Open Access Article
Open Access ArticleEfficient biocatalytic stereoselective reduction of methyl acetoacetate catalyzed by whole cells of engineered E. coli
Y. H. Cuia,
P. Weiac,
F. Penga,
M. H. Zongac and
W. Y. Lou *abc
*abc
aLab of Applied Biocatalysis, School of Food Science and Technology, South China University of Technology, Guangzhou 510640, Guangdong, China. E-mail: wyLou@scut.edu.cn; Tel: +86-20-22236669
bGuangdong Province Key Laboratory for Green Processing of Natural Products and Product Safety, South China University of Technology, Guangzhou 510640, Guangdong, China
cSchool of Chemistry and Chemical Engineering, South China University of Technology, Guangzhou 510640, Guangdong, China
First published on 9th March 2018
Abstract
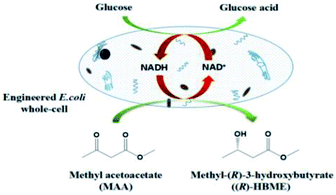
Asymmetric synthesis of chiral β-hydroxy esters, the key building blocks for many functional materials, is currently of great interest. In this study, the biocatalytic anti-Prelog reduction of methyl acetoacetate (MAA) to methyl-(R)-3-hydroxybutyrate ((R)-HBME) was successfully carried out with high enantioselectivity using the whole cell of engineered E. coli, which harbored an AcCR (carbonyl reductase) gene from Acetobacter sp. CCTCC M209061 and a GDH (glucose dehydrogenase) gene from Bacillus subtilis 168 for the in situ regeneration of the coenzyme. Compared with the corresponding wild strain, the engineered E. coli cells were proved to be more effective for the bio-reduction of MAA, and afforded much higher productivity. Under the optimized conditions, the product e.e. was >99.9% and the maximum yield was 85.3% after a reaction time of 10 h, which were much higher than those reported previously. In addition, the production of (R)-HBME increased significantly by using a fed-batch strategy of tuning pH, with a space-time yield of approximately 265 g L−1 d−1, thus the issue in previous research of relatively low substrate concentrations appears to be solved. Besides, the established bio-catalytic system was proved to be feasible up to a 150 mL scale with a large-scale relatively high substrate concentration and selectivity. For further industrial application, these results open a way to use of whole cells of engineered E. coli for challenging higher substrate concentrations of β-ketone esters enantioselective reduction reactions.
1. Introduction
Currently, asymmetric reduction of β-ketone esters with whole-cell biocatalysts, especially using engineered E. coli cells, has attracted much attention from the viewpoint of many recognized journals.1,2 Compared with asymmetric chemical processes, biotransformation processes of chiral β-hydroxy esters production afford several advantages, such as milder reaction operation conditions (e.g., ambient temperature and pressure), no need to use toxic chemicals and expensive heavy metal catalysts.3 Also, chiral β-hydroxy esters can be obtained by isolated carbonyl reductase,4,5 but expensive reducing co-enzymes are necessary in the reaction. These drawbacks restrict its application in industrial-scale production.Chiral β-hydroxy esters are the key building blocks for many fine chiral pharmaceuticals, agrochemicals, flavors and other functional materials.6 For example, it was demonstrated that methyl-3-hydroxybutyrate (HBME) could be a better ingredients for anti-AD drugs,7 and it was reported that dementia was the fourth largest fatal disease, globally.8 In addition, some researchers also found that HBME improved the learning and memory of the mice, then it should be further developed as novel drug candidates against memory decline.9 Besides, it also has been reported that HBME has the ability to inhibit osteoporosis induced by ovariectomization, and it also has been proved to be safe and effective for anti-osteoporosis.10 Moreover, HBME has a great potential to be developed as a biodegradable implant material for its polymers was listed as one of the most promising biodegradable implant material.11,12
The asymmetric bio-reduction of 3-oxo esters using resting whole cells, especially for engineered E. coli cells, is well accepted to be a safe, economical, and mild method to prepare for chiral β-hydroxy ester.13,14 The strategy of adding carbohydrate during the reaction can be adopted for in situ regeneration of co-enzymes with whole cells. Compared with the wild strain, the engineered E. coli offers many advantages for its higher expression level, cheaper medium components, sample enzyme systems and purification procedures. Here a constructing genetic engineering microorganism, which harboring an AcCR (EC 1.1.1.184) gene from Acetobacter sp. CCTCC M209061 and a GDH (EC 1.1.147) gene from Bacillus subtilis 168 for the in situ regeneration of the coenzyme, was applied for catalytic reduction of β-ketone esters.15,16 In our previous study, we found that the application of the engineered E. coli constructed by our group was a little restricted by its relatively low tolerance toward high concentrations of β-ketone esters, which limited its industrial production apparently.16 Aiming at that problem, the biocatalytic reduction of MAA as a model substrate was carried out in the following study and a series of strategy was adopted for improving the productivity.
