Open Access Article
Open Access ArticleExploring the trans-membrane dynamic mechanisms of single polyamidoamine nano-drugs via a “force tracing” technique†
Siyuan Zhoua,
Boyu Yanga,
Yang Chena,
Qingrong Zhanga,
Mingjun Caib,
Haijiao Xub,
Guocheng Yanga,
Hongda Wang *bc and
Yuping Shan*a
*bc and
Yuping Shan*a
aSchool of Chemistry and Life Science, Advanced Institute of Materials Science, Changchun University of Technology, Changchun 130012, China. E-mail: shanyp@ciac.ac.cn
bChangchun Institute of Applied Chemistry, Chinese Academy of Science, Renmin St 5625, Changchun, Jilin 130022, China. E-mail: hdwang@ciac.ac.cn
cUniversity of Chinese Academy of Sciences, Beijing, 100049, China
First published on 27th February 2018
Abstract
Considerable technological success has been achieved in the drug delivery of nano-drugs for chemotherapy, but the main obstacles in understanding the drug delivery dynamic mechanisms for nano-drug applications stem from technical limitations. In this paper, we explored the trans-membrane dynamic processes of polyamidoamine nano-drugs via a “force tracing” technique.
Nanocarriers have been used as intelligent drug delivery systems (DDSs) to significantly improve the bioavailability of poorly soluble drugs and the efficacy of drugs for clinical treatment.1–6 Among the many nanocarriers, polyamidoamine (PAMAM) dendrimers undoubtedly show a superior performance, attracting researchers’ attention,7 since they have the advantages of non-immunogenicity and a high transport efficiency for carrying bioactive agents.8 In addition, as PAMAM is sensitive to pH, the drug can be released from the interior of the macromolecule via diffusion, so as to achieve a sustained release and enhance the drug efficacy. In contrast to other nanocarriers, PAMAM dendrimers are highly branched synthetic macromolecules obtained via iterative stepwise reaction sequences,9 and the highly branched structure ensures the facile modification of the dendrimer nanoparticle surface with different therapeutic drugs, diagnostic agents, and targeting ligands.10 The branched structure also makes it easy to conjugate with fluorescein isothiocyanate for imaging live cells and tracking the drug carriers.11 The internal molecular cavity of PAMAM provides the ability to physically encase small molecule drugs through electrostatic interactions and/or hydrogen bonding interactions.12
Usually, covalent conjugation and encapsulation are the main methods for loading drugs on PAMAM. E. W. Meijer and coworkers reported that guest molecules were captured within the internal cavities of the dendritic nanoparticles, and that the diffusion of guest molecules out of the carriers into solution was slow because of the close packing of the shell.13 In order to understand the effect of drug loading on the structural properties of the dendrimer, V. Jain et al. used molecular dynamic (MD) simulations to characterize the molecular models of dendrimer-nateglinide encapsulation complexes, and the MD analysis revealed that both the pH value of the solution and the terminal groups of the dendrimer play a part in drug encapsulation efficiency.14 Another effective approach used in drug loading is the covalent conjugation of the drug to the terminal functional groups of the dendrimers.12 Y. Gao et al. successfully conjugated ursolic acid (UA) and folic acid (FA) to PAMAM, and a subsequent cellular uptake study indicated that the presence of FA enhanced the uptake of dendrimeric prodrugs by HeLa cells with over-expressed folate receptors (FRs). This effect could be attributed to FR-mediated endocytosis.15 R. Mohammad et al. developed PAMAM-pullulan conjugates and investigated their targeting activity in delivering genes into liver cells. Furthermore, they assessed the cytotoxicity and transfection efficiency of the new derivatives.16 Although studies into dendrimers as drug and gene carriers have been widely performed, these bulk experiments lack the microenvironment to investigate the effects of heterogeneous living cell membranes. In order to improve chemotherapy with DDSs, it is very important to understand the trans-membrane mechanism of nano-drugs at the single nanoparticle level. A technique with high temporal–spatial resolution for tracking the trans-membrane dynamic process of DDSs at the single nanoparticle level is highly desirable.
