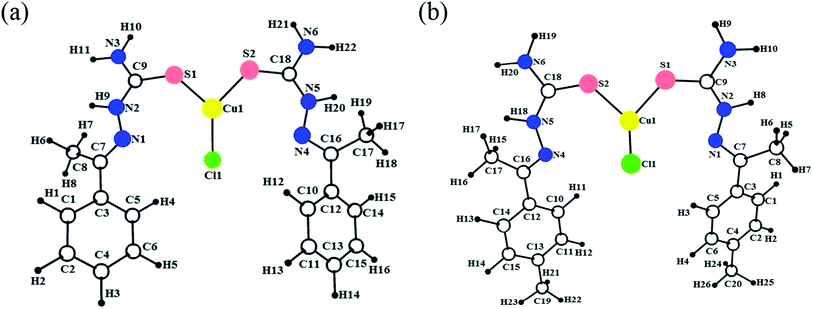
The molecular structure of the ligand L4 is shown in Fig. 2, and the selected bond lengths and bond angles are listed in Table S2.† In the crystal structure, the phenyl ring is fused with the thiosemicarbazone moiety. The phenyl ring and thiosemicarbazone moiety lie in a plane, which is evidenced by the torsion angle value C1–C7–N1–N2 = −176.7°. The thiosemicarbazone moiety with an extended conformation can be seen from the torsion angle values C7–N1–N2–C9 = −167.6° and N1–N2–C9–S1 = −174.1°. The best plane passing through the thiosemicarbazone group makes a dihedral angle of 43.63(1)° with the phenyl ring. The methyl group (C8) is oriented syn-periplanar to C2 [C2–C1–C7–C8 = 28.1°] and anti-periplanar to C6 [C6–C1–C7–C8 = −151.6°]. The O1 atom deviates from the phenyl ring by −0.043(1) Å. The details of hydrogen bonding of ligand L4 is given in Table S3.† The ligand L4 has N3–H3⋯N1 intramolecular hydrogen bonds forming an S(5) ring motifs as shown in Fig. 2. In the crystal structure of L4, the N3–H3B⋯S1 intermolecular interactions generating R22(8) ring motif, N2–H2⋯S1 intermolecular interactions forming R22(8) ring motif, and C8–H8A⋯S1 & N2–H2⋯S1 interactions constitute a pair of bifurcated acceptor bonds generating R21(7) ring motif viewed down 'b' axis as shown in Fig. S8a.† The S1 atom is involved in well defined trifurcated acceptor hydrogen bonds with the N2–H2⋯S1, N3–H3B⋯S1 and C8–H8A⋯S1 as shown in Fig. S8a.† The N3–H3A⋯O1 intermolecular interaction generating R22(22) ring motif viewed down 'a' axis as shown in Fig. S8b.† The intermolecular O1–H1⋯S1 hydrogen bonds forming a C(11) chain running along c axis as shown in Fig. S8c.† The two molecules are also held together by π–π interaction with the distance 3.8108(2) Å between the centroids of two adjacent 4-hydroxybenzene rings (symmetry code is Cg1 = 1 − x, 1 − y, 1 − z) as shown in Fig. S8d.†
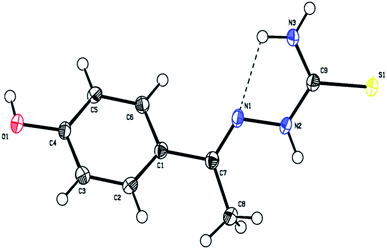 | ||
| Fig. 2 The molecular structure of the ligand L4 showing the atom labelling scheme. The displacement ellipsoids are drawn at 50% probability level. | ||
Geometry optimization
The DFT calculation gives more suitable information about structural features, in the absence of crystal data, in addition to the energy minimized conformation, such as coordination behaviour in metal complexes and HOMO–LUMO energy gap.35 Hereby, all the synthesized copper(I) complexes (1–6) were optimized at B3LYP/GEN and B3LYP/LANL2DZ levels in gas phase and the optimized ground state geometry structures are shown in Fig. 3. The calculated bond lengths and bond angles of the complexes are given in Table S4† along with the reported experimental values.15,16 The calculated values of bond length/angles obtained by using B3LYP/LANL2DZ level are more appropriate than the B3LYP/GEN level. Therefore, DFT/B3LYP/LANL2DZ level was preferred to B3LYP/GEN level for determining the molecular structure. The three coordinated Cu(I) complexes (1–6), coordinate through two sulphur and one chlorine atom. The observed Cu–S bond length values (Cu–S1 = 2.210–2.219 Å; Cu–S2 = 2.219–2.236 Å) and the Cu–Cl bond length values (2.301–2.312 Å) of the complexes are more consistent with the previously reported work.15 It has been noticed that many bond lengths and bond angles obtained from the optimized structures were fitted with the experimentally observed data with a slight differences. The differences in calculated and experimental bond length values of Cu–S1, Cu–S2 and Cu–Cl are found in the range 0.001–0.008. The difference in calculated and experimental bond angles of (S2)–Cu1–(S1), (S2)–Cu1–(Cl1) and (S1)–Cu1–(Cl1) are found in the range 0.17–2.34. The negligible differences were observed between the calculated and experimental bond length and bond angles values. The B3LYP method in combination with the LANL2DZ basis gives an excellent estimation of Cu–S and Cu–Cl bond distances (Cu–S1, Cu–S2 and Cu–Cl).Biological significance
