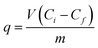Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Cadmium specific proteomic responses of a highly resistant Pseudomonas aeruginosa san ai†
Lidija Izrael-Živković‡
 a,
Milena Rikalović‡b,
Gordana Gojgić-Cvijovićc,
Saša Kazazić
a,
Milena Rikalović‡b,
Gordana Gojgić-Cvijovićc,
Saša Kazazić d,
Miroslav Vrviće,
Ilija Brčeskie,
Vladimir Beškoskie,
Branka Lončarevićc,
Kristina Gopčevića and
Ivanka Karadžić
d,
Miroslav Vrviće,
Ilija Brčeskie,
Vladimir Beškoskie,
Branka Lončarevićc,
Kristina Gopčevića and
Ivanka Karadžić *a
*a
aDepartment of Chemistry, Faculty of Medicine, University of Belgrade, Visegradska 26, Belgrade, Serbia. E-mail: ivanka.karadzic@med.bg.ac.rs; Tel: +381-113607067
bFaculty of Applied Ecology Futura, University of Singidunum, Belgrade, Serbia
cInstitute of Chemistry, Technology and Metallurgy, Department of Chemistry, University of Belgrade, Studentski trg 12, Belgrade, Serbia
dRuđer Bošković Institute, Bijenička cesta 54, Zagreb, Croatia
eFaculty of Chemistry, University of Belgrade, Studentski trg 12-16, Belgrade, Serbia
First published on 16th March 2018
Abstract
Pseudomonas aeruginosa san ai is a promising candidate for bioremediation of cadmium pollution, as it resists a high concentration of up to 7.2 mM of cadmium. Leaving biomass of P. aeruginosa san ai exposed to cadmium has a large biosorption potential, implying its capacity to extract heavy metal from contaminated medium. In the present study, we investigated tolerance and accumulation of cadmium on protein level by shotgun proteomics approach based on liquid chromatography and tandem mass spectrometry coupled with bioinformatics to identify proteins. Size exclusion chromatography was used for protein prefractionation to preserve native forms of metalloproteins and protein complexes. Using this approach a total of 60 proteins were observed as up-regulated in cadmium-amended culture. Almost a third of the total numbers of up-regulated were metalloproteins. Particularly interesting are denitrification proteins which are over expressed but not active, suggesting their protective role in conditions of heavy metal exposure. P. aeruginosa san ai developed a complex mechanism to adapt to cadmium, based on: extracellular biosorption, bioaccumulation, the formation of biofilm, controlled siderophore production, enhanced respiration and modified protein profile. An increased abundance of proteins involved in: cell energy metabolism, including denitrification proteins; amino acid metabolism; cell motility and posttranslational modifications, primarily based on thiol-disulfide exchange, were observed. Enhanced oxygen consumption of biomass in cadmium-amended culture versus control was found. Our results signify that P. aeruginosa san ai is naturally well equipped to overcome and survive high doses of cadmium and, as such, has a great potential for application in bioremediation of cadmium polluted sites.
1. Introduction
In order to survive heavy metal exposure some microorganisms evolved specific mechanisms of resistance that make them key players in bioremediation strategies.1 The use of resistant microbes in the remediation of heavy metals contamination is efficient, relatively inexpensive and reliable.1,2 Basically, the mechanisms allowing the microorganisms to cope with metals in the environment are complexation and active efflux of heavy metals. Pseudomonas is well-known for their capacity to tolerate high concentrations of several heavy metals including a highly toxic cadmium.3–5 Recent reports have highlighted the potential of new isolates of P. aeruginosa for application in bioremediation of water and soil contaminated with cadmium.6,7 To tolerate heavy metals, Pseudomonas demonstrates numerous survival adaptations such as: energy-dependent efflux of metal ions, extracellular biosorption and cellular sequestration, biofilm formation, resistance to oxidative stress, and preservation of iron acquisition and homeostasis.2–4,8–13The key to our understanding of adaptation strategies and mechanisms of Pseudomonas heavy metal resistance lies in proteomic changes, which correlate with metabolic paths enabling microbes to survive and persist in heavy metal contaminated environments. However, the most common proteomic studies of Pseudomonas strains are related to zinc, copper14 and chromium,8 while the studies of proteomic fluctuations in response to cadmium are rare and data show unclear and complex responses.9,11,12 Comparative proteomics is a powerful technique to identify alterations in protein expression caused by changing environmental conditions and it was used to determine the molecular mechanisms of cadmium (Cd) tolerance in a Pseudomonas exposed to cadmium.8,9,11,12 In order to enrich metalloproteins in an analyzed proteome, size exclusion chromatography (SEC) has been used for protein prefractionation.15 SEC has been reliable in separation of metalloproteins and protein complexes maintaining them in a native state.15,16
P. aeruginosa san ai, an environmental isolate from mineral cutting oil, was previously used in various studies in our laboratory related to rhamnolipid,17 exopolysaccharide (EPS),18 the extracellular enzymes, lipase and protease,19,20 and their application in detergents21 as well as in the study related to the organism's capacity for chromium removal.22 In this study, we investigated the distribution of cadmium, its uptake by extracellular polymers and effects of intracellular cadmium on protein expression in P. aeruginosa san ai. We used a global proteomic approach based on SEC protein prefractionation coupled with tandem mass spectrometry and bioinformatics to identify specific protein response to cadmium. Proteomics technique was already successfully applied to P. putida,2,9 P. fluorescens,11,12 and P. brassicacearum, but never before to any environmental isolate of P. aeruginosa. Furthermore, in order to understand the complex mechanism of response to cadmium, besides proteins we have analyzed the content of: exopolysaccharide, siderophores, nitrate and nitrite, as well the rates of O2 consumption.
2. Material and methods
Growth conditions of P. aeruginosa san ai
P. aeruginosa san ai was isolated from industrial mineral metal-cutting oil.19 The nucleotide sequences of 16S rDNA of P. aeruginosa san has been submitted in GenBank with accession number JQ012798. Microorganism was deposited in the National Collection of Agricultural and Industrial Microorganisms (NCAIM), Faculty of Food Sciences, Corvinus University of Budapest, Budapest, Hungary, under the label NCAIM (P) B 001380. Microorganism is available from the corresponding author. Genome sequencing and assembly is deposited as NCBI BioProject, accession: PRJNA195719.P. aeruginosa san ai was activated on nutrient agar (Torlak, Belgrade, Serbia) at 30 °C for 24 h and transferred to a 500 mL Erlenmeyer flask, containing 100 mL of Luria broth (LB) medium, g L−1 (NaCl – 5, yeast extract – 5 and tryptone – 10) to obtain a preculture. The flask was incubated at 30 °C for 20 hours and shaken at 250 cycles per min with a horizontal shaker (Kuhner, Basel, Switzerland). Actively grown preculture (1 mL) was used to inoculate 100 mL of LB with cadmium and control in 500 mL Erlenmeyer flasks to obtain initial cellular concentrations of approximately 5 × 105 cells per mL and agitated at 250 cycles per min with Kuhner horizontal shaker. LB medium with increasing concentrations of cadmium (mM): 0.45, 0.9, 1.8, 2.7, 3.6, 4.5, 5.4, 6.3 and 7.2, at pH 7.2 was prepared using CdCl2·H2O stock solution, concentration of 100 mM. Two controls were utilized: non-inoculated controls – LB with appropriate concentrations of cadmium but no P. aeruginosa san ai and control of bacterial growth – LB inoculated with P. aeruginosa san ai but without cadmium. Bacterial growth was monitored in cadmium supplemented broth and control, as a change of optical density at 580 nm using spectrophotometer Shimadzu UV-2600 (Kyoto, Japan). Measurements of OD580 were conducted in triplicate.