To date, the biocatalytic anti-Prelog stereoselectivity reduction of MAA to (R)-HBME using engineered E. coli cells has remained largely untapped, with only few published accounts covered it, where some biocatalysts including Baker's yeast,17 Acetobacter pasteurianus GIM1.158 cells,18 carbonyl reductase from Acetobacter sp. CCTCC M209061 were tested and achieved product e.e. and yields of (R)-HBME were unsatisfactory.19 In the case of the baker's yeast as biocatalyst, although the optimum substrate concentration was around 100 mM, the product was (S)-HBME, the yield was only 32.8% and the product e.e. only recorded 86.3%. In addition, the whole cells of engineered E. coli (the alcohol dehydrogenase of L. brevis and the NAD+-dependent format dehydrogenase gene from Mycobacterium vaccae N10 were coexpressed) gave a relatively low optical substrate concentration (around 40 mM) with (R)-HBME was produced.31 The Acetobacter pasteurianus GIM1.158 cells afforded the product yield of 92.9% with over 97.2% product e.e. in the biocatalytic reduction, but the substrate-tolerant concentration was still as low as 10 mM. For industrial-scale application, the yield and enantioselelctivity of (R)-HBME need to be further improved in the aqueous phase (Scheme 1).
In the present study, a wild strain of Acetobacter sp. CCTCC M209061 and an engineered E. coli mentioned above were compared for their potential for bio-reduction of MAA to valuable (R)-HBME. The engineered E. coli cells were found to be more active in catalyzing the bioreduction of MAA, and for the first time were applied for the asymmetric synthesis of the prochiral β-hydroxy ester deeply. The effects of several crucial variables on the microbial cell-based bio-reduction were explored systematically. Besides, for the production of a high yield of (R)-HBME, the glucose acid as a by-product was explored and a fed-batch strategy of tuning pH by the corresponding alkaline compound was adopted with an excellent effectiveness. Also, the efficient biocatalytic process for MAA reduction to (R)-HBME with whole cells of engineered E. coli was evaluated on a preparative scale, and showing a huge potential application in industrialization.
2. Results and discussion
2.1 Effects of key reaction conditions on the biocatalytic reduction of MAA
For further improving the results of the biocatalytic reduction including the initial reaction rate, the yield and the enantioselectivity, it is of great significant to study the influences of some key conditions on the reaction, such as buffer pH, reaction temperature, and the biocatalyst dosage etc. In addition, the comparison of the time course of the bioreduction between resting E. coli whole-cells and the corresponding wild strain under optimized conditions was also conducted.As illustrated in Fig. 1a, buffer pH exerted a notable impact on both the initial reaction rate and the maximum yield. The initial rate and the yield increased with pH changes from 4.0 to 6.5, the maximal initial reaction rate was appeared on pH 7.0, and the maximal yield of 99.0% were achieved within pH from 6.5 to 7.0 after a reaction time of 8.5 h, however, it decreased slightly with the rise of pH from 7.0 to 8.0. Overall, the engineered E. coli cells exhibited good catalytic performances within the range of pH from 6.5–8.0, indicating that there was no problem with the activity of carbonyl reductase and glucose dehydrogenase within that pH range. Moreover, excellent selectivities (>99.9%) were observed within the assayed buffer pH, indicating that the pH has no effect on its enantioselectivity. Taking into account the initial rate and yield, the optimum buffer pH for the bioreduction of MAA to (R)-HBME was 7.0.
