Open Access Article
Open Access ArticleO-GlcNAcylation confers protection against Staphylococcus aureus infection in Caenorhabditis elegans through ubiquitination†
Loganathan Vigneshwari,
Boopathi Balasubramaniam,
Sivasamy Sethupathy ,
Shunmugiah Karutha Pandian
,
Shunmugiah Karutha Pandian and
Krishnaswamy Balamurugan
and
Krishnaswamy Balamurugan *
*
Department of Biotechnology, Science Campus, Alagappa University, Karaikudi 630 003, Tamil Nadu, India. E-mail: bsuryar@yahoo.com; Fax: +91 4565 229334; Tel: +91 4565 225215
First published on 26th June 2018
Abstract
Glycosylation is one of the most prevalent post-translational modifications in biological systems. In Caenorhabditis elegans, O-GlcNAcylation has been shown to be actively involved in the regulation of dauer formation and detoxification of toxins secreted by invading pathogens. On this backdrop, the present study is focused on understanding the role of O-GlcNAcylation in C. elegans during Staphylococcus aureus infection using a gel based proteomic approach. Results of time course killing assays with wild-type and mutants of glycosylation and comparison of results revealed an increase in the survival of the C. elegans oga-1 mutant when compared to wild-type N2 and the ogt-1 mutant. Increased survival of C. elegans N2 upon S. aureus infection in the presence of O-(2-acetamido-2-deoxy-D-glucopyranosylidenamino) N-phenylcarbamate (PUGNAc-an OGA inhibitor) further confirmed the involvement of O-GlcNAcylation in protecting C. elegans from infection. The two-dimensional gel-based proteomic analysis of the control and S. aureus infected C. elegans oga-1 mutant followed by mass spectrometric identification of differentially expressed proteins has been carried out. The results revealed that key proteins involved in ubiquitination such as Cullin-1 (CUL-1), Cullin-3 (CUL-3), BTB and MATH domain-containing protein 15 (BATH-15), ubiquitin-conjugating enzyme E2 variant 3 (UEV-3) and probable ubiquitin-conjugating enzyme E2 7 (UBC-7) are upregulated. Real-time PCR analysis further confirms the upregulation of genes encoding the above-mentioned proteins which are involved in the ubiquitin-mediated pathways in C. elegans. In addition, treatment of C. elegans wild-type N2 and the oga-1 mutant with PUGNAc + suramin and suramin (an ubiquitination inhibitor), respectively has resulted in increased sensitivity to S. aureus infection. Hence, it is presumed that upregulation of proteins involved in the ubiquitination pathway could be the key regulatory mechanism responsible for the enhanced survival of the oga-1 mutant during S. aureus infection.
1. Introduction
Host–pathogen interaction is a wide spread mechanism in which a pathogen maintains itself inside the host at a population level which either directs infection or protection depending on the susceptibility of the host, virulence factor, route of transmission as well as environmental factors.1 Caenorhabditis elegans is one of the best preclinical in vivo models to study host–pathogen interactions.2 Understanding host–pathogen interactions in C. elegans at the molecular level reveals the nature of infection and uncovers diagnostic and curative targets to overcome harmful infectious diseases and helps to identify target pathways and mechanisms conserved in higher organisms.3 C. elegans has been used as a successful model system to study the pathogenesis of Gram-positive4 and Gram-negative pathogens and fungal pathogens5 as well as the role of toxins, siderophores6 and other virulence determinants during host–pathogen interactions. C. elegans responds to pathogens through an array of immune pathways such as DBL-1/TGF-β,7 DAF-2/DAF-16,8 MAP kinase,9 ERK and toll-like receptors10 which are shown to be conserved in human and other model systems. It has recently been reported that C. elegans can also be used to study virus–host interactions.11,12 Host–pathogen interaction leads to alterations in both genomic and subsequently in proteomic levels. From the perspective of the OMICs approach, proteomics is an important phase as it reveals the changes in active players in the system.13 Due to its genetic amenability and availability of mutants, investigation of C. elegans at the post-translational level during host–pathogen interaction could pave a way for identification of novel drug targets and immune pathways. Recently, huge attention has been paid to studying the role of different post-translational modifications (PTMs) which could regulate the innate immune response of C. elegans during host–pathogen interactions via modulating the protein–protein interactions.13 There are more than 200 PTMs that happen in biological systems. Among these, glycosylation is one of the most prevalent PTMs, in which carbohydrate molecules are covalently attached to amino acid functional groups.14 Glycosylation plays an important role in prognosis and various disease conditions and is considered as a target for therapy response.15,16 Glycosylated proteins/glycan compositions were used for the development of protein biomarkers.17 O-GlcNAc cycling has been reported to regulate several biological processes in C. elegans. For instance, storage of nutrients, dauer formation, longevity,18–20 protein homeostasis,21 neuroprotection,22 glucose stress response,23 innate immune response24 and detoxification of toxins secreted by pathogens.25 Researchers have extensively used ogt-1 (ok430) and oga-1 (ok1207) mutant strains to investigate the role of O-GlcNAcylation in regulating the above mentioned biological processes. In addition, O-GlcNAcylation also modulates the function of the proteasome, chaperone and intracellular calcium level and interacts with other post translational modifications such as phosphorylation and ubiquitination to regulate certain cellular functions.26 O-GlcNAcylation is an intracellular protein modification that links a single N-acetylglucosamine moiety to serine and threonine residues in various target proteins. O-GlcNAc (O-linked β-N-acetylglucosamine) is regulated by two key enzymes namely O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) which catalyse the addition and removal of the N-acetylglucosamine moiety to the target protein, respectively.27 O-GlcNAc is a reversible regulatory modification which happens often on cytoplasmic and nuclear proteins. Uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) is the end product of the hexosamine biosynthetic pathway, which assimilates glucose, amino acid, fatty acid, and nucleotide metabolism to synthesize the substrate for O-GlcNAcylation.28 Catalytic activity and substrate specificity of OGT greatly depend on the UDP-GlcNAc level in the cell, making O-GlcNAcylation a nutrient responsive process.29 Hence the UDP-GlcNAc biosynthesis pathway is termed a nutrient responsive/sensitive pathway. O-GlcNAcylation has an impact on the nutrient and cellular signalling processes.30 O-(2-Acetamido-2-deoxy-D-glucopyranosylidene)amino-N-phenylcarbamate (PUGNAc) is a potent inhibitor of O-GlcNAc-β-N-acetylglucosaminidase and is normally used to inhibit OGA, the enzyme which is responsible for the removal of O-GlcNAc from target proteins in biological settings to study the effects of elevated O-GlcNAc levels in nuclear and cytoplasmic proteins.31 Increasing the cellular level of O-GlcNAc by glucosamine or PUGNAc has been shown to enhance the ubiquitination.32,33Staphylococcus aureus is a Gram-positive, versatile, widespread pathogen and also lives as a commensal organism in the human respiratory tract.34 S. aureus causes a range of infections from skin infections to life-threatening diseases such as endocarditis, pneumonia and Toxic Shock Syndrome (TSS).35 It has been previously shown that, structural proteins, proteins involved in protein folding and energy production were differentially expressed in C. elegans upon S. aureus infection.36 Previously our group has reported the role of the innate immune response of C. elegans and its signalling pathways, functional proteins against Vibrio alginolyticus,37 Pseudomonas aeruginosa,38 Protease mirabilis39 and Klebsiella pneumoniae40 using proteomic approaches. However, proteomic analysis involving the study of the role of post translational modifications especially O-GlcNAcylation still remains scanty. Against this backdrop, the present investigation is focused on delineating the role/involvement of O-GlcNAcylation in C. elegans during S. aureus infection using two-dimensional gel-based proteomic analysis followed by MALDI-ToF/ToF identification of differentially regulated proteins.