Culture broth was centrifuged at 4000 × g for 20 min (Sorvall, rotor SS-1, New Town, USA) and the biomass and supernatant obtained were further analyzed.
All spectrophotometric measurements were done on Shimadzu UV-2600 (Kyoto, Japan).
Evaluation of cadmium resistance
The minimal inhibitory concentration (MIC) of cadmium was determined in LB medium supplemented with cadmium concentrations ranging from 0.5 to 10 mM. Tubes containing growth medium and increasing concentrations of cadmium were inoculated with 24 h culture to obtain an initial optical density of approximately 0.06 at 580 nm. Turbidity was measured after 48 h of growth. The minimum concentration of the metal inhibiting complete growth was taken as the MIC.23Cadmium determination and distribution
Cadmium was quantified in: biomass, culture supernatant, EPS, cell homogenate and cell debris after homogenization (see below) spectrophotometrically using an iCAP 6500 Duo ICP (Thermo Fisher Scientific, Cambridge, UK) at 214.438 nm.24 Biomass, culture supernatant, EPS, homogenate and cell debris were digested with HNO3 and H2O2 in a digester DS-6, Tecator, (Sweden) prior to analysis of cadmium content. Cadmium content in biomass and supernatant was analyzed every 24 h, while EPS, homogenate and cell debris were analyzed after 48 h of bacterial growth. Eluates from gel size exclusion chromatography were measured directly, without prior digestion of the samples.25 The amount of cadmium removed by biosorbent at time t, was calculated according to:26where q is cadmium uptake (mM g−1), V – sample volume (L), Ci – initial cadmium concentration (mM), Cf – final cadmium concentration (mM), m – mass of dry biosorbent (g)
Nitrate and nitrite determination
Nitrate and nitrite levels in culture medium were determined by ion chromatography on an Ionchrom analyzer (Dionex, Sunnyvale, USA) with conductivity detector (Dionex, Sunnyvale, USA), SRS controller and autosampler AS 3000 (Thermo Scientific, USA), column Dionex IonPac TM AS14A (4 × 250 mm), eluent: 8.0 mM Na2CO3/1 mM NaHCO3, flow rate: 1 mL min−1, applied current: 43 mA, injection volume: 50 μL, standard mix solution of: fluoride, chloride, bromide, nitrite, nitrate, phosphate, sulfate, concentration 0.01 g L−1.27Siderophore determination
Pyoverdin was monitored in culture supernatant by measuring the absorbance at 400 nm. The concentration was calculated using a molar absorption coefficient ε (400) = 2 × 104 M−1 cm−1.28 The concentration of pyochelin was determined spectrophotometrically at 310 nm according to Sokol and calculated by ε (310) = 4.2 × 103 M−1 cm−1.29 Determinations were conducted in triplicate.Exopolysaccharide determination
Exopolysaccharide was measured in culture supernatant after precipitation with three volumes of cold ethanol. EPS was collected by centrifugation at 5000 × g for 20 min, air-dried for 24 hours and quantified by measuring mass.30Respiration measurement
Respiration activity of P. aeruginosa san ai was measured using a twelve-channel Micro-Oxymax® respirometer (Columbus Instruments, Columbus, USA) connected to a PC. The experiments were performed in MicroOxymax light-proof 500 mL bottles (Duran, Wertheim, Germany) containing 100 mL of LB medium and stirred constantly (200 rpm) with magnetic stirrer (Heidolph, Schwabach, Germany) at 29 ± 2 °C for four days. Respiration rates (μL min−1), as well as cumulative oxygen consumed (μL) were determined. Cell respiration was measured every 240 min for four days. A sterile medium with cadmium and a Pseudomonas – inoculated medium without cadmium were used as growth controls. All experiments were performed in three replicates.Protein isolation and identification
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g at 4 °C using Beckman Coulter L5-65, rotor SW 28 (Beckman, Indianopolis, USA).31 Three volumes of acetone were added to supernatant and precipitated proteins were collected by centrifugation at 12
000 × g at 4 °C using Beckman Coulter L5-65, rotor SW 28 (Beckman, Indianopolis, USA).31 Three volumes of acetone were added to supernatant and precipitated proteins were collected by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min (Microcentrifuge Force 12, Denver Instruments, New York, USA).25 Precipitate was dissolved in a minimal volume of buffer A and applied to Sephadex G-100 column (2.5 × 70 cm) equilibrated in buffer A. Fractions (volumes of 5.5 mL) were collected and absorbance at 280 nm was recorded. Fractions were pooled according to absorbance. Cadmium was determined in pooled fractions. Pooled fractions were lyophilized and kept in the refrigerator prior to use, when solutions of desired concentrations in water were prepared. The protein concentration was determined by the Bradford method.32 Two temporary independent column separations were done.
000 × g for 10 min (Microcentrifuge Force 12, Denver Instruments, New York, USA).25 Precipitate was dissolved in a minimal volume of buffer A and applied to Sephadex G-100 column (2.5 × 70 cm) equilibrated in buffer A. Fractions (volumes of 5.5 mL) were collected and absorbance at 280 nm was recorded. Fractions were pooled according to absorbance. Cadmium was determined in pooled fractions. Pooled fractions were lyophilized and kept in the refrigerator prior to use, when solutions of desired concentrations in water were prepared. The protein concentration was determined by the Bradford method.32 Two temporary independent column separations were done.Peptide mixtures obtained after tryptic digestion were applied to the precolumn at a flow rate of 5 μL min−1 in 2% (v/v) acetonitrile with 0.1% (v/v) formic acid. Peptides were eluted by a linear gradient of A (water, 0.1% formic acid) and B (acetonitrile, 0.1% formic acid), as follows: 0 min – A (98%), B (2%); 50 min – A (5%), B (95%); 50–55 min – A (5%), B (95%) with flow rate of 400 nL min−1. Ion source conditions were optimized with a calibration solution according to the instrument provider. All MS survey scans were performed from m/z 400–1400 with enhanced resolution. Data analysis was performed by selection of the five most abundant precursors rejecting singly charged ions.
Metallothionein identification was additionally attempted using the Mascot database search tool (version 2.3.02) by querying MS data against a custom metallothionein database. The custom database was assembled out of 2518 protein sequences found to be related to metallothioneins and is listed within the Uniprot knowledge base. Mascot search parameters were set as follows: enzyme –trypsin, missed cleavages – 6, modifications – cysteine carbamidomethylation (global) and methionine oxidation (variable), precursor mass tolerance – 1.2 Da, MS/MS mass tolerance – 0.6 Da, ions with monoisotopic m/z value and +2 and +3 charge state were tested. Only top hit compounds with significance threshold of p < 0.01 were considered.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD008020. Project name: Cadmium specific proteomic responses of Pseudomonas aeruginosa san ai.