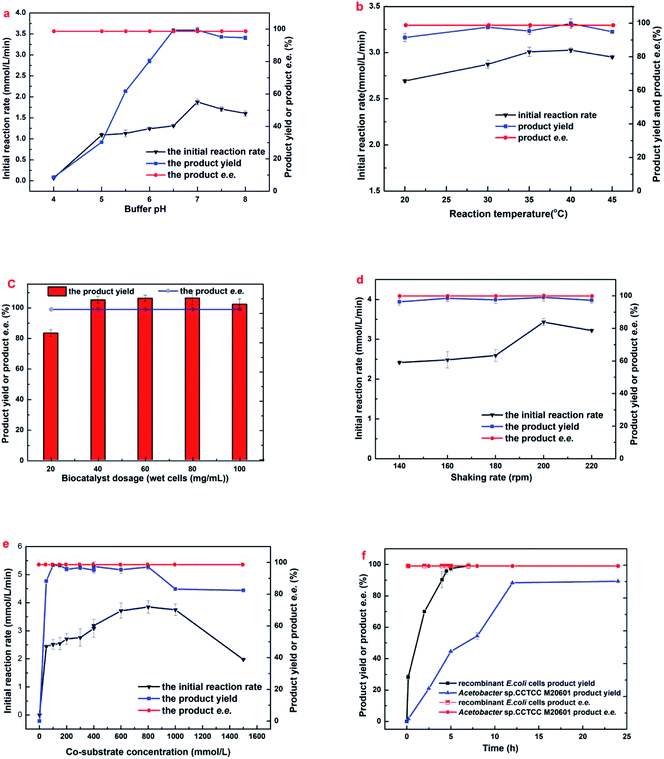 | ||
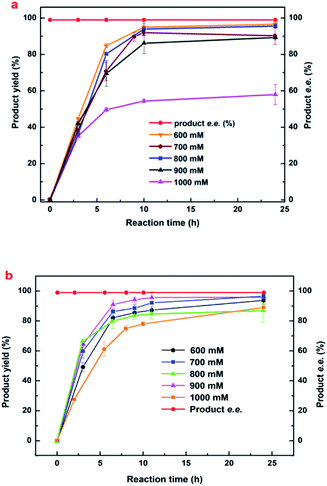
| Fig. 1 Effects of (a) initial buffer pH, (b) biocatalyst dosage, (c) temperature, (d) shaking rate, (e) cosubstrate concentration and (f) reaction time on (R)-HBME synthesis. The following general conditions applied unless otherwise stated: 150 mM MAA, 400 mM glucose, 60 mg mL−1 microbial cells, 4 mL phosphate buffer (200 mM, pH 7.0), 40 °C, 180 rpm; (a) buffers used: pH 4.0–6.0 citrate buffer (200 mM), pH 6.0–8.0 phosphate buffer (200 mM), 30 °C, 7 h; (b) gradual increase of temperature from 20 to 45 °C with corresponding periods of 24, 12, 10, 6, and 6.5 h; (c) gradual increase of cell dosage from 20 to 100 mg mL−1 microbial cells (wet weight) with corresponding periods of 24, 8, 6, 5, and 4 h; (d) gradual increase of shaking rate from 140 to 220 rpm with corresponding periods of 10, 8, 6, 4.5, and 6.5 h; (e) gradual increase of co-substrate concentration from 0 to 1500 mg mL−1 glucose, 200 rpm, and 500 mM glucose was added every 2 h in the case of 1000 mM and 1500 mM glucose when product yield and e.e. was recorded; (f) the two strains subjected to its optimized conditions, respectively: engineered E. coli at pH 7.0, 40 °C, 200 rpm and Acetobacter sp. CCTCC M209061 strain at pH 5.5, 35 °C, 200 rpm.27 | ||
2.2 Substrate and exogenous-product inhibition and toxicity toward engineered E. coli cells
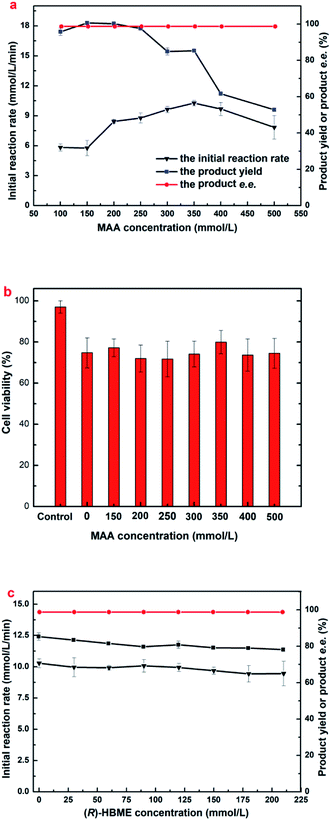
The tolerance of E. coli cells toward MAA was evaluated (Fig. 2a). As shown in Fig. 2a, the reaction rate accelerated markedly when substrate concentration increased from 100 to 350 mM, and the highest initial reaction rate (0.617 mol L−1 h−1) was observed at 350 mM MAA in the presence of 800 mM glucose and a yield of 85.3% was obtained. Further increasing the substrate concentration leading to a clear drop in the initial rate and the yield. However, the maximum yield showed no significantly change when substrate increased from 100 to 300 mM and about 95.5 to 99.0% were obtained within 3–10 h. Continue to increase the substrate concentration to 400 mM resulted in a clear decline to a yield of 61.7%. Microbial cells were capable of smoothly reducing MAA in the concentration up to 500 mM to a yield of 53.1% after 10 h. The effects suggested that the significant inhibition of substrate occurs at a relatively high MAA concentration (above 350 mM). To date, this is the highest MAA concentration of microbial cells ever reported.