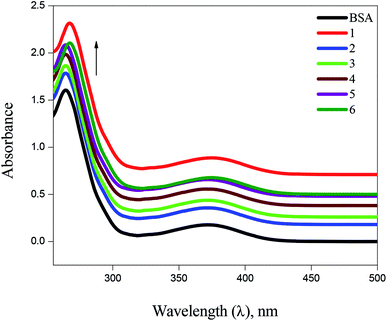
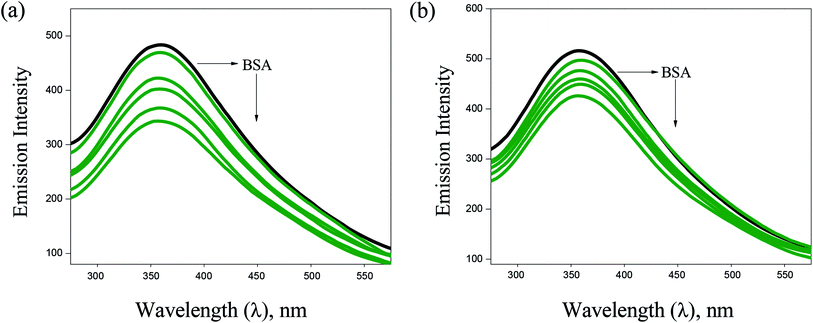
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values between 0.76 ± 34 and 1.43 ± 11, which means higher solubility of complexes in n-octanol (lipophilic phase) than in water (hydrophilic phase). The observed lipophilicity values are higher than those for the previously reported Ag(I), Au(I), Ru(II), Ga(III) and Pt(II) complexes.46–49
P values between 0.76 ± 34 and 1.43 ± 11, which means higher solubility of complexes in n-octanol (lipophilic phase) than in water (hydrophilic phase). The observed lipophilicity values are higher than those for the previously reported Ag(I), Au(I), Ru(II), Ga(III) and Pt(II) complexes.46–49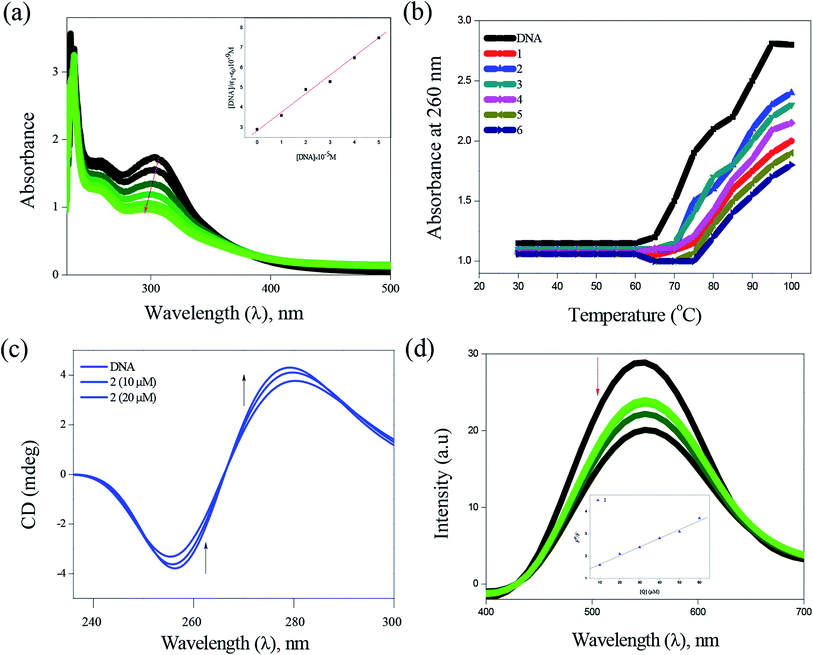 | ||
| Fig. 4 DNA binding of complexes with CT-DNA: absorption spectra of complex 2 (a), thermal denaturation of complexes 1–6 (b), CD spectra of 2 (c) and emission spectra of complex 2 (d). | ||
| Complexes | Kb × 105 (M−1) | Tm (°C) | Kapp × 105 (M−1) | Kq × 105 (M−1) | Kbin × 105 (M−1) | ‘n’ value |
|---|---|---|---|---|---|---|
| 1 | 4.91 ± 0.11 | 68.7 ± 0.82 | 2.3 ± 0.16 | 2.87 ± 1.12 | 2.33 ± 0.18 | 1.31 |
| 2 | 6.01 ± 0.48 | 69.9 ± 0.11 | 3.0 ± 0.52 | 3.48 ± 1.13 | 3.28 ± 1.32 | 1.75 |
| 3 | 5.93 ± 0.12 | 69.7 ± 0.19 | 2.9 ± 0.57 | 3.36 ± 0.22 | 3.17 ± 0.91 | 1.64 |
| 4 | 5.42 ± 0.65 | 69.1 ± 0.31 | 2.6 ± 1.02 | 3.11 ± 0.48 | 2.79 ± 0.75 | 1.48 |
| 5 | 4.44 ± 0.27 | 68.1 ± 0.16 | 1.4 ± 1.18 | 2.49 ± 0.74 | 2.13 ± 0.88 | 1.21 |
| 6 | 2.81 ± 0.79 | 66.5 ± 0.19 | 1.1 ± 1.04 | 2.06 ± 0.97 | 1.54 ± 0.52 | 1.09 |
| Cisplatin | 0.32 ± 0.15 | — | — | — | — | — |
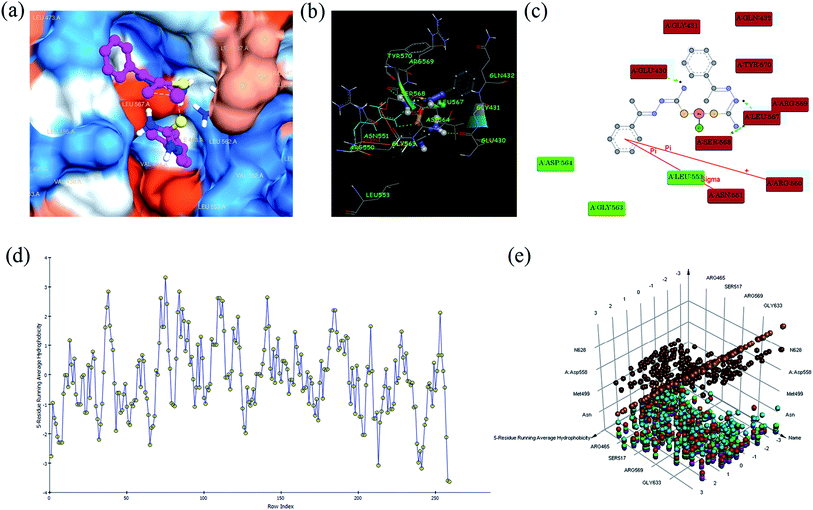
All the complexes (1–6) are located in the hydrophobic cavities in subdomains II and the binding score are listed in Table S6.† All the complexes were strongly interact with FAK via π–π, σ–π, hydrogen bonding, electrostatic and van der Waals interactions (Fig. 8). The complex 1 shows one π–π interaction formed between the phenyl ring and ARG 550 (bond length: 3.6 Å), which also shows one σ–π interaction formed between phenyl ring and ASN 551 (bond length: 3.3 Å). This complex was stabilized by four hydrogen bonding interaction, first one was formed between oxygen atom of GLU 430 and hydrogen atom (–NH) of the ligand (bond length: O⋯H = 3.4 Å), second one was formed between oxygen atom of LEU 567 and hydrogen atom (–NH) of the ligand (bond length: O⋯H = 4.0 Å), third one was formed between oxygen atom of SER 568 and hydrogen atom of –NH2 group present in the ligand (bond length: O⋯H = 3.2 Å), and fourth one was formed between oxygen atom of LEU 567 and hydrogen atom of –NH2 group present in the ligand (bond length: O⋯H = 4.1 Å). Besides, the binding model was enhanced by electrostatic interaction formed between the complex and the residues GLU 430, GLY 431, GLN 432, ARG 550, ASN 551, LEU 567, SER 568, ARG 569 and TYR 570, and van der Waals interaction formed between the complex and the residues LEU 553, GLY 563 and ASP 564 of FAK kinase receptor.