3. Results and discussion
Effects of cadmium on P. aeruginosa san ai growth
The minimal inhibitory concentration of 7.2 mM of cadmium classifies P. aeruginosa san ai in a group of highly resistant Pseudomonas.35–37 As concentrations which affect metabolic changes in Pseudomonas species, but do not induce non-culturability, were reported to be about 15% of MIC,9,11 a concentration of 0.9 mM of cadmium was selected for further analysis of the P. aeruginosa san ai response to cadmium exposure.Extracellular cadmium binding
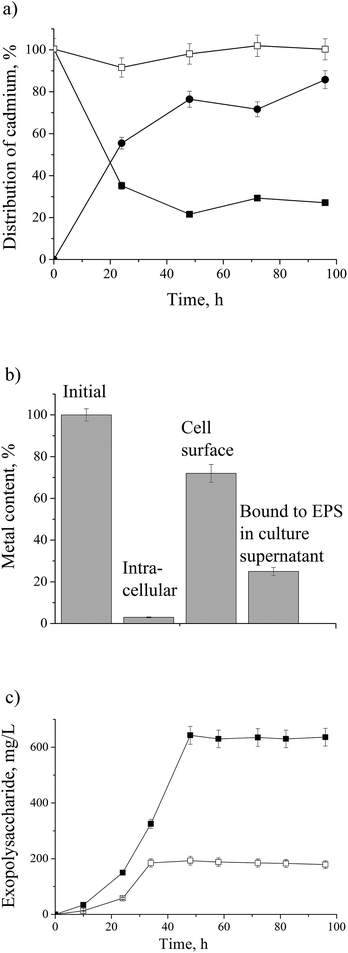
The dynamics of cadmium removal from the culture supernatant and its assimilation by the P. aeruginosa san ai are shown in Fig. 1a and b. In the course of exposure to initial concentration of 0.9 mM, 75% of cadmium was accumulated in biomass. P. aeruginosa san ai removes 0.307 mM of cadmium per g of biomass after 48 hours, implying a large biosorption potential of leaving biomass. The remaining 25% of cadmium was bound to EPS in the culture supernatant. Extracellular polysaccharides organized around cells or excreted in growth medium greatly contribute to defense mechanisms in Pseudomonas.1,38 The sorption cadmium uptake by EPS was 0.375 mmol g−1. Furthermore, cadmium concentrations found in the cell homogenate was very low (3%) suggesting that main part of cadmium was bound to the cell surface. Concentrations of EPS in the culture supernatant were determined and approximately three times higher production of EPS in cadmium-amended culture was found (Fig. 1c). The alteration of EPS production under the exposure of heavy metal chromium was reported.38 The changes in EPS production are related to its protective roles for microbial community such as prevention of dehydration, adhesion, and preserve against environmental stress including antibiotics and toxins, which all together are incorporated in process of biofilm formation. Furthermore, EPS acts as a protective layer by restricting diffusion of some antimicrobial agents into the biofilm by being an ion exchanger.1Protein response to cadmium
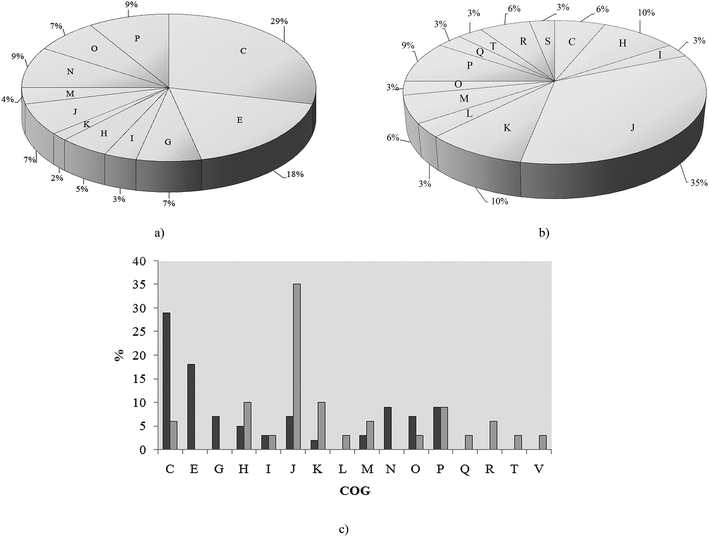
As we aimed to analyze metalloproteins and metabolic pathways related to cadmium exposure in P. aeruginosa san ai we used SEC prior to MS analysis. Global analyses of metalloproteins and protein complexes using the gel-free methodology, based on size exclusion chromatography (SEC) has been recently validated.15 Proteins were separated in fractions: FI, FII and FIII (ESI, Fig. 1†) and the cadmium content in fractions was determined as (mg L−1): 0.144, 0.005 and 0.001, respectively, while no cadmium was found in the control sample (with no added cadmium). Proteins from fractions FI, FII and FIII were analyzed by liquid chromatography coupled with tandem mass spectrometry. Identified proteins in cadmium-amended and control samples were compared and only those proteins which were uniquely found in cadmium-amended samples, but not appeared in control sample were considered as differentially expressed under cadmium exposure.There was a total of 60 proteins differentially expressed in Cd-amended culture which were classified according to COG data base (https://www.ncbi.nlm.nih.gov/COG/) into 11 groups (Fig. 2a) and 32 proteins differentially expressed in control classified into 13 groups (Fig. 2b). Only one protein was found in common in both samples (porin F). Most of the proteins in Cd-amended culture functionally belong to: energy production and conservation, amino acids metabolism, posttranslational modifications and cell motility, while in control prevailed proteins of translation and transcription. The comparison of protein functions in Cd-amended and control culture reveals quite different patterns (Fig. 2c). List of proteins with identified peptide sequences in cadmium-amended and control samples are shown in ESI (Tables 1, 2 and 3†).
Metalloproteins
Cadmium resistance of P. aeruginosa is accompanied by up-regulation of metalloproteins uniquely found in cadmium-amended culture, most of which are located in the outer membrane or periplasm – the first line of the cell defense. The crystal ionic radius of Cd2+ (in pm) of 109, being close to the radii of Hg2+ (116) and Ca2+ (114),39 significantly determines the specificity of the cadmium response in bacterial cells. We have identified 16 metalloproteins that represent almost 30% of all over-expressed proteins (ESI, Table 1†). The greatest part of metalloproteins observed in our study belongs to two COG categories – inorganic ion transport and metabolism (P) and energy production and conversion (C).P. aeruginosa san ai is a denitrificator and as such it possesses dissimilatory nitrate respiration system19 (GenBank accession number JMKR00000000) composed of metalloproteins: azurin (AZUR) – copper containing, cytochrome C551 (CY551) – iron containing, nitrite reductase (NIRS) – iron containing, and nitrous-oxide reductase (NOSZ) – copper and calcium containing.40 We believe that cadmium replaced the mentioned metals in denitrification proteins. Copper substitution by cadmium in the blue copper redox metalloprotein azurin (AZUR) was already reported and it is based on similar metal (cadmium and copper) coordination and binding sites containing cysteine and histidine.41 Also, cadmium competition with iron in heme and Fe–S proteins was already observed.42,43 Exchange of essential metals (Fe, Cu, Ca, etc.) with cadmium in metalloproteins with non-redox function will probably lead to a change, but not to a lost of their metabolic activity, while replacing redox metal with Cd will certainly lead to the complete inactivation of a particular protein. However, in conditions of overproduction of metalloproteins, and at relatively low concentration of cadmium, it is possible that part of the metalloproteins completely saved their metal ions and thus the function. Low sub-lethal concentrations of cadmium, influences fine-tuning of metabolic and energy system in process of adaptation. On the other hand, high concentrations of cadmium cause a massive change in the total metabolic picture (data not shown). In order to confirm the thesis that denitrification system is not active we determined the content of nitrate and nitrite in culture filtrate and respiration rate of culture. Very low content of 0.125 mM nitrate and 0.005 mM nitrite was far less than concentrations reported as necessary for denitrification.44 Oxygen respiration rate (Fig. 3) clearly showed increased oxygen consumption in a cadmium-amended culture implying that P. aeruginosa san ai did not activate its machinery for denitrification in given conditions, thus these proteins seemingly serve as a metal ion depot. Proteins of denitrification cascade which is not active, obviously, can protect microbial cells against heavy metals. In accordance with our observation Gui et al.45 found that aerobic denitrification was inhibited with heavy metals, including cadmium. Based on the similarity of ionic radii, Cd2+ probably replaced mercury in mercuric transport protein (MERP). Similar to our finding, Jain et al.46 reported the expression of mercuric resistance protein (MERD) in P. monteilii in the presence of cadmium.