2. Materials and methods
2.1. Bacterial strains and growth conditions
Staphylococcus aureus ATCC 11632 and Escherichia coli OP50 (obtained from the Caenorhabditis Genetics Center (CGC)) were maintained in tryptic soy agar and Luria Bertani (LB), respectively. Both the cultures were grown at 37 °C in Luria Bertani (LB) broth for killing assay and infections.2.2. Caenorhabdiis elegans maintenance
C. elegans N2 wild-type, and deletion mutant strains RB1169 [oga-1 (ok1207)] and RB653 [ogt-1 (ok430) III] were obtained from the Caenorhabditis Genetics Center (CGC), Minnesota, USA and maintained in standard Nematode Growth Medium (NGM) plates.41,42 NGM plates were seeded with E. coli OP50 as a food source and maintained at 20 °C. The age synchronized L4 stage C. elegans were used for all the experimental assays. Age synchronized L4 stage C. elegans were obtained by bleaching the egg stage hermaphrodite animals with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of commercial bleach and 5 M KOH.
1 ratio of commercial bleach and 5 M KOH.
2.3. Worm liquid-killing assay
Approximately 10 numbers of wild-type N2 and oga-1 (ok1207) and ogt-1 (ok430) mutant C. elegans were transferred to a 24 well culture plate containing sterile M9 buffer along with S. aureus (20% inoculum with OD 600 = 0.1). Then the plates were incubated at 20 °C and the survival of the worms was monitored for every 2 hours until the complete death of the worms. In addition, the killing assay was also carried out in the presence of PUGNAc (0.5 mM). Animals were considered to be dead if there is no response to touch by a platinum loop or there is invisible contraction of the pharynx. C. elegans fed on E. coli OP50 served as a control.432.4. Colony forming unit assay
C. elegans N2, oga-1 and ogt-1 mutants were infected with S. aureus for 48 h. After preinfection, approximately 10 infected worms were taken and washed thoroughly with M9 buffer containing 1 mM sodium azide to avoid the exclusion of invaded bacteria from the worms’ intestine. To the washed worms, 400 mg of 1 mM silicon carbide particles (HiMedia, India) was added and vortexed for 20 min to disturb the worms so that they release the colonized bacterial cells and the resulting suspension was serially diluted and plated on the Hichrome S. aureus agar (Himedia) to determine the colony forming unit (CFU).432.5. Confocal laser scanning microscopic (CLSM) analysis
To confirm the intestinal colonization, C. elegans N2, oga-1 and ogt-1 mutants were exposed to S. aureus for 72 h. After infection, worms were washed thrice with M9 buffer containing 1 mM sodium azide and stained with 0.1% acridine orange solution for 5 min. After staining, the worms were washed twice with M9 buffer to remove left-over stain. Then, the worms were placed on microscopic slides, covered with cover slips and visualized under CLSM [(LSM 710, Carl Zeiss, Germany) (10×, dry, EC Plan-Neofluar objective lens with 0.3 NA and 10× optical lens)] using 488 nm argon laser to excite the acridine orange and a 500 to 600 nm band pass emission filter for the detection of the fluorescence signal as mentioned in our previously published reports.39,44 The fluorescence image acquisition was done using Zen 2009 image software (Carl Zeiss, Germany) in which the scan speed was set to 2 s, bit depth 12 and pin hole was set 1 Airy Unit (AU) as the parameters.45 The gain master, digital gain and digital offset were set constant for each sample. Further, the Transmitted-Photo Multiplier Tube detector (T-PMT) mode was also enabled in the software to acquire the bright field images. The presence of acridine orange fluorescence in the pharyngeal and intestinal region indicates the colonisation of S. aureus.2.6. Pharyngeal pumping assay
To analyze the pharyngeal pumping rate of C. elegans during S. aureus exposure, wild-type N2, oga-1 and ogt-1 worms were exposed to S. aureus for different time intervals (54, 60, 66 and 72 h) and transferred to NGM plates and their pumping rate was monitored carefully for twenty consecutive seconds. C. elegans fed with E. coli OP50 were used as a control.39 In addition, control and S. aureus infected worms were observed under bright field microscope at 200× magnification (NIKON SMZ, Japan) to observe the pharyngeal damage.462.7. Two-dimensional gel electrophoresis (2-DGE)
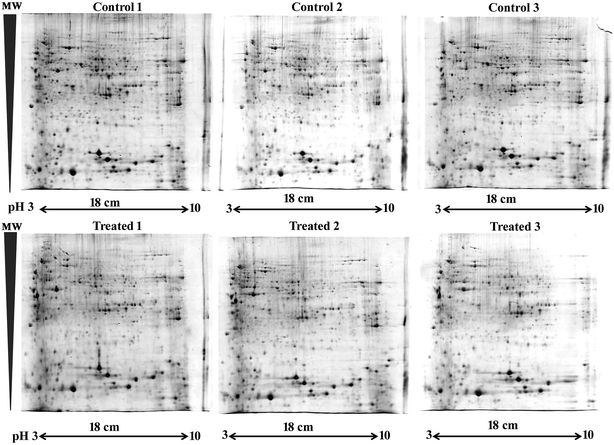
2-DGE was carried out in biological triplicate with minor modifications.40 The total proteins (1 mg) of C. elegans mutant oga-1 fed with E. coli OP50 (control) and oga-1 mutant infected with S. aureus for 72 h were purified using a 2D clean-up kit. The resulting pellets were dissolved in urea–thiourea sample buffer and loaded on to the Immobiline DryStrip gel strips (18 cm, non-linear, pH 3–10 cm) which were subjected to rehydration for 12 h at 20 °C. The rehydrated strips were isoelectrically focused using an Ettan™ IPGphor 3 isoelectric focusing system with standard parameters. Then, the strips were reduced and alkylated with dithiothreitol (DTT) and iodoacetamide (IAA), respectively. Then the strips were placed on 12% sodium dodecyl sulphate-polyacrylamide gels (SDS-PAGE) containing a traceable amount of tracking dye bromophenol blue for the separation of the second dimension of the proteins. After electrophoresis, the gels were stained with colloidal Coomassie Brilliant Blue (CBB) G-250 staining for 12 h and destained with double distilled water and images were captured using a gel scanner.402.8. Image analysis and in gel trypsin digestion of differentially expressed proteins
The processed 2-DGE gel images were analysed using Image Master 2D Platinum 7.0 software (GE Healthcare). The differentially expressed protein spots were subjected to trypsin digestion for MALDI-ToF/ToF analysis. Briefly, protein spots with 1.5 fold down or upregulation were excised from the gel and washed with MilliQ H2O and destained with the destaining solution containing 50% acetonitrile (ACN) containing 25 mM ammonium bicarbonate (NH4HCO3) to remove the CBB. Then the destained gels were dehydrated using 100% acetonitrile and vacuum dried. The dehydrated gel plugs were rehydrated with trypsin digestion buffer (10 mM NH4HCO3 in 10% ACN) containing 400 ng of trypsin and incubated in ice for 45 min. After rehydration, gel plugs were overlayed with 40 mM NH4HCO3 in 10% ACN and incubated at 37 °C for 16 h for tryptic digestion. After digestion, the peptides were extracted with extraction buffer containing (0.1% trifluoroacetic acid (TFA) in 60% ACN). The extraction step was repeated twice to increase the yield and pooled peptides were vacuum dried and purified using C18 zip tips (Merck Millipore) as per the manufacturer’s directions and the peptides were analysed using MALDI-ToF/ToF (AXIMA Performance, SHIMADZU BIOTECH). The MALDI MS software generated mono isotopic peak list (m/z range of 700–3000 Da) was analysed using Swiss Prot (taxonomy-C. elegans) database by MASCOT with the typical protein detection constraints.472.9. STRING and gene ontology analysis