The complex 2 was stabilized by two hydrogen bonding interactions, first one was formed between hydrogen atom of GLU 430 and nitrogen atom (–N) of the ligand (bond length: H⋯N = 3.6 Å), and second one was formed between hydrogen atom of GLU 430 and nitrogen atom (–N) of the ligand (bond length: H⋯N = 3.7 Å). Besides, the binding model was enhanced by electrostatic interaction formed between this complex and residues GLU 430, ARG550, LEU 567 and ARG 569, and van der Waals interaction formed between the complex and the residues GLY 431, GLN 432, PHE 433, VAL 436, ALA 452, GLU 471, GLU 500, CYS 502, LEU 553 and GLY 566 of FAK kinase receptor.
The complex 3 shows one π–π interaction formed between the 4-methoxybenzene ring and ARG 550 (bond length: 3.7 Å), which is also stabilized by three hydrogen bonding interactions, first one was formed between hydrogen atom of ASP 564 and oxygen atom of methoxy group present in the ligand (bond length: H⋯O = 3.8 Å), second one was formed between hydrogen atom of GLU 430 and nitrogen atom (–NH) of the ligand (bond length: H⋯N = 4.1 Å), and third one was formed between oxygen atom of LUE 567 and hydrogen atom (–NH) of the ligand (bond length: O⋯H = 4.0 Å). Besides, the binding model was enhanced by electrostatic interaction formed between this complex and residues GLU 430, GLN 432, ASN 551, ASP 564, LEU 567, ARG 569, TYR 570 and MET 571, and van der Waals interaction formed between the complex and the residues GLY 431, GLU 506, LEU 553, GLY 563 and GLY 566 with FAK kinase receptor.
The complex 4 shows one π–π interaction formed between the 4-hydroxybenzene ring and ARG 550 (bond length: 3.6 Å), which is also stabilized by four hydrogen bonding interaction, first one was formed between hydrogen atom of ARG 569 and oxygen atom of 4-hydroxybenzene ring of the ligand (bond length: H⋯O = 3.2 Å), second one was formed between oxygen atom of GLY 566 and hydrogen atom (–OH) of the ligand (bond length: O⋯H = 3.9 Å), third one was formed between oxygen atom of ARG 550 and hydrogen atom of –NH2 group present in the ligand (bond length: O⋯H = 3.7 Å), and forth one was formed between oxygen atom of LEU 500 and hydrogen atom (–OH) of the ligand (bond length: O⋯H = 2.9 Å). Besides, the binding model was enhanced by electrostatic interaction formed between complex and residues GLN 432, GLU 471, GLU 500, LEU 501, CYS 502, ARG 550, ASN 551, GLY 566, LEU 567 and ARG 569, and van der Waals interaction formed between by complex and residues ILE 428, GLU 430, GLY 431, ALA 452, TYR 570, MET 571, VAL 484, MET 499, LEU 553 and ASP 564 with FAK kinase receptor.
The complex 5 shows one π–π interaction formed between the 4-nitrobenzene ring and ARG 550 (bond length: 3.7 Å), which also shows one σ–π interaction formed between 4-nitrobenzene ring and LEU 567 (bond length: 3.5 Å). This complex was stabilized by six hydrogen bonding interaction, first one was formed between hydrogen atom of ARG 569 and oxygen atom of –NO2 group present in the ligand (bond length: H⋯O = 3.1 Å), second one was formed between hydrogen atom of ARG 569 and nitrogen atom of –NO2 group present in the ligand (bond length: H⋯N = 3.0 Å), third one was formed between hydrogen atom of ARG 569 and oxygen atom of –NO2 group present in the ligand (bond length: H⋯O = 3.4 Å), fourth one was formed between hydrogen atom of ASP 564 and oxygen atom of –NO2 group present in the ligand (bond length: H⋯O = 3.9 Å), fifth one was formed between oxygen atom of ARG 550 and hydrogen atom of –NH2 group present in the ligand (bond length: O⋯H = 3.7 Å), and sixth one was formed between hydrogen atom of GLU 430 and nitrogen atom of –NH group present in the ligand (bond length: H⋯N = 5.3 Å). Besides, the binding model was enhanced by electrostatic interaction formed between the complex and the residues GLU 430, GLN 432, GLU 506, ARG 550, ASN 551, ASP 564, GLY 566, LEU 567, ARG 569 and TYR 570, and van der Waals interaction formed between the complex and the residues GLY 431, VAL 484, LEU 553, GLY 563 and MET 571 with FAK kinase receptor.