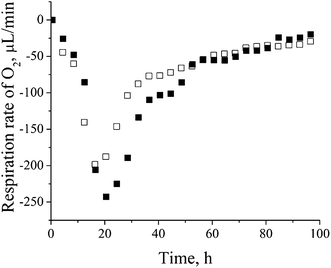 | ||
| Fig. 3 Oxygen respiration rate of control versus cadmium-amended culture of P. aeruginosa san ai: □ – control, ■ – cadmium-amended. | ||
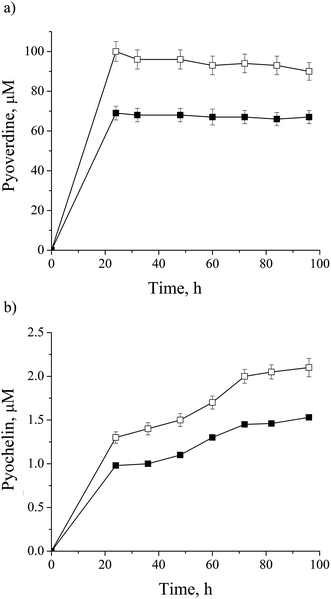
Most of the metalloproteins found to be impacted by cadmium are iron containing: NIRS, CY551, catalase (CATA), superoxide dismutase (SODF), bacterioferritin (BFR), Fe(3+)-pyochelin receptor (FPTA) and molybdenum cofactor biosynthesis protein (MOAA1) as a Fe–S cluster protein (ESI, Table 1†). As already mentioned, iron could be substituted by cadmium.42,43 On the other side, iron concentration is essential for cell homeostasis, so some microorganisms synthesize siderophores to control iron bioavailability. P. aeruginosa san ai produces pyoverdine, a highly iron-specific siderophore, and pyochelin, a siderophore with a broad specificity,14,47 and they were both monitored in the culture supernatant and depicted in Fig. 4a and b, respectively. We observed the down-regulation of both siderophores which was in agreement with results reported by Teitzel et al.14 and Poirier et al.12 In addition we identified ferric uptake regulation protein (FUR) responsible for negative regulation of siderophore biosynthetic process and an iron storage protein BFR, similar to Poirier et al.11 These results clearly show the effect of cadmium on the iron homeostasis.
Expected metallothioneins were not found in this investigation. Possible reasons for their absence include their time-dependent production and low concentration, along with the problematic digestion and preparation procedures for MS analysis, which have been already reported.9
Cell energy metabolism
Up-regulation of protein biosynthesis – ESI† (Table 2) caused by cadmium in P. aeruginosa san ai expectedly had an increased energy requirement.8 Up-regulation of electron transfer flavoprotein, ETFA/B involved in electron transport to the main respiratory chain suggested enhanced respiration, whereas up-regulation of ATP synthase implied an increased production and consumption of ATP (Fig. 5). Over-expression of the dihydrolipoyl dehydrogenase component of the α-ketoacid dehydrogenases complex, together with components of α-ketoglutarate dehydrogenase (2-oxoglutarate dehydrogenase) and pyruvate dehydrogenase complexes, produce NADH and provide electrons for the respiratory chain. So, the activity of these enzymes is of crucial importance in the stimulation of the respiratory chain and consequently in ATP supply under conditions of an increased ATP demand in cadmium exposed cells. In addition, produced keto acids might contribute cadmium chelation, preventing heavy metal stress. Cadmium-amended culture in exponential phase showed oxygen consumption of 7.14 μmol × min−1 g−1 versus 5.19 μmol × min−1 g−1 in control. An increased O2 consumption of 37% in exponential phase and 30% in early stationary phase well document a more intense respiration in cadmium-amended culture than in control (Fig. 3).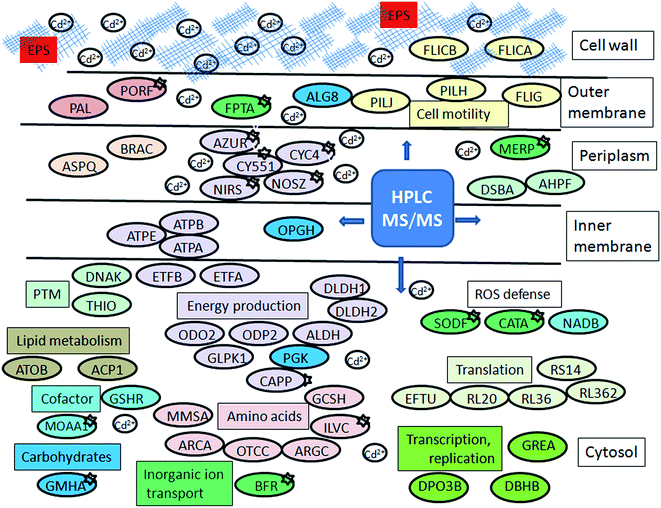 | ||
| Fig. 5 Schematic model of P. aeruginosa san ai response to cadmium. Protein abbreviations are the same as in Tables 1 and 2. Proteins which belong to the same COG are labeled by the same colour. | ||
Amino acid metabolism
As Table 2 shows, an intensive amino acid metabolism took place when P. aeruginosa san ai was exposed to cadmium. Arginine, lysine, glycine, histidine, asparagine, and valine metabolism, as well as the transport of branched amino acids were affected by cadmium (Table 2, Fig. 5 and ESI,† Table 2). Increased amino acid metabolism is certainly connected to protein overexpression. In addition, up regulation of the glycine cleavage system, GCSH which is highly sensitive to alterations in the oxidation–reduction state of the respiratory chain (e.g. oxidizing conditions stimulate while reducing conditions strongly inhibit this process), was observed.48 Arginine deiminase and catabolic ornithine carbamoyltransferase, enzymes of arginine degradation by the arginine deiminase (ADI) pathway were identified. ADI is a non-redox, ATP producing process which can be induced under various stress conditions.49,50 Actually, the amino acid catabolism is the main effect of cadmium and can be considered in relation to: improved energy production, biosynthesis of basic building blocks for other structural molecules, or particular amino acid like cysteine for cadmium sequestering.| Identified protein | Entry nameb | Easy Prot score | Number of matched peptides | Sequence coverage, % | Metal (according to Uniprot) | Molecular mass kDa | COGa | SECc fraction |
|---|---|---|---|---|---|---|---|---|
| a According to https://www.ncbi.nlm.nih.gov/COG/.b Matching microorganism sources are: PSEAE – P. aeruginosa ATCC 15692/PAO1, PSEAI – P. aeruginosa, and PSEA8 – P. aeruginosa LESB58.c Size exclusion chromatography fraction. | ||||||||
| Azurin | AZUR_PSEAE | 116 | 4 | 42 | Cu | 13.9 | C | I |
| Nitrous-oxide reductase, [chain 0] | NOSZ_PSEAE | 152 | 5 | 18 | Ca, Cu | 65.8 | C | I |
| Nitrite reductase/cytochrome cd1 | NIRS_PSEAE | 58 | 4 | 9 | Fe | 62.6 | C | I |
| Cytochrome c-551 | CY551_PSEAE | 33 | 2 | 27 | Fe | 10.9 | C | I |
| Cytochrome c4 [chain 0] | CYC4_PSEAE | 23 | 2 | 7 | Fe | 18.6 | C | I |
| Phosphoenolpyruvate carboxylase (PEPCase) | CAPP_PSEAE | 20 | 2 | 4 | Mg | 97.8 | C | I |
| Malate synthase G | MASZ_PSEAE | 79 | 5 | 10 | Mg | 78.5 | C | I |
| Bacterioferritin | BFR_PSEAE | 34 | 2 | 12 | Fe | 17.9 | P | I |
| Fe(3+)-pyochelin receptor | FPTA_PSEAE | 68 | 4 | 5 | Fe | 76 | P | I |