It is worth noting that a yield only 85.3% was obtained when the MAA concentration was 350 mM, which could not be explained rationally by the initial reaction rate, because that results showed no substrate and product inhibition occurs at 350 mM MAA. Continue to increase the dosage of E. coli cells at 350 mM of MAA, there was no boost in the yield (dada not shown). It has been demonstrated that some β-ketone esters might be toxic to microbial cells.27 Therefore, cell viability was assayed after the engineered E. coli cells incubated in the presence of various MAA concentrations (Fig. 2b). It was found that more than 79.95% of the cells remained alive after in an incubation period of 12 h when the substrate concentration was from 150 mM to 500 mM, which was comparable to the control group that in the absence of MAA, but the fresh cell survival rate was 97.05% yet. That results suggested that MAA almost exerted no toxic effect on the cells within the range of concentration investigated, and higher substrate concentration just slows down the activity of the enzyme, but there was no death of the most cells. Small part of the cells death could be caused by mechanical damage under 200 rpm speed, and led to a cell lysis occurred in the reaction solution.
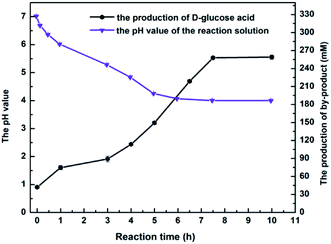
The above results on MAA inhibition and toxicity suggested that engineered E. coli cells could be tolerant to substrate concentrations up to 350 mM, but the substrate didn't convert into the product completely. It is well known that the glucose oxidase oxidizes glucose into glucose acid, which is exhibited weak acidity. Accordingly, the changes of the pH in reaction solution were monitored as the reaction processed at 350 mM concentration of MAA, and it was found that the value of pH in the solution reduced to about 3.7 after an incubation for 10 h. Therefore, we reasoned that the yield was 85.3% rather than approaches 100% might be owed to the D-glucose oxidized to D-glucuronic acid and leading to an acidic environment in the solution. All the data was recorded in the presence of 800 mM glucose, and the product e.e. kept constantly above 99.9% within the tested range of MAA concentration. Obviously, under the present reaction conditions, the optimum concentration of the substrate was up to 350 mM.
2.3 Improved synthesis of (R)-HBME
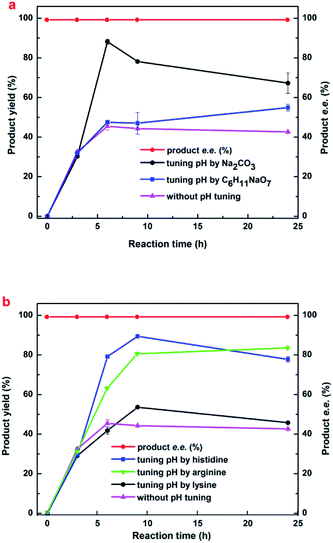
Maximizing the substrate concentration furtherly is critical for efficient synthesis of (R)-HBME from MAA. Additionally, a high concentration of the product accumulation in the reaction solution is highly desired. As we all know, the glucose dehydrogenase oxidizes D-glucose to convert it into D-glucuronic acid in the reaction system, as a result, a dramatically drop in pH on the level may be aroused in the reaction solution. Besides, as illustrated in Fig. 1a, the dropped pH in the reaction solution reduced the reaction rates and inhibited the activities of double-enzyme seriously. Inspired by this consequences, the pH values was monitored in the process of reaction and a fed-batch feeding of the alkaline compound to reaction solution was conducted in the biocatalytic synthesis of (R)-HBME.2.4 Preparative scale biocatalytic reduction of MAA to (R)-HBME
To show the applicability of the biocatalytic enantioselective reduction of MAA to (R)-HBME catalyzed by E. coli cells, we also carried out the bioreduction on a 150 mL preparative scale. The reaction process was monitored by GC analysis and the product was extracted from the reaction mixture with ethyl acetate upon the exhaustion of the substrate. Although slightly lower than that obtained on the 4 mL scale, the isolated yield (85%) was much higher than that reported previously and the product e.e.was very high (>99.9%). Hence the biocatalytic reduction of MAA to (R)-HBME with engineered E. coli cells is promising and competitive.3. Experimental section