Protein–protein interactions (PPI) between the list of regulated proteins identified from 2-DGE were carried out by STRING 10.5 (http://https://string-db.org/) analysis with a medium confidence score of 0.4. The interacting partners and their role in biological activities were identified by Gene Ontology (GO).372.10. Killing assay with ubiquitination inhibitor
To confirm the role of ubiquitination in the survival of C. elegans N2, oga-1 (ok1207) and ogt-1 (ok430) mutants during S. aureus infection, a killing assay was carried out as mentioned previously in the presence of PUGNAc (0.5 mM), suramin (100 μM) and a combination of both cullin-RING E3 ubiquitin ligases inhibitor suramin (100 μM) + PUGNAc (0.5 mM) respectively.2.11. Total RNA isolation, RT-PCR and quantitative PCR analysis
The total RNA was isolated from the C. elegans oga-1 mutant infected with S. aureus and with the respective control E. coli OP50 for 72 h by the TRIZOL method according to the manufacturer’s instruction and then the isolated RNA was converted into cDNA using MultiScribe™ Reverse Transcriptase enzyme (Applied Biosystems Inc., USA). Quantitative PCR analysis was performed to study the expression of specific protein genes (cul-1, cul-3, bath-15, uev-3, ubc-7 [Table 1]) involved in C. elegans during S. aureus infection using Power SYBR Green PCR Master Mix (Applied Biosystems). The C. elegans β-actin gene was used as an internal control. Gene expression levels were calculated by 2−ΔΔct values. Gene expression analysis was carried out in biological triplicate.40| Genes | Primer sequences |
|---|---|
| cul-1 fp | 5′-AATCTCGTCGAAGGTGATGG-3′ |
| cul-1 rp | 5′-TATCGGCTGTTGATCATGGA-3′ |
| cul-3 fp | 5′-GGAATTTGTGGCCCTCTACA-3′ |
| cul-3 rp | 5′-AAGAGACGTTTGGCGAGGTA-3′ |
| bath-15 fp | 5′-TTCGCACGAAAGATCAACTG-3′ |
| bath-15 rp | 5′-CCAAATCCATCGAAACATCC-3′ |
| uev-3 fp | 5′-TGTAATAATTGATGGACCTGTTGG-3′ |
| uev-3 rp | 5′-TTTCCATATTTCCCAAGATTCG-3′ |
| ubc-7 fp | 5′-TGCATCTCGATTCTTCACGA-3′ |
| ubc-7 rp | 5′-CAACGTTTGCAGGAGATTCA-3′ |
2.12. Statistical analysis
All the experiments were conducted in triplicate and the results were analysed using one-way analysis of variance with a statistical significance p-value of <0.05, using the SPSS tool (Chicago, IL, USA).3. Results and discussion
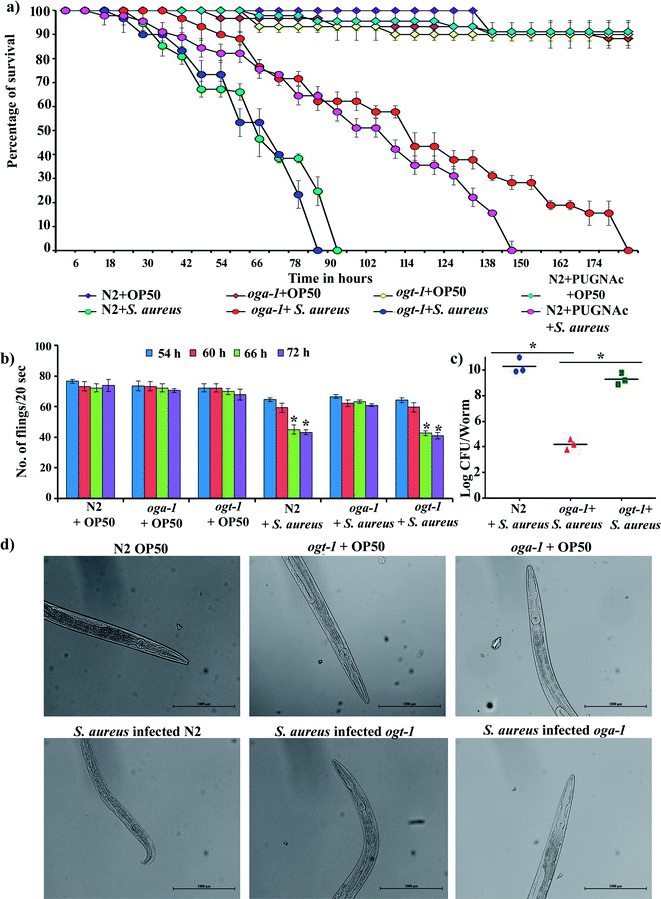
3.1. Effect of S. aureus infection on the survival and pharyngeal pumping of C. elegans N2, oga-1 mutant and ogt-1 mutant
The availability of O-GlcNAcylation mutants such as oga-1 (ok1207) and ogt-1 (ok430) deletion mutant is beneficial for the study of its role during host–pathogen interactions. The liquid killing assay was carried out in the presence of FUDR (5-fluorodeoxyuridine) to inhibit progeny production. L4 stage oga-1, ogt-1 mutants and wild-type N2 C. elegans were challenged with S. aureus and the complete mortality of N2 and ogt-1 mutant were observed at 90 and 80 h, respectively. In contrast, a relatively increased survival of the oga-1 mutant was observed during S. aureus infection. Even after 168 h of infection, 20% of oga-1 mutant worms had survived (Fig. 1a). Thus, the increased survival could be attributed to the O-GlcNAcylation. To further confirm the involvement of O-GlcNAcylation in the increased survival of the C. elegans oga-1 mutant, wild-type N2 C. elegans was infected with S. aureus in the presence of PUGNAc (0.5 mM) which is known to inhibit the activity of O-GlcNAcase (OGA). Survival of PUGNAc treated wild type nematodes upon S. aureus infection was found to be similar to that of oga-1 mutant. These results further substantiated that the enhanced O-GlcNAcylation in the oga-1 mutant is a possible factor responsible for the increase in survival of C. elegans during S. aureus infection. The complete killing of wild type C. elegans N2 upon infection with S. aureus observed in the present study corroborates well with the previous report wherein complete killing of wild-type N2 worms infected with S. aureus was observed at 90 h.43 In addition, to further confirm that the susceptibility of the ogt-1 mutant to S. aureus infection is due to lack of O-GlcNAcylation and not due to lack of nutrition, the killing assay was carried out in the presence of heat killed E. coli OP50. Survival of ogt-1 in the presence and absence of a food source is found to be similar during S. aureus infection (ESI Fig. 1†). Hence, it is confirmed that the susceptibility of ogt-1 mutant to S. aureus can be attributed to the lack of O-GlcNAcylation. It has been previously reported that O-GlcNAc plays a key role in eliciting a pathogen specific immune response in C. elegans.24 Briefly, the ogt-1 mutant was shown to be highly sensitive to S. aureus infection. Whereas, the sensitivity of C. elegans wild type and oga-1 mutant to S. aureus NCTC8325 infection was shown to be similar.24 In contrast to these findings, in the current study we observed the increase in the survival of oga-1 mutant during S. aureus (ATCC 11632) infection. However, in the present study also the ogt-1 mutant remains highly sensitive to S. aureus infection. Merkx-Jacques et al., have shown that the sensitivity of C. elegans to environmental E. coli isolates differs as a result of variation in the virulence.48 Recent reports from our group also revealed a difference in survival during infection with different strains of S. flexeneri (such as S. flexneri 2b and S. flexneri M9OT).49,50 In addition, P. aeruginosa PA14 has already been reported as more pathogenic to C. elegans than P. aeruginosa PAO1.51 Hence, it is hypothesized that the difference in the survival of the oga-1 mutant against S. aureus infection observed in the present investigation when compared to the results of Bond et al., could be attributed to a variation in the virulence of the S. aureus strain.Most Gram-negative and Gram-positive bacteria have been reported to escape from the grinder and antimicrobial peptides present in the C. elegans pharynx and colonize in the microvilli brush border and intestine. The pharynx morphology defective mutant (phm-2) C. elegans is hypersensitive to pathogenic infection.52 Bacterial pathogens have been known to affect the pharyngeal pumping rate during the course of infection in C. elegans39 and hence, the pharyngeal pumping rate of N2, oga-1 and ogt-1 during S. aureus infection was observed at 54, 60, 66 and 72 h (Fig. 1b). The results showed profound alterations in the pharyngeal pumping of N2 and ogt-1 worms upon S. aureus infection. Similar to the liquid killing assay, in the oga-1 mutant, no significant alteration in pharyngeal pumping was observed. When the bacterial exposure time increased, the pharyngeal pumping rate decreased gradually in the N2 and ogt-1 mutant C. elegans. This could be due to damage caused by S. aureus in the pharyngeal bulb during infection. These results suggested that the involvement of enhanced glycosylation in the oga-1 mutant C. elegans has prevented the infection caused by S. aureus when compared to the wild-type and the ogt-1 mutant worms.21 For further confirmation of pharyngeal bulb damage, bright field microscopic analysis was carried out. Light micrographs clearly shows the pharyngeal damage in S. aureus infected N2 and ogt-1 and not in oga-1 worms (Fig. 1d).