The complex 6 shows one π–π interaction formed between the 4-chlorobenzene ring and ARG 550 (bond length: 3.6 Å), which is also stabilized by one hydrogen bonding interaction formed between oxygen atom of GLU 506 and hydrogen atom of –NH2 group present in the ligand (bond length: H⋯O = 4.1 Å). Besides, the binding model was enhanced by electrostatic interaction formed between this complex and residues GLU 430, GLN 432, ASP 564, LEU 567, ARG 569 and TYR 570, and van der Waals interaction formed between the complex and the residues GLY 431, MET 499, GLU 506, LEU 553 and MET 571 with FAK kinase receptor.
The results demonstrate FAK kinase receptor as the potent inhibitor for all the complexes, because all the complexes (1–6) were retained strongly by the binding pocket of FAK. Among the complexes, the complex 2 binds to FAK stronger than the other complexes, which is consistent with the experimental DNA/BSA binding studies. The obtained results of in silico studies indicated that the interaction between complexes and FAK was dominated by electrostatic, van der Waals, hydrogen bonding, π–π and σ–π interactions.
| Complexes | IC50 values (μM)a | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MCF-7 | At 95% confidence interval | R2 | HeLa | At 95% confidence interval | R2 | Hep-2 | At 95% confidence interval | R2 | EAC | At 95% confidence interval | R2 | NHDF | L6 | |
| a Average of three independent determinations; results are expressed as mean ± SD. | ||||||||||||||
| 1 | 16.8 ± 0.53 | 11.43–19.08 | 0.9881 | 14.08 ± 0.66 | 10.32–16.85 | 0.9675 | 15.3 ± 0.32 | 12.08–18.11 | 0.9801 | 14.6 ± 0.37 | 11.87–17.08 | 0.9932 | >100 | >100 |
| 2 | 10.9 ± 0.12 | 07.21–14.01 | 0.9987 | 9.89 ± 0.82 | 06.13–13.17 | 0.9895 | 9.92 ± 1.26 | 07.92–12.64 | 0.9913 | 8.32 ± 1.03 | 05.23–11.05 | 0.9854 | >100 | >100 |
| 3 | 11.2 ± 0.27 | 08.65–15.13 | 0.9911 | 9.91 ± 1.01 | 07.98–13.39 | 0.9802 | 10.1 ± 0.31 | 08.87–12.17 | 0.9788 | 8.91 ± 0.54 | 06.03–11.19 | 0.9863 | >100 | >100 |
| 4 | 13.9 ± 0.41 | 10.04–17.71 | 0.9894 | 12.1 ± 0.17 | 08.47–16.86 | 0.9789 | 12.4 ± 0.82 | 09.74–15.54 | 0.9884 | 11.1 ± 0.83 | 08.33–14.67 | 0.9786 | >100 | >100 |
| 5 | 18.9 ± 1.09 | 14.09–21.18 | 0.9796 | 16.05 ± 0.52 | 11.95–18.22 | 0.9691 | 18.4 ± 0.43 | 15.06–21.23 | 0.9972 | 15.89 ± 1.09 | 12.63–18.19 | 0.9854 | >100 | >100 |
| 6 | 21.7 ± 0.19 | 18.24–23.32 | 0.9869 | 22.94 ± 0.18 | 17.54–25.61 | 0.9601 | 23.9 ± 1.06 | 20.76–26.19 | 0.9846 | 18.08 ± 0.84 | 16.58–21.94 | 0.9809 | >100 | >100 |
| Cisplatin | 12.1 ± 0.84 | 09.06–14.87 | 0.9912 | 10.48 ± 0.12 | 08.77–13.03 | 0.9879 | 14.6 ± 0.81 | 11.02–17.27 | 0.9975 | 11.46 ± 0.86 | 09.92–15.78 | 0.9892 | >100 | >100 |
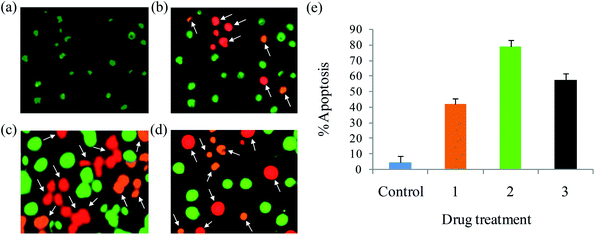 | ||
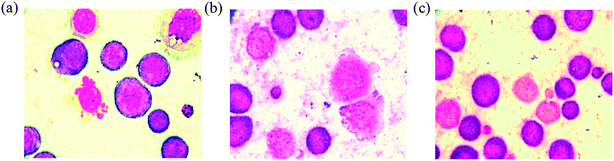
| Fig. 9 AO/EB staining of EAC cells for 24 h: control (a), complexes 1 (b), 2 (c) and 3 (d), and scar bar diagram (e). | ||
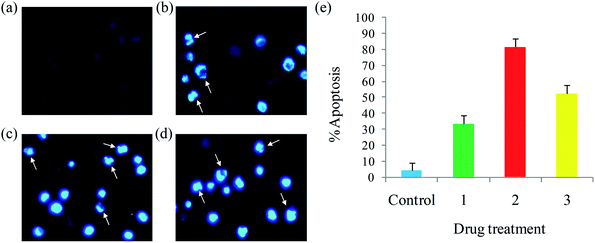 | ||
| Fig. 10 Hoechst 33258 staining of EAC cells for 24 h: control (a), complexes 1 (b), 2 (c) and 3 (d), and scar bar diagram (e). | ||
| Design of treatment | MST (in days) | ILS (%) |
|---|---|---|
| a N = 6; data were expressed as mean ± SEM.b P < 0.001 vs. tumour control.c P < 0.001 vs. 5-FU.d P < 0.01 vs. complex 2; data were analysed by using one-way ANOVA followed by Tukey–Kramer multiple comparison test. | ||
| Tumour control | 18.17 ± 0.87 | — |
| 5-FU | 34.83 ± 0.91c,b | 91.68 |
| 2 | 33.5 ± 1.12d,b | 84.36 |
| 3 | 27.7 ± 0.88b | 52.45 |
| Design of treatment | Day15 | Day20 | Day25 | Day30 |
|---|---|---|---|---|
| a N = 6; data were expressed as mean ± SEM.b P < 0.001 vs. tumour control.c P < 0.001 and.d P < 0.01 vs. complex 2; data were analysed by using one-way ANOVA followed by Tukey–Kramer multiple comparison test. | ||||
| Tumour control | 1.29 ± 0.096 | 1.69 ± 0.096 | 1.94 ± 0.12 | 2.26 ± 0.14 |
| 2 | 0.944 ± 0.096b | 0.99 ± 0.098d,b | 1.06 ± 0.097c,b | 1.13 ± 0.098c,b |
| 3 | 1.02 ± 0.097b | 1.16 ± 0.083b | 1.29 ± 0.081b | 1.79 ± 0.088b |