3.1 Biological and chemical materials
The engineered E. coli strain was previously reconstructed by our group and preserved in our laboratory.15,16Methyl acetoacetate (99.9% purity, MAA) was from Sigma-Aldrich (USA). Methyl (S)-3-hydroxybutyrate (98% purity, (S)-HBME), methyl (R)-3-hydroxybutyrate (98% purity, (R)-HBME), n-decane (>99.9% purity), His, Arg, Lys, Na2CO3 and C6H11NaO7 (>99.9% purity) were also obtained from Sigma-Aldrich (USA). All other reagents and solvents were purchased from commercial sources and were of analytical pure grade.
3.2 Cultivation of the engineered E. coli cells
The cultivation of the engineered E. coli cells were described in our previous article and within that a difference was the volume and the pH of the cultivation medium was 50 mL and 7.0, respectively.15,16 The cells were at the late exponential growth phase and were harvested by centrifugation (9000g, 5 min, 4 °C), washed twice with 0.85% NaCl solution, and collected by centrifugation (8000g, 5 min, 4 °C). After centrifugation the supernatant was discarded clearly, then the biomass of the wet cells was given by a precision balance of another millimeter.3.3 General procedure for asymmetric reduction of MAA
Typically, 4 mL phosphate buffer (200 mM, pH 7.0) containing 150 mM MAA, 400 mM glucose and 60 mg mL−1 (wet weight) microbial cells was added to a 10 mL stoppered flask capped with a septum. The reaction mixture firstly was incubated at 30 °C and 180 rpm. During this period, equal amount of samples (50 μL) were withdrawn from the reaction mixtures at specified time intervals, the samples were extracted with 200 μL ethyl acetate containing 15.1 mmol L−1 n-decane for GC analysis. The details of the buffer pH, reaction temperature, co-substrate concentration, substrate concentration and exogenous product concentration were specified for each case.The initial reaction rate was calculated as the increase of (R)-HBME concentrations in the first 10 minutes reaction stage. The yield was conducted as the ratio of the measured product amount to the theoretical product amount at the end of the reaction.
3.4 The procedure for improved synthesis of (R)-HBME
Biocatalytic synthesis of (R)-HBME was conducted by fed-batch feeding of D-glucose and corresponding alkaline compounds. After glucose was almost used up, 200 mM glucose were supplemented. Tuning pH by a serious of alkaline compound, including Na2CO3, C6H11NaO7, His, Arg and Lys (added in fed-batch mode). The reactions were carried out at 60 mg mL−1 (wet weight) microbial cells, 200 rpm and 35 °C for 24 h. The substrate concentration was specified for each case.3.5 Preparative scale biotransformation
The preparative scale biocatalytic reduction of MAA to (R)-HBME was carried out by adding 60 mg mL−1 of E. coli cells and 1000 mM of MAA to 150 mL of the co-solvent system of phosphate buffer (200 mM, pH 7.0) containing 1000 mM glucose at 35 °C and 180 rpm. Tuning pH of the reaction mixture to approximately 7.0 every 1.5 h by His–Na2CO3. The reaction was terminated when no substrate was detectable by GC analysis. The products were extracted from the reaction mixture with ethyl acetate. The product e.e. and the isolated yield were also determined by GC analysis.3.6 GC analysis of MAA
The reaction mixtures were analyzed using a columns named CP-Chiralsil-Dex-CB (USA) by a Shimadzu GC2010, which equipped with a flame ionization detector.Split ratio of sample injection was 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and the temperature of the injector, the detector and the column were kept at 170 °C, 170 °C and 90 °C, respectively. The whole process lasts 5.2 minutes. The employed insert gas was nitrogen, to carry the volatile MAA and (R)-HBME through the column. The insert gas flow rate in the column was 1.45 mL min−1. The retention-times for n-decane, MAA and (R)-HBME were 2.6, 3.5 and 4.5 min, respectively.