3.2. Evaluation of intestinal colonization of S. aureus in C. elegans N2, oga-1 mutant and ogt-1 mutant
Intestinal colonization of bacterial pathogens is one of the important mechanisms required to cause infection under in vivo conditions. For further confirmation of S. aureus intestinal colonization in C. elegans N2, oga-1 and ogt-1 mutants, a CFU assay was performed. The CFU of S. aureus in the N2 and ogt-1 C. elegans after 48 h infection was found to be high, when compared to oga-1 (Fig. 1c). The intestinal colonization of S. aureus in wild type N2 C. elegans observed in the present study corroborates well with our previous report.43 C. elegans is a tractable model system to visualize the bacterial intestinal colonization. Hence, to visualize the intestinal colonization of S. aureus infected C. elegans wild-type N2, oga-1 mutant and ogt-1 mutant are observed under CLSM and the results are presented in (Fig. 2). From the CLSM micrographs, it is evident that the intestinal colonization of S. aureus was found to be similar in the N2 and ogt-1 mutant. Whereas, the wild-type N2+ PUGNAc (0.5 mM) and oga-1 mutant showed reduced intestinal colonization of S. aureus.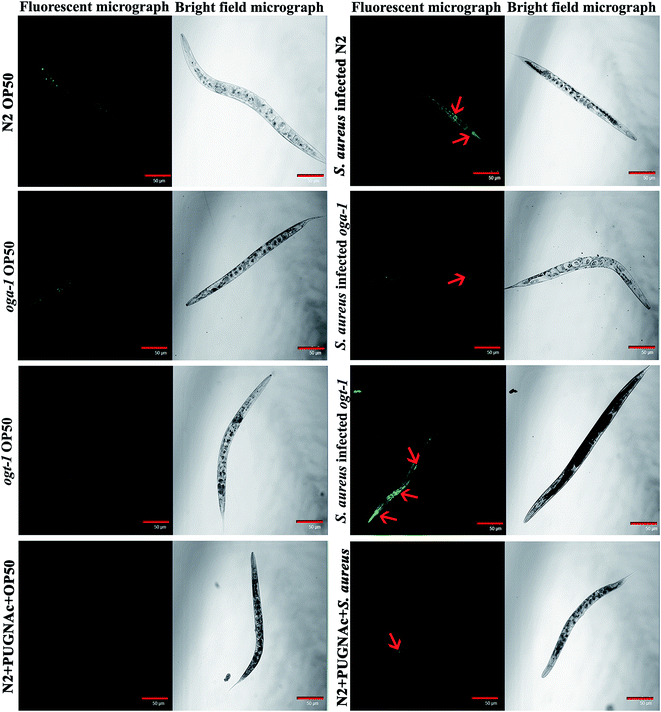 | ||
| Fig. 2 Representative CLSM micrographs (fluorescence and bright field) depicting the intestinal colonization of S. aureus in C. elegans N2, oga-1 mutant and ogt-1 mutant. Scale bar indicates 50 μM. | ||
3.3. Proteomic analysis of C. elegans oga-1 mutant upon S. aureus infection
To decipher the molecular mechanism responsible for the increased survival of the C. elegans oga-1 mutant during S. aureus infection 2-DGE was performed and the protein spots were stained with CBB and images were captured and analysed using ImageMaster 2D platinum software to detect the differentially expressed protein spots (Fig. 3). In total 536 spots were matched between control and treated gels and among them, 17 and 22 spots were upregulated and downregulated, respectively (more than 1.5 fold with an ANOVA value of 0.05) and were selected for MALDI-ToF/ToF analysis. A list of differentially identified proteins identified using MALDI-ToF/ToF is presented in Table 2.| S. No. | Regulation | UniProtKB ID | Protein name | Gene name | MASCOT score | No. of peptides matched | Sequence coverage | ANOVA |
|---|---|---|---|---|---|---|---|---|
| 1 | Down | ATPB_CAEEL | ATP synthase subunit beta | atp-2 | 57 | 10 | 30% | 0.02 |
| 2 | Up | GRDN_CAEEL | Girdin homolog | grdn-1 | 41 | 9 | 7% | 0.04 |
| 3 | UP | CUL3_CAEEL | Cullin-3 | cul-3 | 55 | 13 | 23% | 0.05 |
| 4 | Down | FAR1_CAEEL | Fatty-acid and retinol-binding protein 1 | far-1 | 61 | 12 | 61% | 0.04 |
| 5 | Down | HSP7D_CAEEL | Heat shock 70 kDa protein D | hsp-4 | 59 | 9 | 19% | 0.06 |
| 6 | Up | MDHM_CAEEL | Probable malate dehydrogenase | mdh-2 | 57 | 9 | 30% | 0.06 |
| 7 | Up | CUL1_CAEEL | Cullin-1 | cul-1 | 68 | 26 | 29% | 0.04 |
| 8 | Down | COX6A_CAEEL | Probable cytochrome c oxidase subunit 6A | tag-174 | 55 | 6 | 52% | 0.00 |
| 9 | Up | BCAT_CAEEL | Branched-chain-amino-acid aminotranferase | bcat-1 | 89 | 19 | 27% | 0.01 |
| 10 | Up | EPG2_CAEEL | Ectopic P granules protein 2 | epg-2 | 74 | 28 | 23% | 0.03 |
| 11 | Up | UCR1_CAEEL | Cytochrome b-c1 complex subunit 1 | ucr-1 | 69 | 12 | 33% | 0.01 |
| 12 | Down | ACOX5_CAEEL | Probable peroxisomal acyl-coenzyme a oxidase 5 | acox-5 | 58 | 12 | 12% | 0.03 |
| 13 | Down | EI2BG_CAEEL | Probable translation initiation factor eIF-2B subunit γ | ppp-1 | 43 | 4 | 15% | 0.02 |
| 14 | Down | IPYR_CAEEL | Probable inorganic pyrophosphatase 1 | pyp-1 | 88 | 11 | 25% | 0.03 |
| 15 | Down | CYP5_CAEEL | Peptidylprolyl cistrans isomerase 5 | cyn5 | 118 | 9 | 29% | 0.02 |
| 16 | Down | RN207_CAEEL | Probable RING finger protein | F47G9.4 | 47 | 10 | 13% | 0.05 |
| 17 | Up | HSP12_CAEEL | Heat shock protein Hsp-16.2 | hsp-16.2 | 29 | 4 | 22% | 0.02 |
| 18 | Down | LEC1_CAEEL | 32 kDa beta-galactoside-binding lectin | lec1 | 86 | 17 | 48% | 0.05 |
| 19 | Down | VIT6_CAEEL | Vitellogenin-6 | vit-6 | 115 | 43 | 26% | 0.02 |
| 20 | Down | CLAP3_CAEEL | Protein CLASP-3 | cls-3 | 35 | 7 | 11 | 0.04 |
| 21 | Up | TFDP1_CAEEL | Transcription factor dpl-1 | dpl-1 | 52 | 10 | 21% | 0.04 |
| 22 | Up | SNB5_CAEEL | Synaptobrevin-like protein 5 | snb-5 | 33 | 4 | 29% | 0.02 |
| 23 | Up | ENO_CAEEL | Enolase | enol-1 | 61 | 11 | 31% | 0.05 |
| 24 | Up | BAT15_CAEEL | BTB and MATH domain-containing protein 15 (fragment) | bath-15 | 59 | 8 | 31% | 0.02 |
| 25 | Down | GLS2_CAEEL | Putative glutaminase 2 | glna-2 | 46 | 11 | 18% | 0.02 |
| 26 | Down | GPA1_CAEEL | Guanine nucleotide-binding protein alpha-1 subunit | gpa-1 | 37 | 7 | 21% | 0.00 |
| 27 | UP | UB2V3_CAEEL | Ubiquitin-conjugating enzyme E2 variant 3 | uev-3 | 51 | 12 | 27% | 0.02 |
| 28 | Up | UBC7_CAEEL | Probable ubiquitin-conjugating enzyme E2 7 | ubc-7 | 35 | 6 | 18% | 0.02 |