Conclusions
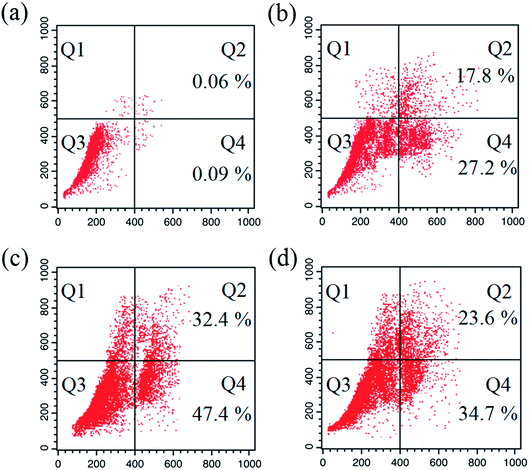
A series of six bis(thiosemicarbazone)copper(I) complexes (1–6) were synthesized and acknowledged as potent anticancer agents. All the complexes adopted trigonal planar 'Y′ shaped geometry. The complexes strongly bind with CT-DNA via intercalative mode, which also show pronounced cleavage activity against pBR322 DNA. All the complexes significantly interact with BSA via static quenching mode and interact with focal adhesion kinase (FAK) receptor via π–π, σ–π, hydrogen bonding, electrostatic and van der Waals interactions. In vitro cytotoxicity of the complexes was tested against four cancer (MCF-7, HeLa, Hep-2 and EAC) and two normal (NHDF and L6 myotubes) cell lines by MTT assay with respect to commercially used metal-based anticancer drug cisplatin. Complexes powerfully kills EAC tumour cell line compared to other tested cancer cell lines and all the complexes proved their non-toxicity against normal cell lines. The complexes exhibit induced apoptosis, which is confirmed by AO/EB, Hoechst 33258 and PI (flow cytometry) staining assays along with superior ROS generation. In the cell cycle arrest, the complexes effectively block the EAC cells in S phase. The cellular uptake study shows that the complexes can go inside the cytoplasm and accumulate in the cell nuclei. Western blot analysis also proved induced apoptosis due to the complexes via mitochondria-dysfunction pathway. In addition, the complexes significantly reduce the solid tumor volume in female Swiss albino mice.Conflicts of interest
There are no conflicts to declare.References
- M. Whitnall, J. Howard, P. Ponka and D. R. Richardson, A class of iron chelators with a wide spectrum of potent antitumor activity that overcomes resistance to chemotherapeutics, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 14901–14906 CrossRef CAS PubMed.
- A. E. Stacy, D. Palanimuthu, P. V. Bernhardt, D. S. Kalinowski, P. J. Jansson and D. R. Richardson, Zinc(II)–thiosemicarbazone complexes are localized to the lysosomal compartment where they transmetallate with copper ions to induce cytotoxicity, J. Med. Chem., 2016, 59, 4965–4984 CrossRef CAS PubMed.
- http://www.clinicaltrials.gov.
- A. L. Vavere and J. S. Lewis, Cu-ATSM: a radiopharmaceutical for the PET imaging of hypoxia, Dalton Trans., 2007, 4893–4902 RSC.
- P. J. Crouch, L. W. Hung, P. A. Adlard, M. Cortes, V. Lal, G. Filiz, K. A. Perez, M. Nurjono, A. Caragounis, T. Du, K. Laughton, I. Volitakis, A. I. Bush, Q. X. Li, C. L. Masters, R. Cappai, R. A. Cherny, P. S. Donnelly, A. R. White and K. J. Barnham, Increasing Cu bioavailability inhibits Aβ oligomers and tau phosphorylation, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 381–386 CrossRef CAS PubMed.
- B. K. Bhuyan and T. Betz, Studies on the mode of action of the copper(II) chelate of 2-keto-3-ethoxybutyraldehyde-bis(thiosemicarbazone), Cancer Res., 1968, 28, 758–763 CAS.
- C. H. Chan-Stier, D. Minkel and D. H. Petering, Reactions of bis(thiosemicarbazonato) copper(II) complexes with tumor cells and mitochondria, Bioinorg. Chem., 1976, 6, 203–217 CrossRef CAS PubMed.
- M. A. Cater, H. B. Pearson, K. Wolyniec, P. Klaver, M. Bilandzic, B. M. Paterson, A. I. Bush, P. O. Humbert, S. La Fontaine, P. S. Donnelly and Y. Haupt, Increasing intracellular bioavailable copper selectively targets prostate cancer cells, ACS Chem. Biol., 2013, 8, 1621–1631 CrossRef CAS PubMed.
- L. W. Hung, V. L. Villemagne, L. Cheng, N. A. Sherratt, S. Ayton, A. R. White, P. J. Crouch, S. Lim, S. L. Leong, S. Wilkins, J. George, B. R. Roberts, C. L. Pham, X. Liu, F. C. Chiu, D. M. Shackleford, A. K. Powell, C. L. Masters, A. I. Bush, G. O'Keefe, J. G. Culvenor, R. Cappai, R. A. Cherny, P. S. Donnelly, A. F. Hill, D. I. Finkelstein and K. J. Barnham, The hypoxia imaging agent Cu II (ATSM) is neuroprotective and improves motor and cognitive functions in multiple animal models of Parkinson's disease, J. Exp. Med., 2012, 209, 837–854 CrossRef CAS PubMed.