1, and the temperature of the injector, the detector and the column were kept at 170 °C, 170 °C and 90 °C, respectively. The whole process lasts 5.2 minutes. The employed insert gas was nitrogen, to carry the volatile MAA and (R)-HBME through the column. The insert gas flow rate in the column was 1.45 mL min−1. The retention-times for n-decane, MAA and (R)-HBME were 2.6, 3.5 and 4.5 min, respectively.
The detection of product e.e. was assayed using a same column and with derivatization of (R)-HBME by TBDMCS (tert-butyldimethylsilyl chloride).20 Except for the column temperature 70 °C for 15 min and the flow rate in the column 0.5 mL min−1 and other parameters were under the GC detection conditions described above, the retention-times for (R)-HBME and (S)-HBME were 9.8 and 11.2 min, respectively.
All experiments were performed at least in duplicate and the average error for this determination was less than 1.0%, and the values were expressed as the standard deviations.
3.7 Flow cytometry assay of cell viability
The viability of engineered E. coli cells was assayed after incubated for 12 h in phosphate buffer system (200 mM, pH 7.0), containing 800 mM glucose under the designated conditions. The beads of engineered E. coli cells were withdrawn from the reaction solution and then added to phosphate buffer (200 mM, pH 7.0) to dissolve and wash the beads and diluted to 105 colony-forming units per mL and dyed with 50 μg mL−1 PI for 15 min in the dark.21 A Gallios flow cytometer (Beckman Coulter Inc., USA) was used for determining the cell viability, and the fluorescence emission was observed at 488 nm and was recorded at 630 nm. A software of Kaluza were adopted for data processing. The cell viability was expressed as the ratio of the cells unstained with PI to the total cells.3.8 HPLC analysis of CH2OH(CHOH)4COOH
Firstly, an adequate amount of NaOH were added to the reaction mixtures until the pH of the solution was 10.0, and then the analysis of the glucose acid was converted to analyze the sodium gluconate. The next, the sodium gluconate analyzed using an Eclipse XDB-C18 column (4.6 mm × 250 mm, 5 μm, Agilent, USA) by reversed-phase HPLC equipped with a Waters 996 photodiode array detector (Waters, USA). The mobile phase was the mixture of methanol, ultrapure water and 1.0% H3PO3 solution (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, v/v/v) with a flow rate of 0.6 mL min−1. The retention times of sodium gluconate were 6.3 min.
50, v/v/v) with a flow rate of 0.6 mL min−1. The retention times of sodium gluconate were 6.3 min.
4. Conclusions
A selective and high-efficient biocatalytic approach for the anti-Prelog asymmetric reduction of β-ketone esters using engineered E. coli resting cells has been developed. That engineered E. coli cells had good catalytic performances over a wide range of conditions. Furthermore, the results described here clearly showed that the whole-cell biocatalytic process is feasible up to a 150 mL scale. If further scale-up is possible, the biocatalytic reduction of β-ketone esters by the engineered E. coli whole-cells was promising for the development of an industrially sound biocatalytic process for production.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge the National Natural Science Foundation of China (21336002; 21376096; 21676104), and the Program of State Key Laboratory of Pulp and Paper Engineering (2017ZD05) for partially funding this work.Notes and references
- Z. W. Luo and S. Y. Lee, Nat. Commun., 2017, 8, 1–8 CrossRef PubMed.
- K. Nakamura, K. Kitano, T. Matsuda and A. Ohno, Tetrahedron Lett., 1996, 27, 1629–1632 CrossRef.
- S. M. Thomas, R. Dicosimo and V. Nagarajan, Trends Biotechnol., 2002, 20, 238–242 CrossRef CAS PubMed.
- T. Ema, H. Yagasaki, N. Okita, M. Takeda and T. Sakai, Tetrahedron, 2006, 62, 6143–6149 CrossRef CAS.
- M. Wada, H. Kawabata, M. Kataoka, Y. Yasohara, N. Kizaki, J. Hasegawa and S. Shimizu, J. Biosci. Bioeng., 1999, 87, 144–148 CrossRef CAS PubMed.
- D. Gamenara and d. M. P. Dominguez, Biotechnol. Adv., 2009, 27, 278–285 CrossRef CAS PubMed.