| 29 | Down | DAF25_CAEEL | Dauer abnormal formation protein 25 | daf-25 | 59 | 8 | 13% | 0.42 |
| 30 | Up | GLC7A_CAEEL | Serine/threonine-protein phosphatase PP1-alpha | gsp-1 | 39 | 5 | 18% | 0.07 |
| 31 | Down | NDUAC_CAEEL | Probable NADH dehydrogenase [ubiquinone] 1 alpha sub complex subunit 12 | Y94H6A.8 | 36 | 3 | 24% | 0.04 |
| 32 | Up | YQR2_CAEEL | Uncharacterized protein F19C6.2 | F19C6.2 | 54 | 9 | 14% | 0.04 |
| 33 | Up | AP2M_CAEEL | AP-2 complex subunit mu | dpy-23 | 62 | 8 | 24% | 0.04 |
| 34 | Down | YNP4_CAEEL | Uncharacterized protein T05G5.4 | T05G5.4 | 57 | 5 | 31% | 0.49 |
| 35 | Down | DDX3_CAEEL | ATP-dependent RNA helicase laf-1 | laf-1 | 35 | 6 | 9% | 0.05 |
| 36 | Down | KBP3_CAEEL | Kinetochore-binding protein 3 | kbp-3 | 47 | 5 | 35% | 0.05 |
| 37 | Up | NDUS2_CAEEL | Probable NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial | gas-1 | 33 | 5 | 12% | 0.05 |
| 38 | Down | TBB2_CAEEL | Tubulin beta-2 chain | tbb-2 | 74 | 11 | 18% | 0.01 |
| 39 | Down | HM21_CAEEL | Homeobox protein ceh-21 | ceh-21 | 62 | 9 | 22% | 0.05 |
Protein–protein interaction networks of the differentially expressed proteins upon S. aureus infection in the oga-1 mutant were predicted using STRING analysis with a medium confidence score of 0.4. Interestingly, the STRING network revealed that the upregulated proteins are able to interact with the proteins involved in the ubiquitin-mediated pathways in C. elegans (Fig. 4a). Gene ontology analysis of upregulated proteins revealed their involvement in protein ubiquitination, receptor-mediated endocytosis, vesicle-mediated transport, multicellular organismal development, endocytosis, determination of adult lifespan, multicellular organismal aging, generation of precursor metabolites and energy, establishment of localization, anatomical structure development, nematode larval development, cellular metabolic process, embryo development ending in birth/egg hatching, primary metabolic process and single organism reproductive process (Fig. 4b). Whereas, down-regulated proteins are found to be involved in nematode larval development, single-organism developmental process, multicellular organismal development, molecular function, macromolecular complex, respiratory chain, mitochondrial part, mitochondrial inner membrane, protein complex, mitochondrial envelope and intracellular organelle part (Fig. 4c and d).
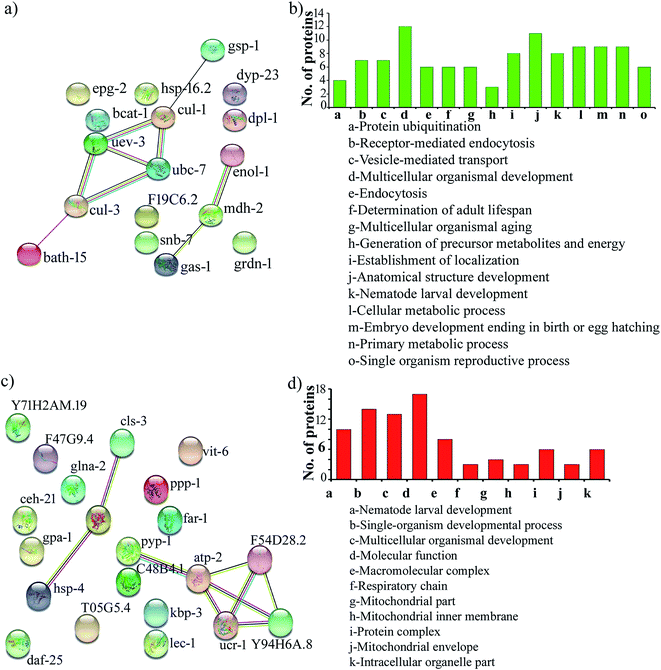 | ||
| Fig. 4 Protein–protein interaction networks and gene ontology of the upregulated proteins (a & b) and down regulated proteins (c & d). | ||
In the present investigation, proteins involved in the ubiquitination of C. elegans such as, Cullin-1 (CUL-1), Cullin-3 (CUL-3), BTB and MATH domain-containing protein 15 (BATH-15), ubiquitin-conjugating enzyme E2 variant 3 (UEV-3) and probable ubiquitin-conjugating enzyme E2 7 (UBC-7) were found to be upregulated upon S. aureus infection in the C. elegans oga-1 mutant when compared to the control. Upregulation of the above-mentioned proteins hinted at enhanced ubiquitination in the S. aureus infected C. elegans oga-1 mutant. Ubiquitination is well known for its involvement in degradation of proteins by the Ubiquitin-Proteasome System (UPS). Previously, it has been reported that a ubiquitination-mediated response is required for the survival of C. elegans during Orsay virus and Nematocida parisii infection.53 In eukaryotes, ubiquitin ligases are the group of proteins mainly involved in ubiquitination, which requires the Skp1-Cul1-F-box protein (SCF) to form a multi-subunit RING-finger type ligase.54 CUL-1 and CUL-3 are the important hydrophobic proteins serve as molecular scaffolds of SCF.55 Upregulation of Cullin-6 an ortholog of cullin protein has been shown to be important in C. elegans to target the nematode killer N. parisii in its intestine. Hence, upregulation of CUL-1 and CUL-3 could be responsible for the enhanced survival of the C. elegans oga-1 mutant during S. aureus infection. F-box and MATH-BTB family proteins are known to have a conserved cullin binding and a variable substrate binding domain.56 It has already been hypothesized that MATH-BTB proteins are one of the ubiquitin-dependent proteasome adapters that recognize and target foreign proteins, especially proteins, from bacterial and viral origin for proteolysis.55 Hence, it is presumed that the upregulation of BATH-15 in the S. aureus infected C. elegans oga-1 mutant is responsible for the degradation of proteins involved in the virulence and adherence of the S. aureus. Furthermore, it is also hypothesised that the reduction in the intestinal colonization of S. aureus in oga-1 mutant C. elegans could also be attributed to the upregulation of proteins involved in the ubiquitination. The possible role of ubiquitination in protecting the C. elegans oga-1 mutant from S. aureus is presented in (Fig. 5).