- B. M. Zeglis, V. Divilov and J. S. Lewis, Role of metalation in the topoisomerase IIα inhibition and antiproliferation activity of a series of α-heterocyclic-N4-substituted thiosemicarbazones and their Cu(II) complexes, J. Med. Chem., 2011, 54, 2391–2398 CrossRef CAS PubMed.
- A. N. Kate, A. A. Kumbhar, A. A. Khan, P. V. Joshi and V. G. Puranik, Monitoring cellular uptake and cytotoxicity of copper(II) complex using a fluorescent anthracene thiosemicarbazone ligand, Bioconjugate Chem., 2014, 25, 102–114 CrossRef CAS PubMed.
- M. F. Zaltariov, M. Hammerstad, H. J. Arabshahi, K. Jovanović, K. W. Richter, M. Cazacu, S. Shova, M. Balan, N. H. Andersen, S. Radulović, J. Reynisson, K. K. Andersson and V. B. Arion, New iminodiacetate-thiosemicarbazone hybrids and their copper(II) complexes are potential ribonucleotide reductase R2 inhibitors with high antiproliferative activity, Inorg. Chem., 2017, 56, 3532–3549 CrossRef CAS PubMed.
- M. C. Rodriguez-Arguelles, L.-S. Ee, J. Sanmartin, P. Pelagatti and F. Zami, Copper complexes of imidazole-2-, pyrrole-2- and indol-3-carbaldehyde thiosemicarbazones: inhibitory activity against fungi and bacteria, J. Inorg. Biochem., 2005, 99, 2231–2239 CrossRef PubMed.
- A. R. Cowley, J. R. Dilworth, P. S. Donnelly and J. M. White, Copper complexes of thiosemicarbazone-pyridylhydrazine (THYNIC) hybrid ligands: a new versatile potential bifunctional chelator for copper radiopharmaceuticals, Inorg. Chem., 2006, 45, 496–498 CrossRef CAS PubMed.
- P. M. Krishna and K. H. Reddy, Synthesis, single crystal structure and DNA cleavage studies on first 4N-ethyl substituted three coordinate copper(I) complex of thiosemicarbazone, Inorg. Chim. Acta, 2009, 362, 4185–4190 CrossRef CAS.
- N. González-Ballesteros, D. Pérez-Álvarez, M. S. C. Henriques, B. F. O. Nascimento, M. Laranjo, K. Santos, J. Casalta-Lopes, A. M. Abrantes, M. F. Botelho, M. Pineiro, J. A. Paixão and M. C. Rodríguez-Argüelles, Copper(I) complexes of methyl 4-aryl-6-methyl-3,4-dihydropyrimidine-2(1H)-thione-5-carboxylates. Synthesis, characterization and activity in human breast cancer cells, Inorg. Chim. Acta, 2015, 438, 160–167 CrossRef.
- N. González-Ballesteros, D. Pérez-Álvarez, M. C. Rodríguez-Argüelles, M. S. C. Henriques, J. A. Paixão and S. Prado-López, Synthesis, spectral characterization and X-ray crystallographic study of new copper(I) complexes. Antitumor activity in colon cancer, Polyhedron, 2016, 119, 112–119 CrossRef.
- P. R. Chetana, B. S. Srinatha, M. N. Somashekar and R. S. Policegoudra, Synthesis, spectroscopic characterisation, thermal analysis, DNA interaction and antibacterial activity of copper(I) complexes with N,N′-disubstituted thiourea, J. Mol. Struct., 2016, 1106, 352–365 CrossRef CAS.
- G. Meloni, V. Sonois, T. Delaine, L. Guilloreau, A. Gillet, J. Teissié, P. Faller and M. Vasak, Metal swap between Zn7-metallothionein-3 and amyloid-β-Cu protects against amyloid-β toxicity, Nat. Chem. Biol., 2008, 4, 366–372 CrossRef CAS PubMed.
- C. F. Bell, K. A. K. Lott and N. Hearn, Copper complexes of pyridine 2-aldehyde and 2-acetylpyridine thiosemicarbazones, Polyhedron, 1987, 6, 39–44 CrossRef CAS.
- K. Wong, A. R. Morgan and W. Parachych, Controlled cleayage of phage R17 RNA within the virion by treatment with ascorbate and copper(II), Can. J. Biochem., 2001, 52, 950–958 CrossRef.
- M. K. Rauf, I. -ud-Din, A. Badshah, M. Gielen, M. Ebihara, D. de Vos and S. Ahmed, Synthesis, structural characterization and in vitro cytotoxicity and anti-bacterial activity of some copper(I) complexes with N,N′-disubstituted thioureas, J. Inorg. Biochem., 2009, 103, 1135–1144 CrossRef CAS PubMed.
- M. Yang, A. S. Jalloh, W. Wei, J. Zhao, P. Wu and P. R. Chen, Biocompatible click chemistry enabled compartment-specific pH measurement inside, Nat. Commun., 2014, 5, 4981–4991 CrossRef CAS PubMed.
- A. K. Boal and A. C. Rosenzweig, Structural biology of copper trafficking, Chem. Rev., 2009, 109, 4760–4779 CrossRef CAS PubMed.
- V. Gandin, A. Trenti, M. Porchia, F. Tisato, M. Giorgetti, I. Zanusso, L. Trevisi and C. Marzano, Homoleptic phosphino copper(I) complexes with in vitro and in vivo dual cytotoxic and anti-angiogenic activity, Metallomics, 2015, 7, 1497–1507 RSC.
- V. P. Singh, P. Singh and A. K. Singh, Synthesis, structural and corrosion inhibition studies on cobalt(II), nickel(II), copper(II) and zinc(II) complexes with 2-acetylthiophene benzoylhydrazone, Inorg. Chim. Acta, 2011, 379, 56–63 CrossRef CAS.