- J. Zhang, Q. Cao, S. Li, X. Lu, Y. Zhao, J. S. Guan, J. C. Chen, Q. Wu and G. Q. Chen, Biomaterials, 2013, 34, 7552–7562 CrossRef CAS PubMed.
- D. L. Hoyert and J. Xu, Deaths: Preliminary Data for 2011, National Center for Health Statistics, 2012, vol. 61, pp. 1–51 Search PubMed.
- X. H. Zou, H. M. Li, S. Wang, M. Leski, Y. C. Yao, X. D. Yang, Q. J. Huang and G. Q. Chen, Biomaterials, 2009, 30, 1532 CrossRef CAS PubMed.
- X. Q. Xiao, Y. Zhao and G. Q. Chen, Biomaterials, 2007, 28, 3608–3616 CrossRef CAS PubMed.
- G. Tian, Q. Wu, S. Sun, I. Noda and G. Q. Chen, Appl. Spectrosc., 2001, 55, 888–893 CrossRef CAS.
- Q. Wu, Y. Wang and G. Q. Chen, Artif. Cells, Blood Substitutes, Immobilization Biotechnol., 2009, 37, 1–12 CrossRef CAS PubMed.
- V. D. Silva, B. U. Stambuk and M. D. G. Nascimento, J. Mol. Catal. B: Enzym., 2012, 77, 98–104 CrossRef CAS.
- Y. Yasohara, N. Kizaki, J. Hasegawa, S. Takahashi, M. Wada, M. Kataoka and S. Shimizu, Appl. Microbiol. Biotechnol., 1999, 51, 847–851 CrossRef CAS PubMed.
- P. Wei, Y. H. Cui, M. H. Zong, P. Xu, J. Zhou and W. Y. Lou, Bioresour. Bioprocess, 2017, 4, 39 CrossRef PubMed.
- P. Wei, J. X. Gao, G. W. Zheng, H. Wu, M. H. Zong and W. Y. Lou, J. Biotechnol., 2016, 230, 54–62 CrossRef CAS PubMed.
- H. N. And and S. Yao, Ind. Eng. Chem. Res., 2007, 46, 7921–7926 CrossRef.
- P. X. Du, P. Wei, W. Y. Lou and M. H. Zong, Microb. Cell Fact., 2014, 13, 84 CrossRef PubMed.
- X. H. Chen, P. Wei, X. T. Wang, M. H. Zong and W. Y. Lou, PLoS One, 2014, 9, e94543 Search PubMed.
- J. H. Shi, X. W. Cheng, L. Zhou and W. Yan, Chin. J. Anal. Chem., 2007, 35, 714–718 CAS.
- X. Y. Zhang, M. H. Zong and N. Li, Green Chem., 2017, 10, 372–378 Search PubMed.
- J. L. Guo, X. Q. Mu and Y. Xu, Bioprocess Biosyst. Eng., 2010, 33, 797–804 CrossRef CAS PubMed.
- N. A. Salvi and S. Chattopadhyay, ChemInform, 2005, 36, 3397–3400 CrossRef.
- F. Luo, D. Lu and Y. Gong, J. Mol. Catal. B: Enzym., 2011, 70, 101–107 CrossRef CAS.
- M. Goretti, C. Ponzoni, E. Caselli, E. Marchegiani, M. R. Cramarossa, B. Turchetti, L. Forti and P. Buzzini, Bioresour. Technol., 2011, 102, 3993–3998 CrossRef CAS PubMed.
- W. Lou, W. Wang, T. J. Smith and M. Zong, Green Chem., 2009, 11, 1377–1384 RSC.
- X. T. Wang, X. H. Chen, Y. Xu, W. Y. Lou, H. Wu and M. H. Zong, J. Biotechnol., 2013, 163, 292–300 CrossRef CAS PubMed.
- C. H. Ra, G. T. Jeong, M. K. Shin and S. K. Kim, Bioresour. Technol., 2013, 140, 421–425 CrossRef CAS PubMed.
- Y. Zhang, B. Han and T. C. Ezeji, N. Biotechnol., 2012, 29, 345–351 CrossRef CAS PubMed.
- Y. Ni and J. H. Xu, Biotechnol. Adv., 2012, 30, 1279–1288 CrossRef CAS PubMed.
- M. Ernst, B. Kaup and S. Bringer-Meyer, Appl. Microbiol. Biotechnol., 2005, 66, 629–634 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2018 |