| Catalase | CATA_PSEAE | 73 | 4 | 8 | Fe | 55.5 | P | I |
| Superoxide dismutase [Fe] | SODF_PSEAE | 14 | 1 | 6 | Fe | 21.3 | P | I |
| Mercuric transport protein periplasmic component | MERP_PSEAI | 77 | 3 | 55 | Hg | 9.3 | P | II |
| Phosphoheptose isomerase | GMHA_PSEAE | 55 | 3 | 19 | Zn | 21.4 | G | I |
| Molybdenum cofactor biosynthesis protein A 1 | MOAA1_PSEAE | 36 | 4 | 9 | Fe, 4Fe–4S | 36.6 | H | I |
| Ketol-acid reductoisomerase (NADP(+)) | ILVC_PSEA8 | 101 | 4 | 15 | Mg | 36.4 | H | I |
| Ferric uptake regulation protein | FUR_PSEAE | 92 | 3 | 3 | Fe, Zn | 15 | P | II |
| Outer membrane porin F | PORF_PSEAE | 111 | 6 | 28 | Ca | 37.6 | M | I |
| Identified protein | Entry nameb | Easy Prot score | Peptide matched | Coverage (%) | Molecular mass (kDa) | COGa | SECc fraction |
|---|---|---|---|---|---|---|---|
| a According to https://www.ncbi.nlm.nih.gov/COG/.b Matching microorganism sources are: PSEAE – P. aeruginosa ATCC 15692/PAO1, PSEAB – P.aeruginosa, UCBPP-PA14, PSEAI – P. aeruginosa, PSEA7 – P. aeruginosa PA7, PSEA8 – P. aeruginosa PA8 and PSEOL – P. oleovorans.c Size exclusion chromatography fraction. | |||||||
| Electron transfer flavoprotein subunit, β | ETFB_PSEAE | 101 | 4 | 23 | 26.3 | C | I |
| Electron transfer flavoprotein subunit, α | ETFA_PSEAE | 92 | 5 | 16 | 31.4 | C | I |
| ATP synthase subunit alpha | ATPA_PSEA8 | 75 | 4 | 10 | 55.3 | C | I |
| ATP synthase subunit beta | ATPB_PSEAE | 39 | 3 | 7 | 49.5 | C | I |
| ATP synthase epsilon chain | ATPE_PSEAE | 35 | 2 | 16 | 14.7 | C | I |
| Dihydrolipoyl dehydrogenase1 | DLDH1_PSEAE | 160 | 7 | 25 | 48.6 | C | I |
| Aldehyde dehydrogenase | ALDH_PSEOL | 34 | 4 | 9 | 52.8 | C | I |
| Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex | ODO2_PSEAE | 50 | 3 | 10 | 42.9 | C | I |
| Glycerol kinase | GLPK1_PSEAE | 190 | 9 | 22 | 55.9 | C | I |
| Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex | ODP2_PSEAE | 32 | 3 | 9 | 56.7 | C | I |
| Arginine deiminase | ARCA_PSEAE | 27 | 2 | 9 | 46.4 | E | I |
| Ornithine carbamoyltransferase, catabolic | OTCC_PSEAE | 68 | 4 | 13 | 38 | E | I |
| N-acetyl-gamma-glutamyl-phosphate reductase | ARGC_PSEAE | 30 | 2 | 8 | 36.7 | E | I |
| Glycine cleavage system H protein 1 | GCSH1_PSEAE | 30 | 2 | 16 | 13.7 | E | II |
| Glycine cleavage system H protein 2 | GCSH2_PSEAE | 28 | 2 | 15 | 13.6 | E | II |
| Glutaminase-asparaginase | ASPQ_PSEAE | 32 | 2 | 13 | 38.6 | E | II |
| Leucine-, isoleucine-, valine-, threonine- and alanine-binding protein | BRAC_PSEAE | 58 | 3 | 23 | 39.7 | E | I |
| Methylmalonate-semialdehyde dehydrogenase | MMSA_PSEAE | 80 | 4 | 12 | 53.5 | E | I |
| Phosphoglycerate kinase | PGK_PSEAE | 64 | 4 | 16 | 40.4 | G | II |
| Glycosyltransferase alg8 | ALG8_PSEAE | 19 | 2 | 5 | 56.5 | G | I |
| Glucans biosynthesis glucosyltransferase H | OPGH_PSEA7 | 32 | 4 | 4 | 96 | G | I |
| Acetyl-CoA acetyltransferase | ATOB_PSEAE | 20 | 2 | 3 | 40.3 | I | II |
| Acyl carrier protein 1 | ACP1_PSEAE | 50 | 2 | 19 | 8.7 | I | III |
| L-Aspartate oxidase | NADB_PSEAE | 17 | 2 | 4 | 60 | H | I |
| Glutathione reductase | GSHR_PSEAE | 22 | 3 | 5 | 49 | H | I |
| Transcription elongation factor GreA | GREA_PSEAE | 77 | 3 | 25 | 17.1 | K | I |
| DNA-binding protein HU-beta | DBHB_PSEAE | 44 | 2 | 32 | 9 | K | III |
| DNA polymerase III subunit beta | DPO3B_PSEAE | 51 | 3 | 8 | 40.6 | L | I |
| Elongation factor Tu | EFTU_PSEAB | 38 | 2 | 5 | 43.3 | J | III |
| 50S ribosomal proteins L36 2 | RL362_PSEAB | 42 | 3 | 86 | 5.9 | J | III |
| 50S ribosomal proteins L 20 | RL20_PSEAE | 22 | 2 | 13 | 13.3 | J | III |
| 30S ribosomal proteins S14 | RS14_PSEAE | 21 | 2 | 21 | 11.5 | J | III |
| 50S ribosomal proteins L36 | RL36_PSEAE | 24 | 2 | 58 | 4.4 | J | III |
| Peptidoglycan-associated lipoprotein | PAL_PSEAE | 68 | 3 | 31 | 15.8 | M | II |
| Protein PilJ | PILJ_PSEAE | 26 | 2 | 4 | 72.5 | N | I |
| Protein PilH | PILH_PSEAE | 72 | 3 | 23 | 13.2 | N | II |
| A-type flagellin [chain 0] | FLICA_PSEAI | 82 | 4 | 13 | 39.9 | N | II |
| B-type flagellin | FLICB_PSEAE | 30 | 2 | 5 | 49 | N | I |
| Flagellar motor switch protein FliG | FLIG_PSEAE | 28 | 3 | 9 | 37 | N | II |
| Thiol:disulfide interchange protein DsbA | DSBA_PSEAB | 35 | 2 | 12 | 23.3 | O | II |
| Thioredoxin | THIO_PSEAE | 86 | 4 | 43 | 11.8 | O | II |
| Chaperone protein DnaK | DNAK_PSEAE | 123 | 6 | 11 | 68.3 | O | I |
| Alkyl hydroperoxide reductase subunit/thioredoxin peroxidase | AHPF_PSEAE | 22 | 3 | 12 | 20.5 | O | II |
Protein biosynthesis and posttranslational modifications
The excess of cadmium caused up-regulation of protein biosynthesis in P. aeruginosa san ai (Table 2 and ESI, Table 2†). The transcription elongation factor, GREA regulates transcription, while the factor EFTU and several 30S and 50S ribosomal proteins contribute to protein translation. Damaged proteins were repaired by chaperone DNAK. Posttranslational modifications are mainly related to protein redox processing based on the thiol group from cysteine. The thiol-containing molecules can serve to chelate heavy metal and thus prevent binding of cadmium to functionally important proteins. Cadmium interaction with biomolecules, in particular its reactivity toward the thiol group is based on a very low solubility product of CdS, (Ksp = 1 × 10−29). Once transported into the cytoplasm, cadmium cation binds to free thiol groups disrupting protein structure and function.10,14 As redox exchange of cysteine residue cannot exist in isolation several redox-active proteins were identified as up-regulated. Revealed redox