- Bruker, APEX2, SAINT and SADABS, Bruker AXS Inc., Madison, Wisconsin, USA, 2009 Search PubMed.
- G. M. Sheldrick, A short history of SHELX, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, A64, 112–122 CrossRef PubMed.
- G. M. Sheldrick, Crystal structure refinement with SHELXL, Acta Crystallogr., Sect. A: Found. Adv., 2015, C71, 3–8 CrossRef PubMed.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. K. Howard and H. Puschmann, OLEX2: a complete structure solution, refinement and analysis program, J. Appl. Crystallogr., 2009, 42, 339–341 CrossRef CAS.
- M. P. Nardelli, PARST95-an update to PARST: a system of fortran routines for calculating molecular structure parameters from the results of crystal structure analyses, J. Appl. Crystallogr., 1995, 28, 659 CrossRef CAS.
- A. L. Spek, Structure validation in chemical crystallography, Acta Crystallogr., Sect. A: Found. Crystallogr., 2009, D65, 148–155 CrossRef PubMed.
- L. J. Farrugia, WinGX and ORTEP for Windows: an update, J. Appl. Crystallogr., 2012, 45, 849–854 CrossRef CAS.
- D. Mahendiran, R. S. Kumar, V. Viswanathan, D. Velmurugan and A. K. Rahiman, Targeting of DNA molecules, BSA/c-Met tyrosine kinase receptors and anti-proliferative activity of bis(terpyridine)copper(II) complexes, Dalton Trans., 2016, 45, 7794–7814 RSC.
- D. Mahendiran, R. S. Kumar and A. K. Rahiman, Heteroleptic silver(I) complexes with 2,2′:6′,2′′-terpyridines and naproxen: DNA interaction, EGFR/VEGFR2 kinase, growth inhibition and cell cycle arrest studies, Mater. Sci. Eng. C., 2017, 76, 601–615 CrossRef CAS PubMed.
- D. Mahendiran, P. Gurumoorthy, K. Gunasekaran, R. S. Kumar and A. K. Rahiman, Structural modeling, in vitro antiproliferative activity, and the effect of substituents on the DNA fastening and scission actions of heteroleptic copper(II) complexes with terpyridines and naproxen, New J. Chem., 2015, 39, 7895–7911 RSC.
- D. Mahendiran, R. S. Kumar, V. Viswanathan, D. Velmurugan and A. K. Rahiman, In vitro and in vivo anti-proliferative evaluation of bis(4′-(4-tolyl)-2,2′:6′,2′′-terpyridine)copper(II) complex against Ehrlich ascites carcinoma tumors, J. Biol. Inorg Chem., 2017, 22, 1109–1122 CrossRef CAS PubMed.
- Organization for Economic Co-operation and Development (OECD) guidelines for testing of chemical, Acute oral toxicity Acute toxic class method, Paris, 2001, vol. 423, pp. 1–14 Search PubMed.
- T. S. Lobana, R. Sharma, G. Bawa and S. Khanna, Bonding and structure trends of thiosemicarbazone derivatives of metals-an overview, Coord. Chem. Rev., 2009, 253, 977–1055 CrossRef CAS.
- C. Santini, M. Pellei, V. Gandin, M. Porchia, F. Tisato and C. Marzano, Advances in copper complexes as anticancer agents, Chem. Rev., 2014, 114, 815–862 CrossRef CAS PubMed.
- T. E. Kokina, L. A. Sheludyakova, Y. A. Eremina, E. V. Vorontsova, L. A. Glinskaya, D. A. Piryazev, E. V. Lider, A. V. Tkachev and S. V. Larionov, Complexes of Cu(I) and Pd(I) with (+)-camphor and (−)-cavrone thiosemicarbazones: synthesis, structure, and cytotoxicity of the Pd(II) complex, Russ. J. Gen. Chem., 2017, 87, 2332–2342 CrossRef CAS.
- M. V. Baker, P. J. Barnard, S. J. Berners-Price, S. K. Brayshaw, J. L. Hickey, B. W. Skelton and A. H. White, Cationic, linear Au(I) N-heterocyclic carbene complexes: synthesis, structure and anti-mitochondrial activity, Dalton Trans., 2006, 14, 3708–3715 RSC.
- M. J. McKeage, L. Maharaj and S. J. Berners-Price, Mechanisms of cytotoxicity and antitumor activity of gold(I) phosphine complexes: the possible role of mitochondria, Coord. Chem. Rev., 2002, 22, 127–135 CrossRef.
- J. J. Liu, P. Galettis, A. Farr, L. Maharaj, H. Samarasinha, A. C. McGechan, B. C. Baguley, R. J. Bowen, S. J. Berners-Price and M. J. McKeage, In vitro antitumour and hepatotoxicity profiles of Au(I) and Ag(I) bidentate pyridyl phosphine complexes and relationships to cellular uptake, J. Inorg. Biochem., 2008, 102, 303–310 CrossRef CAS PubMed.
- L. G. Danielsson and Y. H. Zhang, Methods for determining-octanol-water partition constants, Trends Anal. Chem., 1996, 15, 188–196 CAS.
- A. V. Rudnev, L. S. Foteeva, C. Kowol, R. Berger, M. A. Jakupec, V. B. Arion, A. R. Timerbaev and B. K. Keppler, Preclinical characterization of anticancer gallium(III) complexes: solubility, stability, lipophilicity and binding to serum proteins, J. Inorg. Biochem., 2006, 100, 1819–1826 CrossRef CAS PubMed.
- O. Sanchez, S. González, M. Fernández, A. R. H. Padilla, Y. Leon, D. Coll, A. Vidal, P. Taylor, I. Urdanibia, M. C. Goite and W. Castro, Novel silver(I)-and gold(I)-N-heterocyclic carbene complexes. Synthesis, characterization and evaluation of biological activity against tumor cells, Inorg. Chim. Acta, 2015, 437, 143–151 CrossRef CAS.