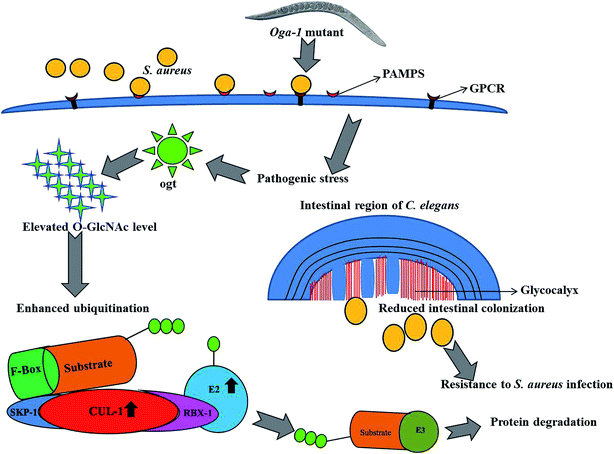 | ||
| Fig. 5 Schematic representation of outcomes from the present study associates the involvement of ubiquitination and O-GlcNAcylation in the C. elegans oga-1 mutant during S. aureus infection. | ||
UEV-3 has been shown to interact with the P38 MAP Kinase family (PMK-3) of the Mitogen-Activated Protein Kinase (MAPK) cascade. Researchers have reported that Proteus mirabilis, Proteus vulgaris and Klebsiella pneumoniae affect the survival of C. elegans by targeting the MAPK pathway.57,58 Hence, the interaction of UEV-3 with PMK-3 is expected to have a role during S. aureus infection in the C. elegans oga-1 mutant.
C. elegans oga-1 and ogt-1 mutants have been shown to have reduced fat levels but increased levels of glycogen and trehalose when compared to wild-type N2.59,60 In the present study, fatty-acid and retinol-binding protein 1 (FAR-1) which is involved in lipid binding was found to be down regulated in the S. aureus infected C. elegans oga-1 mutant. In addition, proteomic analysis revealed the down regulation of Vitellogenin-6 (VIT-6) in the C. elegans oga-1 mutant upon S. aureus infection. Vit-6 RNAi has been shown to increase the intracellular ROS in C. elegans.61 An elevated level of ROS can cause damage to the nucleic acid, proteins and lipids. However, ROS has also been documented for its ability to modulate the innate immune response. For instance, ROS acts as a signalling molecule for the activation of nuclear factor kappa B (NF-κB) and MAPK pathway, which are known to play a protective role in C. elegans during pathogenic infection.62
In addition, proteomic analysis revealed that girdin homolog (GRDN-1) was found to be upregulated upon S. aureus infection. GRDN-1 is an ortholog of mammalian girdin and is a novel signalling/scaffolding protein which plays an important role in multiple cellular processes such as division, mobilization and invasion of cells.63 In C. elegans, cilia are expressed ubiquitously and nucleated by the basal body B. Ciliogenesis is the process in which the cilia are assembled or disassembled during the cell cycle. Girdin acts as a regulator in positioning the basal body and ciliogenesis in both nematodes as well as in human cells. Upregulation of GRDN-1 in the C. elegans oga-1 mutant appears to provide the host with more resistance to S. aureus infection. Furthermore, ectopic P granules protein 2 (EPG-2) was upregulated in the oga-1 mutant upon S. aureus infection. In C. elegans, EPG-2 was involved in the function of autophagosome completion during autophagy.64 Upregulation of EPG-2 in oga-1 mutant could also be one of the components responsible for the resistant to S. aureus infection.
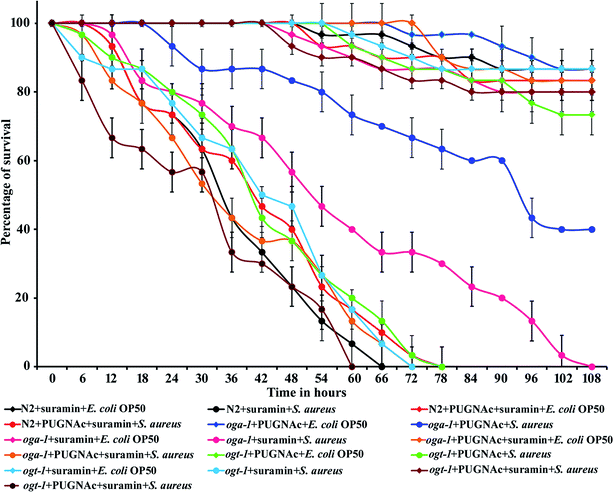
3.4. Effect of PUGNAc and suramin treatment on the survival of C. elegans N2 and oga-1 mutant during S. aureus infection
Suramin is already known to inhibit the cullin-RING E3 ubiquitin ligases.65 Hence, in the present study suramin was used to confirm the role of ubiquitination in increasing the survival of the oga-1 mutant during S. aureus infection. Briefly, C. elegans N2 and oga-1 and ogt-1 mutants were infected with S. aureus in the presence of PUGNAc (0.5 mM), suramin (100 μM) and a combination of PUGNAc + suramin respectively. Interestingly, the susceptibility of C. elegans N2 (treated with PUGNAc + suramin) and oga-1 mutant treated with suramin to S. aureus infection was found to be similar. There is no difference in the survival of oga-1 and PUGNAc treated wild type nematode upon S. aureus infection. PUGNAc + suramin treated ogt-1 mutant worms were found to be more susceptible to S. aureus infection. Based on these results, it is confirmed that the upregulation of proteins involved in ubiquitination in the oga-1 mutant is responsible for its survival during S. aureus infection (Fig. 6).3.5. Confirmation of the upregulation of the ubiquitination pathway genes using real-time PCR
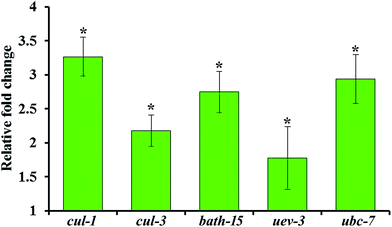
The upregulation of genes involved in ubiquitination such as cul-1, cul-3, bath-15, uev-3 and ubc-7 are further validated by qPCR analysis using the gene-specific primers mentioned in Table 2. Real time PCR analysis revealed that the mRNA level of the selected candidate genes are found to be upregulated during S. aureus infection in the oga-1 mutant (Fig. 7). These results further confirm the role of ubiquitination in protecting oga-1 from S. aureus infection.4. Conclusion