cascade, based on the thiol-disulfide exchange, with oxidation–reduction potentials ranging from −279 mV (thioredoxin, THIO), −243 mV (glutathione reductase, GSHR) to −124 mV (thiol: disulfide interchange protein, DSBA),51 clearly implies that cadmium affects peptides and proteins containing sulfhydryl groups. Delalande et al.52 confirmed that the formation of Cd(GS)2 under physiological conditions is spontaneous. On the other side, the ratio GSH/GSSG is kinetically controlled by GSH-utilizing enzymes,53 from which thioredoxin and glutathione reductase were identified in this study.It has been recently proven that the mechanism of cadmium toxicity involves the production of reactive oxygen species (ROS).54 As cadmium is a redox inactive metal ion, production of free radicals and ROS probably occurs through an indirect mechanism.55 To neutralize ROS, apart from metalloproteins superoxide dismutase SODF and catalase CATA, alkyl hydroperoxide reductase AHPF was up-regulated in the current study (Table 2, Fig. 5). AHPF is a kinetically more potent H2O2 scavenger than catalase, and it is probably the primary scavenger of endogenous H2O2.56 To combat oxidative stress flavoenzyme L-aspartate oxidase (NADB) was up-regulated in P. aeruginosa san ai when exposed to cadmium (Table 1). NADB catalyzes the first step in the biosynthesis of NADP+ and NAD.57
Carbohydrate metabolism
As 2-oxoglutarate and pyruvate can be formed from several amino acids,49 overexpression of 2-oxoglutarate dehydrogenase and pyruvate dehydrogenase could be considered as a result of emphasized amino acid metabolism. 2-Oxoglutarate dehydrogenase involved in Krebs cycle and pyruvate dehydrogenase at Krebs cycle entrance are two main cellular multienzyme complexes, which enable effective energy supply. In the same time, pyruvate is an carbohydrate precursor which can be converted into glucose which is further involved in biosynthesis of lipopolysaccharide and alginate. Indeed, P. aeruginosa san ai exposed to cadmium showed up-regulation of: peptidoglycan-associated lipoprotein-PAL, phospho heptose isomerase GMHA, responsible for lipopolysaccharide biosynthesis, and glycosyltransferase ALG8, involved in alginate biosynthesis (Fig. 5). Over-expression of these proteins suggested an increased biosynthesis of outer membrane polymers according to previous observations.8 These polymers on the cell surface and EPS released to culture supernatant were bound to the main part of cadmium, as shown in Fig. 1c. Exopolysaccharide identified as alginate18 is a linear polymer composed of D-mannuronic acid and L-guluronic acid, and as such greatly contributed to heavy metal binding, reducing cadmium interaction with the microbial cell.Cell motility
Proteins PilH and PilJ as well A- and B-type flagellin, were found up-regulated in this study (Table 2, Fig. 5 and ESI, Table 2†). Cell surface structures composed from these proteins, pilli and flagella, together with EPS and lipopolysaccharides (LPS), are involved in the early stages of P. aeruginosa biofilm formation.58,59 More precisely, pili regulate twitching motility, while flagella has important role in microbial adhesion.60–63 and the use of both, enables P. aeruginosa cells to colonize and rapidly explore microenvironments.60,61 However, Poirier et al.11 reported a down-regulation of flagellin synthesis and a decreased mobility of P. fluorescens BA3SM1 under the exposure to cadmium, zinc and copper, explaining this phenomenon as a bacterial response to economize energy. In addition to motility proteins, an increased amount of EPS alginate and up-regulation of LPS biosynthesis (protein GMHA) were observed – these two molecules are of crucial importance for biofilm formation. Previously in the discussion (section Extracellular cadmium binding) it was notified important role of EPS as protective layer against toxic effect of cadmium acting as an ion exchanger.1 As negatively charged EPS efficiently bound cadmium in supernatant in our study, according to observation of van Hullebush et al.,64 it is obvious that biofilm formation induced by heavy metal presence1 is one more element of the complex metal resistance strategy in P. aeruginosa san ai exposed to cadmium, as it was observed in P. putida KT2440.94. Conclusion
Pseudomonas aeruginosa san ai resists a high concentration of up to 7.2 mM of cadmium and as such is ecologically relevant strain with the potential to be applied in bioremediation of heavy metal polluted environments. Under exposure to cadmium, P. aeruginosa san ai exhibits two main mechanisms: stress and defense response. P. aeruginosa san ai evolved several defense strategies at both intracellular and extracellular levels: (1) extracellular binding of cadmium to exopolysaccharide released in culture broth, (2) binding of cadmium to the cell surface, (3) elevated production of metalloproteins, particularly denitrification proteins, (4) metabolic adaptation to increased concentration of cadmium inside the cell.Biomass of P. aeruginosa san ai exposed to sub-lethal concentration of cadmium (0.9 mM) adsorbed 75% of added cadmium, while remaining 25% in culture supernatant was adsorbed by EPS, implying a large biosorption potential of leaving biomass and EPS. Due to EPS negatively charged chemical groups it has a protective role preventing cadmium entry to the cells. In addition, increased biosynthesis of exopolysaccharide, lipopolysaccharide, pilli and flagellar proteins unambiguously indicates biofilm formation as additional strategy to combat heavy metal outside the cell.