- A. Pastuszko, K. Majchrzak, M. Czyz, B. Kupcewicz and E. Budzisz, The synthesis, lipophilicity and cytotoxic effects of new ruthenium(II) arene complexes with chromone derivatives, J. Inorg. Biochem., 2016, 159, 133–141 CrossRef CAS PubMed.
- I. V. Tetko, H. P. Varbanov, M. Galanski, M. Talmaciu, J. A. Platts, M. Ravera and E. Gabano, Prediction of log
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P for Pt(II) and Pt(IV) complexes: comparison of statistical and quantum-chemistry based approaches, J. Inorg. Biochem., 2016, 156, 1–13 CrossRef CAS PubMed.
P for Pt(II) and Pt(IV) complexes: comparison of statistical and quantum-chemistry based approaches, J. Inorg. Biochem., 2016, 156, 1–13 CrossRef CAS PubMed. - J. Cornillon, L. Campos and D. Guyotat, Focal adhesion kinase (FAK), une protéine aux fonctions multiples, Medecine Sciences, 2003, 19, 743–752 CrossRef PubMed.
- S. K. Mitra and D. D. Schlaepfer, Integrin-regulated FAK-Src signaling in normal and cancer cells, Curr. Opin. Cell Biol., 2006, 18, 516–523 CrossRef CAS PubMed.
- V. Gabarra-Niecko, M. D. Schaller and J. M. Dunty, FAK regulates biological processes important for the pathogenesis of cancer, Cancer Metastasis Rev., 2003, 22, 359–374 CrossRef CAS PubMed.
- N. A. Chatzizacharias, G. P. Kouraklis and S. E. Theocharis, Clinical significance of FAK expression in human neoplasia, Histol. Histopathol., 2008, 23, 629–650 CAS.
- K. B. Dunn, M. Heffler and V. M. Golubovskaya, Evolving therapies and FAK inhibitors for the treatment of cancer, Anti-Cancer Agents Med. Chem., 2010, 10, 722–734 CrossRef CAS PubMed.
- Saswati, A. Chakraborty, S. P. Dash, A. K. Panda, R. Acharyya, A. Biswas, S. Mukhopadhyay, S. K. Bhutia, A. Crochet, Y. P. Patil, M. Nethaji and R. Dinda, Synthesis, X-ray structure and in vitro cytotoxicity studies of Cu(I/II) complexes of thiosemicarbazone: special emphasis on their interactions with DNA, Dalton Trans., 2015, 44, 6140–6157 RSC.
- A. Khan, J. P. Jasinski, V. A. Smoleaski, K. Paul, G. Singh and R. Sharm, Synthesis, structure and cytotoxicity evaluation of complexes of N1-substituted-isatin-3-thiosemicarbazone with copper(I) halides, Inorg. Chim. Acta, 2016, 449, 119–126 CrossRef CAS.
- P. Sathyadevi, P. Krishnamoorthy, R. R. Butorac, A. H. Cowley and N. Dharmaraj, Synthesis of novel heterobimetallic copper(I) hydrazone Schiff base complexes: a comparative study on the effect of heterocyclic hydrazides towards interaction with DNA/protein, free radical scavenging and cytotoxicity, Metallomics, 2012, 4, 498–511 RSC.
- N. Selvakumaran, L. Sandhiya, N. S. P. Bhuvanesh, K. Senthilkumar and R. Karvembu, Structural diversity in aroylthiourea copper complexes-formation and biological evaluation of [Cu(I)(μ-S)SCl]2, cis-Cu(II)S2O2, trans-Cu(II)S2O2 and Cu(I)S3 cores, New J. Chem., 2016, 40, 5401–5413 RSC.
- S. M. Kumar, K. Dhahagani, J. Rajesh, K. Anitha, G. Chakkaravarthi, N. Kanakachalam, M. Marappan and G. Rajagopal, Synthesis, structural analysis and cytotoxic effect of copper(II)-thiosemicarbazone complexes having heterocyclic bases: A selective naked eye sensor for F− and CN−, Polyhedron, 2015, 85, 830–840 CrossRef.
- M. M. Subarkhan, R. N. Prabhu, R. R. Kumar and R. Ramesh, Antiproliferative activity of cationic and neutral thiosemicarbazone copper(II) complexes, RSC Adv., 2016, 6, 25082–25093 RSC.
- C. C. Chou, J. S. Yang, H. S. Lu, S. W. Ip, C. Lo, C. C. Wu, J. P. Lin, N. Y. Tang, J. G. Chung, M. J. Chou, Y. H. Teng and D. R. Chen, Quercetin-mediated cell cycle arrest and apoptosis involving activation of a caspase cascade through the mitochondrial pathway in human breast cancer MCF-7 cells, Arch. Pharmacal Res., 2010, 33, 1181–1191 CrossRef CAS PubMed.
- U. K. Komarnicka, R. Starosta, M. Płotek, R. F. M. de Almeida, M. Jeżowska-Bojczuk and A. Kyzioł, Copper(I) complexes with phosphine derived from sparfloxacin. Part II: a first insight into the cytotoxic action mode, Dalton Trans., 2016, 45, 5052–5063 RSC.
- A. C. Puckett and J. K. Barton, Mechanism of cellular uptake of a ruthenium polypyridyl complex, Biochemistry, 2008, 47, 11711–11716 CrossRef PubMed.
- A. Malugin, P. Kopečková and J. Kopecék, HPMA copolymer-bound doxorubicin induces apoptosis in ovarian carcinoma cells by the disruption of mitochondrial function, Mol. Pharm., 2006, 3, 351–361 CrossRef CAS PubMed.
- J. M. Adams, Ways of dying: Multiple pathways to apoptosis, Genes Dev., 2003, 17, 2481–2495 CrossRef CAS PubMed.
- A. Burlacu, Regulation of apoptosis by Bcl-2 family proteins, J. Cell. Mol. Med., 2003, 7, 249–257 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1475312. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c8ra00954f |
| This journal is © The Royal Society of Chemistry 2018 |