In the present study by using O-GlcNAcylation specific C. elegans mutants, we found that the host acquires more resistance to S. aureus infection. Microscopic and CFU assays revealed the decrease in the colonization of S. aureus in oga-1 when compared to the wild-type N2 and ogt-1 mutant. Treatment of N2 C. elegans with PUGNAc, an OGA inhibitor was also found to increase its survival upon S. aureus infection. This confirms the role of O-GlcNAcylation in protecting C. elegans from pathogenic assault. Proteomic analysis was carried out to further confirm the role of O-GlcNAcylation during the host–pathogen interaction and the results revealed the upregulation of proteins involved in ubiquitination pathway such as CUL-1, CUL-2, CUL-3, BATH-15, UEV-3 and UBC-7. Real-time PCR analysis revealed the upregulation of cul-1, cul-3, bath-15, uev-3 and ubc-7 in S. aureus infected oga-1, which further substantiates the proteomic analysis results. To further confirm the role of ubiquitination in protecting the oga-1 mutant from S. aureus infection, oga-1 mutant C. elegans was treated with suramin (a known cullin-RING E3 ubiquitin ligases inhibitor) and subsequently infected with S. aureus. Suramin treated oga-1 mutant C. elegans was found to be sensitive to S. aureus infection. Furthermore, the sensitivity of C. elegans N2 treated with a combination of PUGNAc and suramin to S. aureus infection was found to be similar to that of untreated C. elegans. Based on the results observed in the current investigation, it is hypothesized that O-GlcNAcylation protects C. elegans oga-1 from S. aureus infection through upregulation of proteins involved in the ubiquitination pathway.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We are grateful to Caenorhabditis Genetics Center, which is funded by the National Institute of Health, National Centre for Research Resources for providing C. elegans N2 wild type, mutant strains and E. coli OP50. L. Vigneshwari acknowledges the Basic Scientific Research Fellowship provided by UGC [grant number F.4-1/2006(BSR)/7-326/2011(BSR)]. The authors sincerely acknowledge the computational and bioinformatics facility provided by the Alagappa University Bioinformatics Infrastructure Facility (funded by DBT, GOI; File No. BT/BI/25/012/2012, BIF). The authors also thankfully acknowledge DST-FIST (Grant No. SR/FST/LSI-639/2015(C)), UGC-SAP (Grant No. F.5-1/2018/DRS-II(SAP-II)) and DST-PURSE (Grant No. SR/PURSE Phase 2/38 (G)) for providing instrumentation facilities.References
- D. Dutta and H. Clevers, Curr. Opin. Immunol., 2017, 48, 15–22 CrossRef PubMed.
- W. Kim, G. L. Hendricks, K. Lee and E. Mylonakis, Expert Opin. Drug Discovery, 2017, 12, 625 CrossRef PubMed.
- T. K. Gerbaba, L. Green-Harrison and A. G. Buret, J. Nematol., 2017, 49, 348 Search PubMed.
- G. J. Yuen and F. M. Ausubel, Virulence, 2018, 9, 683–699 CrossRef PubMed.
- P. Nag, P. R. Aggarwal, S. Ghosh, K. Narula, R. Tayal, N. Maheshwari, N. Chakraborty and S. Chakraborty, Cell Death Discovery, 2017, 3, 17073 CrossRef PubMed.
- D. Kang, D. R. Kirienko, P. Webster, A. L. Fisher and N. V. Kirienko, Virulence, 2018, 9, 804–817 CrossRef PubMed.
- G. V. Mallo, C. L. Kurz, C. Couillault, N. Pujol, S. Granjeaud, Y. Kohara and J. J. Ewbank, Curr. Biol., 2002, 12, 1209–1214 CrossRef PubMed.
- D. A. Garsin, J. M. Villanueva, J. Begun, D. H. Kim, C. D. Sifri, S. B. Calderwood, G. Ruvkun and F. M. Ausubel, Science, 2003, 300, 1921 CrossRef PubMed.
- D. H. Kim, R. Feinbaum, G. Alloing, F. E. Emerson, D. A. GarsinInoue, H. Inoue, M. Tanaka-Hino, N. Hisamoto, K. Matsumoto, M. W. Tan and F. M. Ausubel, Science, 2002, 297, 623–626 CrossRef PubMed.
- M. J. Gravato-Nobre, H. R. Nicholas, R. Nijland, D. O’Rourke, D. E. Whittington, K. J. Yook and J. Hodgkin, Genetics, 2005, 171, 1033–1045 CrossRef PubMed.
- D. B. Gammon, J. Virol., 2017, 91, e00509–e00517 CrossRef PubMed.
- K. Chen, C. J. Franz, H. Jiang, Y. Jiang and D. Wang, BMC Genomics, 2017, 303 CrossRef PubMed.
- P. M. J. Beltran, J. D. Federspiel, X. Sheng and I. M. Cristea, Mol. Syst. Biol., 2017, 13, 922 CrossRef PubMed.
- M. Audagnotto and M. Dal Peraro, Comput. Struct. Biotechnol. J., 2017, 15, 307 CrossRef PubMed.
- G. Lauc, M. Pezer, I. Rudan and H. Campbell, Mechanisms of disease, Biochim. Biophys. Acta, 2016, 1860, 1574 CrossRef PubMed.
- R. J. Solá and K. Griebenow, BioDrugs, 2010, 24, 9 CrossRef PubMed.
- J. Zhao, T. H. Patwa, D. M. Lubman and D. M. Simeone, Curr. Opin. Mol. Ther., 2008, 10, 602 Search PubMed.
- J. A. Hanover and P. Wang, Worm, 2013, 2, 17669–17674 CrossRef PubMed.
- M. E. Forsythe, D. C. Love, B. D. Lazarus, E. J. Kim, W. A. Prinz, G. Ashwell, M. W. Krause and J. A. Hanover, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 11952–11957 CrossRef PubMed.
- S. K. Ghosh, M. R. Bond, D. C. Love, G. G. Ashwell, M. W. Krause and J. A. Hanover, Front. Endocrinol., 2014, 5, 197 Search PubMed.
- P. Wang, B. D. Lazarus, M. E. Forsythe, D. C. Love, M. W. Krause and J. A. Hanover, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 17669–17674 CrossRef PubMed.
- J. A. Hanover, M. E. Forsythe, P. T. Hennessey, T. M. Brodigan, D. C. Love, G. Ashwell and M. A. Krause, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 11266–11271 CrossRef PubMed.
- M. A. Mondoux, D. C. Love, S. K. Ghosh, T. Fukushige, M. Bond, G. R. Weerasinghe, J. A. Hanover and M. W. Krause, Genetics, 2011, 188, 369–382 CrossRef PubMed.
- M. R. Bond, S. K. Ghosh, P. Wang and J. A. Hanover, PLoS One, 2014, 9, e113231 CrossRef PubMed.