Inner cell response on the protein level was investigated by global proteomics approach based on protein prefractionation by size exclusion chromatography which was used to preserve native forms of metalloproteins and protein complexes, followed by liquid chromatography and tandem mass spectrometry coupled with bioinformatics to identify proteins. Almost a third of the total number of 60 differentially expressed proteins in the presence of cadmium were metalloproteins. The fraction in which the metalloproteins prevail has the highest measured cadmium content indicating metal ion replacement in these proteins. Since P. aeruginosa san ai tolerates very high concentrations of cadmium (up to 7 mM), its exposure to 0.9 mM likely did not cause significant loss of biological activity of the cadmium supstituted proteins, presumably because the level of residual essential metal in the proteins remained sufficient for their function.
As in the cytoplasm cadmium cation binds to free thiol groups from proteins and peptides, identified proteins from redox cascade: thioredoxin, glutathione reductase, and thiol: disulfide interchange protein, clearly implies that cadmium affects peptides and proteins containing sulfhydryl groups, as well that P. aeruginosa san ai has a potent mechanisms to maintain cellular –SH redox homeostasis. Replacement of iron by cadmium in metalloproteins alters iron availability, so the down-regulation of siderophores and the over expression of ferric uptake regulation protein responsible for negative regulation of siderophore biosynthesis clearly indicated the effect of cadmium on the iron homeostasis in P. aeruginosa san ai. Accumulation of iron cation which triggers Fenton's reaction elevates expression of superoxide dismutase, catalase, and alkyl hydroperoxide reductase to suppress accumulation of ROS.
Our results point to overexpression of the mercury ion transporter, MERP. As cadmium and mercury (2+) cations physically differ relatively slightly, it seems possible that MERP might be a good candidate for cadmium transport in P. aeruginosa san ai.
Enhanced activities of NAD – dependent dehydrogenases and glycine cleavage system firmly affected respiration, so an enhanced consumption of oxygen in cadmium-amended culture, was measured implying complex cadmium effects on the environment. The present study indicated that the presence of cadmium as a non-essential, non-redox metal induced in P. aeruginosa san ai elevated synthesis of carbohydrates and proteins as the major players in the process of cadmium elimination for P. aeruginosa san ai.
Cadmium resistance of P. aeruginosa san ai is a branched network of powerful extracellular and intracellular adaptation mechanisms, whose harmonic functioning apparently showed a significant potential of the bacteria for its application in environmental heavy metal pollution and co-pollution. Regarding to the observed response the isolate cadmium removal will involve biofilm formation, sequestration by extracellular polymers, and the metal up take by biomass. A deeper understanding of P. aeruginosa san mechanisms involved in cadmium resistance and optimal conditions for efficient biosorption are crucial for its successful application in bioremediation.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was financed by the Ministry of Education, Science and Technological Development, Republic of Serbia, Project III 43004.References
- P. Prabhakaran, M. Aqeel Ashraf and W. Syaidatul Aqma, RSC Adv., 2016, 6, 109862–1098772 RSC.
- G. Haferburg and E. Kothe, Appl. Microbiol. Biotechnol., 2010, 87, 1271–1280 CrossRef CAS PubMed.
- A. Manara, G. DalCorso, C. Baliardini, S. Farinati, D. Cecconi and A. Furini, J. Proteome Res., 2012, 11, 4169–4179 CrossRef CAS PubMed.
- D. Pagès, L. Sanchez, S. Conrod, X. Gidrol, A. Fekete, P. Schmitt-Kopplin, T. Heulin and W. Achouak, Environ. Microbiol., 2007, 9, 2820–2835 CrossRef PubMed.
- X. Wu, J. C. Yalowich and B. B. Hasinoff, J. Inorg. Biochem., 2011, 105, 833–838 CrossRef CAS PubMed.
- B. Muneer, M. J. Iqbal, F. R. Shakoori and A. R. Shakoori, Pak. J. Zool., 2016, 48, 1495–1501 Search PubMed.
- S. Sinha and S. K. Mukherjee, Braz. J. Microbiol., 2009, 40, 655–662 CrossRef CAS PubMed.
- N. K. Kılıç, A. Stensballe, D. E. Otzen and G. Dönmez, Bioresour. Technol., 2010, 101, 2134–2140 CrossRef PubMed.
- C. D. Miller, B. Pettee, C. Zhang, M. Pabst, J. E. McLean and A. J. Anderson, Lett. Appl. Microbiol., 2009, 49, 775–783 CrossRef CAS PubMed.
- D. H. Nies, Appl. Microbiol. Biotechnol., 1999, 51, 730–750 CrossRef CAS PubMed.
- I. Poirier, P. Hammannb, L. Kuhn and M. Bertrand, Aquat. Toxicol., 2013, 128–129, 215–232 CrossRef CAS PubMed.
- I. Poirier, L. Kuhn, A. Demortière, B. Mirvaux, P. Hammann, J. Chicher, C. Caplat, M. Pallud and M. Bertrand, J. Proteomics, 2016, 148, 213–227 CrossRef CAS PubMed.
- G. M. Teitzel and M. R. Parsek, Appl. Environ. Microbiol., 2003, 64, 2313–2320 CrossRef.
- G. M. Teitzel, A. Geddie, S. K. De Long, M. J. Kirisits, M. Whiteley and M. R. Parsek, J. Bacteriol., 2006, 188, 7242–7256 CrossRef CAS PubMed.
- A. Hagège, T. N. S. Huynh and M. Hébrant, TrAC, Trends Anal. Chem., 2015, 64, 64–74 CrossRef.
- U. Aryal, Y. Xiong, Z. McBride, D. Kihara, J. Xie, M. Hall and D. Szymanski, Plant Cell, 2014, 26, 3867–3882 CrossRef CAS PubMed.
- M. Rikalović, A. M. Abdel-Mawgoud, E. Déziel, G. Gojgić-Cvijović, Z. Nestorović, M. Vrvić and I. Karadžić, J. Surfactants Deterg., 2013, 16, 673–682 CrossRef.
- A. Dimitrijević, D. Veličković, M. Rikalović, N. Avramović, N. Milosavić, R. Jankov and I. Karadzić, Carbohydr. Polym., 2011, 83, 1397–1401 CrossRef.
- I. Karadžić, A. Masui and N. Fujiwara, J. Biosci. Bioeng., 2004, 98, 145–152 CrossRef.
- I. Karadžić, A. Masui, L. Izrael-Živković and N. Fujiwara, J. Biosci. Bioeng., 2006, 102, 82–89 CrossRef PubMed.
- S. Grbavčić, D. Bezbradica, L. Izrael-Živković, N. Avramović, N. Milosavić, I. Karadzić and Z. Knežević, Bioresour. Technol., 2011, 102, 11226–11233 CrossRef PubMed.
- N. Avramović, S. Nikolić-Mandić and I. Karadžić, J. Serb. Chem. Soc., 2013, 78, 639–652 CrossRef.