The development of a AFM-based single molecule force spectroscopy (SMFS) technique with an atomic-level high spatial resolution (a Z-resolution of 0.1 nm and an XY-resolution of 0.01 nm) and a sensitivity of 10 pN has become a high priority. SMFS has been widely used in the study of biological fields under near physiological conditions.17–19 Wang’s group developed a “force tracing” technique by improving SMFS, which has been used to track the trans-membrane behaviour of single nanoparticles/molecules.20,21 By applying this technique, the force and duration of single nanoparticles/molecules during the trans-membrane dynamic process could be detected up to the piconewton and millisecond level limits. Here, “force tracing” has been applied to explore the trans-membrane dynamic process of PAMAM related nano-drugs.
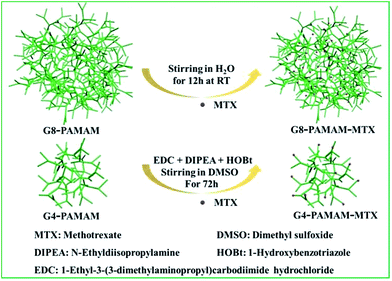
In this study, we selected the chemotherapy drug methotrexate (MTX) as the target drug for delivery, which is one of the drugs that have been delivered using dendrimers. MTX is an antimetabolite and a folate antagonist drug which is widely used in the treatment of various autoimmune diseases, ectopic pregnancy and the induction of medical abortions.22 The trans-membrane dynamic process of the PAMAM-MTX nano-drugs assembled via covalent conjugation and encapsulation were explored respectively. Generation 8 (G8) PAMAM was utilized as a carrier to encapsulate MTX, and generation 4 (G4) PAMAM was utilized as a receptor for the synthesis of the dendrimer conjugates. The two types of PAMAM-MTX nano-drug complex were prepared according to the previous reports.23–25 The schematic of the chemical conjugation and physical encapsulation of MTX with PAMAM is shown in Fig. 1; the cartoon model clearly shows the details of the nano-drugs, with the red balls representing MTX and the green structures representing the PAMAM dendrimer. After G8-PAMAM captured MTX, the MTX molecules were entrapped in the tunable hydrophobic interior cavities of the G8-PAMAM dendrimers, and the surface chemical structure of the newly formed G8-PAMAM-MTX nano-drug complex did not change, still consisting of the amino termini. The products were characterized using an ultraviolet spectrophotometer (ESI Fig. S1†). The lower part of Fig. 1 shows that the MTX molecules are conjugated to the terminal groups of the G4-PAMAM nanoparticles via the reaction of amino groups from the dendrimers with carboxyl groups from the MTX to form amide bonds. The G4-PAMAM-MTX products were characterized using H-NMR (details are shown in ESI Fig. S2 and S3†).
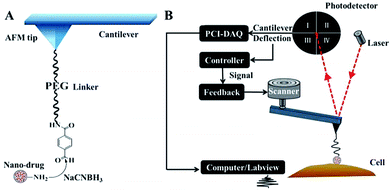
Because of its flat profile, which ensures that stable “force tracing” curves are obtained for the cell membranes, the African Green Monkey Kidney (Vero) cell line was selected as a model for exploring single nano-drug trans-membrane dynamics. In order to perform the single nanoparticle “force tracing” experiment, the PAMAM-MTX nano-drugs were attached to the AFM tip through heterobifunctional PEG linkers (aldehyde-PEG76-NHS), as shown in Fig. 2A. The NHS group was used to connect to the 3-aminopropyltriethoxysilane (APTES)-aminated tip, and the amine group of the PAMAM was bonded with the aldehyde group from the other side of the aldehyde-PEG76-NHS linker. The method of linking a nano-drug to the AFM tip for single molecule “force tracing” tests has been proven effectively.26,27 The main procedure of the “force tracing” based on AFM is shown in Fig. 2B; the deflection of the AFM tip cantilever was indicated by a laser beam detected via a photodetector. The PCI card records the cantilever deflection, and the LabVIEW software generates the “force tracing” curves according to the changes in deflection over time. During the process of the “force tracing” test, the force–distance curve was initially generated to find out the contact point, as the previous report28 (please see the ESI†), and then the PAMAM-MTX nano-drug attached to the AFM tip was moved to the contact point on the cell surface through the AFM feedback system, and while the feedback system was turned off the nano-drug was slightly in contact with the cell membranes. The AFM tip cantilever would bend downwards when the nano-drug was internalized by the cell, and the vertical changes of the cantilever were acquired using the PCI card controlled via LabVIEW.