- B. Cezairliyan, N. Vinayavekhin, D. Grenfell-Lee, G. J. Yuen, A. Saghatelian and F. M. Ausubel, PLoS Pathog., 2013, 9, e1003101 CrossRef PubMed.
- G. W. Hart, C. Slawson, G. Ramirez-Correa and O. Lagerlof, Annu. Rev. Biochem., 2011, 80, 825–858 CrossRef PubMed.
- R. S. Haltiwanger, G. D. Holt and G. W. Hart, J. Biol. Chem., 1990, 265, 2563 Search PubMed.
- A. A. Ramanathan, A. Mehta and C. M. Artlett, Glycobiol. Insights, 2017, 6 Search PubMed.
- Z. V. Wang, Y. Deng, N. Gao, Z. Pedrozo, D. L. Li, C. R. Morales, A. Criollo, X. Luo, W. Tan, N. Jiang and M. A. Lehrman, Cell, 2014, 156, 1179–1192 CrossRef PubMed.
- J. Lee, K. Y. Kim, J. Lee and Y. K. Paik, J. Biol. Chem., 2010, 285, 2930–2939 CrossRef PubMed.
- T. M. Gloster and D. J. Vocadlo, Curr. Signal Transduction Ther., 2010, 5, 74–91 CrossRef.
- C. Guinez, A. M. Mir, V. Dehennaut, R. Cacan, A. Harduin-Lepers, J. C. Michalski and T. Lefebvre, FASEB J., 2008, 22, 2901–2911 CrossRef PubMed.
- G. V. Shrikhande, S. T. Scali, C. G. da Silva, S. M. Damrauer, E. Csizmadia, P. Putheti, M. Matthey, R. Arjoon, R. Patel, J. J. Siracuse and E. R. Maccariello, PLoS One, 2010, 5, e14240 CrossRef PubMed.
- S. Y. Tong, J. S. Davis, E. Eichenberger, T. L. Holland and V. G. Fowler, Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management, Clin. Microbiol. Rev., 2015, 28, 603–661 CrossRef PubMed.
- C. Lin and M. L. Peterson, Expert Rev. Clin. Pharmacol., 2010, 3, 753 CrossRef PubMed.
- A. Bogaerts, I. Beets, L. Temmerman, L. Schoofs and P. Verleyen, Biol. Direct, 2010, 5, 11 CrossRef PubMed.
- S. Durai, N. Singh, S. Kundu and K. Balamurugan, J. Proteomics, 2014, 14, 1820 CrossRef PubMed.
- V. Balasubramanian, D. Sellegounder, K. Suman and B. Krishnaswamy, J. Proteomics, 2016, 145, 141 CrossRef PubMed.
- G. JebaMercy, S. Durai, U. Prithika, S. Marudhupandiyan, P. Dasauni, S. Kundu and K. Balamurugan, J. Proteomics, 2016, 145, 81 CrossRef PubMed.
- A. Kamaladevi and K. Balamurugan, Front. Cell. Infect. Microbiol., 2017, 7, 393 CrossRef PubMed.
- S. Brenner, Genetics, 1974, 77, 71 Search PubMed.
- J. A. Lewis and J. T. Fleming, Methods Cell Biol., 1995, 48, 3 CrossRef PubMed.
- G. JebaMercy, S. K. Pandian and K. Balamurugan, Folia Microbiol., 2011, 56, 373 CrossRef PubMed.
- D. Viszwapriya, G. A. Subramenium, U. Prithika, K. Balamurugan and S. K. Pandian, Pathog. Dis., 2016, 74 Search PubMed.
- M. Hohne, J. Lorscheider, A. von Bardeleben, M. Dufner, M. A. Scharf, M. Godel, M. Helmstädter, E. M. Schurek, S. Zank, P. Gerke and C. Kurschat, Mol. Cell. Biol., 2011, 31, 3241–3251 CrossRef PubMed.
- S. Sethupathy, S. Ananthi, A. Selvaraj, B. Shanmuganathan, L. Vigneshwari, K. Balamurugan, S. Mahalingam and S. K. Pandian, Sci. Rep., 2017, 7, 16328 CrossRef PubMed.
- S. Sethupathy, K. G. Prasath, S. Ananthi, S. Mahalingam, S. Y. Balan and S. K. Pandian, J. Proteomics, 2016, 145, 112–126 CrossRef PubMed.
- A. Merkx-Jacques, A. Coors, R. Brousseau, L. Masson, A. Mazza, Y. C. Tien and E. Topp, Appl. Environ. Microbiol., 2013, 79, 2435–2445 CrossRef PubMed.
- S. Marudhupandiyan, U. Prithika, B. Balasubramaniam and K. Balamurugan, Dev. Comp. Immunol., 2017, 74, 227–236 CrossRef PubMed.
- S. Marudhupandiyan and K. Balamurugan, J. Immunol., 2017, 65, 609–621 Search PubMed.
- B. Vigneshkumar, S. K. Pandian and K. Balamurugan, Arch. Microbiol., 2012, 194, 229–242 CrossRef PubMed.
- M. P. Smith, T. R. Laws, T. P. Atkins, P. C. Oyston, D. I. de Pomerai and R. W. Titball, FEMS Microbiol. Lett., 2002, 210, 181–185 CrossRef PubMed.
- M. A. Bakowski, C. A. Desjardins, M. G. Smelkinson, T. A. Dunbar, I. F. Lopez-Moyado, S. A. Rifkin, C. A. Cuomo and E. R. Troemel, PLoS Pathog., 2014, 10, e1004200 CrossRef PubMed.
- C. M. Xie, W. Wei and Y. Sun, J. Genet. Genome Res., 2013, 40, 97 CrossRef PubMed.
- A. Sarikas, T. Hartmann and Z. Q. Pan, Genome Biol., 2011, 12, 220 CrossRef PubMed.
- J. H. Thomas, Genome Res., 2006, 16, 1017 CrossRef PubMed.
- G. Trujillo, K. Nakata, D. Yan, I. N. Maruyama and Y. Jin, Genetics, 2010, 186, 135 CrossRef PubMed.
- G. JebaMercy, L. Vigneshwari and K. Balamurugan, Cell Host Microbe, 2013, 15, 550 Search PubMed.
- D. C. Love and J. A. Hanover, Sci. STKE, 2005, 312, re13 Search PubMed.
- M. E. Forsythe, D. C. Love, B. D. Lazarus, E. J. Kim, W. A. Prinz, G. Ashwell, M. W. Krause and J. A. Hanover, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 11952 CrossRef PubMed.
- M. Fischer, E. Fitzenberger, R. Kull, M. Boll and U. Wenzel, Genes Nutr., 2014, 9, 414 CrossRef PubMed.
- K. C. McCallum and D. A. Garsin, PLoS Pathog., 2016, 12, e1005923 CrossRef PubMed.
- I. V. Nechipurenko, A. Olivier-Mason, A. Kazatskaya, J. Kennedy, I. G. McLachlan, M. G. Heiman, O. E. Blacque and P. Sengupta, Dev. Cell, 2016, 38, 493 CrossRef PubMed.
- Y. Tian, Z. Li, W. Hu, H. Ren, E. Tian, Y. Zhao, Q. Lu, X. Huang, P. Yang, X. Li and X. Wang, J. Cell Sci., 2010, 141, 1042 Search PubMed.
- W. Kenneth, R. A. Chong, Q. Yu, J. Bai, D. E. Spratt, K. Ching, C. Lee, H. Miao, I. Tappin, J. Hurwitz and N. Zheng, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E2011 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra00279g |
| This journal is © The Royal Society of Chemistry 2018 |