- D. K. Thompson, K. Chourey, G. Wickham, S. B. Thieman, N. C. Ver Berkmoes, B. Zhang, A. T. McCarthy, M. A. Rudisill, M. Shah and R. L. Hettich, BMC Genomics, 2010, 11, 311 CrossRef PubMed.
- US EPA, Trace Elements in Water, Solids, and Biosolids by Inductively Coupled Plasma-Mass Spectrometry- method 200.7, Revision 5.0., 2001, pp. 1–68 Search PubMed.
- J. S. Park, S. Chung, I. S. Park, Y. Kim, C. H. Koh and S. Lee, Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol., 2002, 131, 425–431 CrossRef.
- B. Volesky, Sorption and biosorption, BV Sorbex Inc., Montreal, 2003 Search PubMed.
- J. Pfaff, Method 300.0: Determination of inoraganic anions by ion chromatography, Revision 2.1, United States Environmental Protection Agency, Office of Research and Development, Cincinnati, OH, 1993 Search PubMed.
- J. M. Meyer and M. A. Abdallah, J. Gen. Microbiol., 1978, 107, 319–328 CrossRef CAS.
- P. A. Sokol, J. Clin. Microbiol., 1986, 23, 560–562 CAS.
- J. Jung, J. Noh and W. Park, J. Microbiol., 2011, 49, 208–215 CrossRef CAS PubMed.
- J. Suzuku, N. Kodama, A. Molotkov, E. Aok and C. Tohyama, Biochem. J., 1998, 334, 695–701 CrossRef.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- R. Wang, D. A. Sens, A. Albrecht, S. Garrett, S. Somji, M. A. Sens and X. Lu, Anal. Chem., 2007, 79, 4433–4444 CrossRef CAS PubMed.
- F. Gluck, C. Hoogland, P. Antinori, X. Robin, F. Nikitin, A. Zufferey, C. Pasquarello, V. Fétaud, L. Dayon, M. Müller, F. Lisacek, L. Geiser, D. Hochstrasser, J. C. Sanchez and A. Scherl, J. Proteomics, 2013, 79, 146–160 CrossRef CAS PubMed.
- N. Hu and B. Zhao, FEMS Microbiol. Lett., 2007, 267, 17–22 CrossRef CAS PubMed.
- I. Poirier, N. Jean, J. C. Guary and M. Bertrand, Environ. Eng. Sci., 2009, 26, 1451–1457 CrossRef CAS.
- V. Singh, P. K. Chauhan, R. Kanta, T. Dhewa and V. Kumar, Int. J. Pharm. Sci. Rev. Res., 2010, 3, 164–167 CAS.
- N. K. Kiliç and G. Dönme, J. Hazard. Mater., 2008, 154, 1019–1024 CrossRef PubMed.
- R. D. Shannon, Acta Crystallogr., Sect. A: Cryst. Phys., Diffr., Theor. Gen. Crystallogr., 1976, 32, 751–767 CrossRef.
- D. J. Richardson, Microbiology, 2000, 146, 551–571 CrossRef CAS PubMed.
- H. Engeseth, D. McMillin and J. Otvos, J. Biol. Chem., 1984, 259, 4822–4826 CAS.
- F. Bonomi, M. L. Ganadu, G. Lubinu and S. Pagani, Eur. J. Biochem., 1994, 222, 639–644 CrossRef CAS PubMed.
- A. Schauder, A. Avital and Z. Malik, J. Environ. Pathol., Toxicol. Oncol., 2010, 29, 137–158 CrossRef CAS.
- D. R. Williams, J. J. Rowe, P. Romero and R. G. Eagon, Appl. Environ. Microbiol., 1978, 36, 257–263 CAS.
- M. Gui, Q. Chen, T. Ma, M. Zheng and J. Ni, Appl. Microbiol. Biotechnol., 2017, 101, 1271–1280 Search PubMed.
- S. Jain, A. Rani, S. Marla and R. Goel, Int. J. Adv. Biol. Biomed. Res., 2010, 1, 234–241 Search PubMed.
- T. C. Johnston and E. M. Nolan, Dalton Trans., 2015, 44, 6320–6339 RSC.
- R. Hampson, L. Barron and M. Olson, J. Biol. Chem., 1983, 258, 2993–2999 CAS.
- D. White, The Physiology and Biochemistry of Prokaryotes, 2nd edn, Oxford University Press, New York, 2000 Search PubMed.
- M. Eschbach, K. Schreiber, K. Trunk, J. Buer, D. Jahn and M. Schobert, J. Bacteriol., 2004, 186, 4596–4604 CrossRef CAS PubMed.
- V. Mallikarjun, D. J. Clarke and C. J. Campbell, Free Radical Biol. Med., 2012, 53, 280–288 CrossRef CAS PubMed.
- O. Delalande, H. Desvaux, E. Godat, A. Valleix, C. Junot, J. Labarre and Y. Boulard, FEBS J., 2010, 277, 5086–5096 CrossRef CAS PubMed.
- C. Berndt, C. Lilling and L. Flohe, Front. Pharmacol., 2014, 5, 168 Search PubMed.
- J. Liu, S. Y. Qian, Q. Guo, J. Jiang, M. P. Waalkes, P. Ronald and M. B. Kadiiska, Free Radical Biol. Med., 2008, 45, 475–481 CrossRef CAS PubMed.
- G. Bertin and D. Avarbeck, Biochimie, 2006, 88, 1549–1559 CrossRef CAS PubMed.
- D. C. Seaver and J. A. Imalay, J. Bacteriol., 2001, 183, 7173–7181 CrossRef PubMed.
- R. J. Mailloux, J. Lemire and V. D. Appanna, Anton Leeuw Int. J. G., 2011, 99, 433–442 CrossRef CAS PubMed.
- E. Karunakaran, J. Mukherjee, B. Ramalingam and C. A. Biggs, Appl. Microbiol. Biotechnol., 2011, 90, 1869–1881 CrossRef CAS PubMed.
- M. Rikalovic, N. Avramovic and I. Karadzic, in Structure-Function Relationships of Rhamnolipid and Exopolysacharide Biosurfactants of Pseudomonas aeruginosa as Therapeutic Targets in Cystic Fibrosis Lung Infections, Understanding Cystic Fibrosis, ed Danesh D, Sriramulu, In Tech, Rijeka, Croatia. 2017, ch. 6 Search PubMed.
- C. Toutain, N. Caiazza and G. O'Toole, Molecular basis of biofilm development by Pseudomonads, in Microbial Biofilms, ed G. M. O'Toole, ASM Press, Washington DC, 2004, pp. 43–63 Search PubMed.
- F. J. C. Conrad, M. L. Gibiansky, F. Jin, V. D. Gordon, D. A. Motto and M. A. Mathewson, Biophys. J., 2011, 10, 1608–1616 CrossRef PubMed.
- J. Haiko and B. Westerlund-Wikström, Biology, 2013, 2, 1242–1267 CrossRef PubMed.
- K. Murphy, A. Park, Y. Hao, D. Brewer, J. Lam and C. Khursigaraa, J. Bacteriol., 2014, 196, 1306–1317 CrossRef PubMed.
- E. D. van Hullebush, M. H. Zandvoort and P. N. L. Lens, Environ. Sci. Technol., 2003, 2, 9–33 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra00371h |
| ‡ Authors equally contributed. |
| This journal is © The Royal Society of Chemistry 2018 |