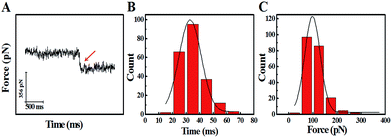
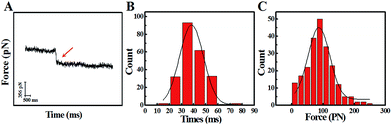
The study of the trans-membrane dynamic process via “force tracing” was first carried out on the G8-PAMAM-MTX nano-drug. Fig. 3A shows the typical “force tracing” curve: the force curves begin from left to right, the x-axis represents time and the y-axis represents the force generated by the deflection change of the AFM tip cantilever. When the G8-PAMAM-MTX attached to the AFM tip is just resting on the cell membrane, there is no apparent force signal and the force curve is level, as shown in the left part of the curve. Once the cellular uptake of the G8-PAMAM-MTX attached to the AFM tip starts occurring, the AFM cantilever suddenly bends downward, and the force signal appears (red arrow). When the transport activity of the G8-PAMAM-MTX is over, the “force tracing” curve recovers to flat, as indicated by the right side of the curve. By positioning the G8-PAMAM-MTX tethered AFM tip at different locations of different cells, thousands of “force tracing” curves were recorded. However, the transport events don’t always happen due to the early characteristics of cell transport activity,29 so the force signals are occasionally found in all the “force tracing” curves. The probability of the force signal (8.53 ± 0.47%) is calculated via random selection from thousands of “force tracing” curves that were collected from at least three independent experiments. As the key parameters of the trans-membrane dynamic process, the duration and force of transporting the G8-PAMAM-MTX nano-drug through the cell membranes can be measured directly from the “force tracing” curves. Fig. 3B shows that the duration of delivery of a single G8-PAMAM-MTX nano-drug molecule into living cells ranges from 11.30 to 68.10 ms, with an average value of 34.38 ± 10.49 ms. The corresponding force distribution is in the range of 45.42–350.87 pN, with an average value of 113.74 ± 45.48 pN, as shown in Fig. 3C.
The average transportation speed of G8-PAMAM-MTX is another key parameter and affects the dynamic trans-membrane mechanism. Therefore the displacement (D) of the G8-PAMAM-MTX during the rapid trans-membrane process needs to be known, and the D value consists of two parts: the bending distance of the cantilever (d) and the extension length of the PEG linker (x).
| D = d + x | (1) |
The force-extension curves of the PEG linker can be described by the extended worm-like chain (WLC) model, which characterizes the force (F)-dependent stretching activity. The equation is as follows:
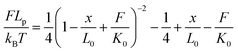 | (2) |
According to the literature,30 kB is the Boltzman constant and T is the temperature in Kelvin (kBT = 4.11 pN nm at 298 K). The persistence length is Lp = 3.8 ± 0.02 Å, the enthalpic correction is K0 = 1561 ± 33 pN, x is the extension of the PEG linker and L0 is the contour length. Given that the PEG unit is 4.2 Å and the terminus group length is 5.25 Å, the contour length L0 of the PEG (76–77 units) used was about 326 Å. The bending distance of the cantilever d could be calculated according to Hooke’s law:
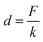 | (3) |
F is the force of transporting G8-PAMAM-MTX through the cell membrane, measured from the “force tracing” curves, and k is the spring constant of the AFM tip cantilever. According to eqn (1), (2), and (3), the displacement D was calculated as 24.70 nm. The average speed of the trans-membrane process could be calculated via D (24.70 nm)/duration (34.38 ms), which is 0.72 μm s−1. These results indicate that transporting a single nano-drug of G8-PAMAM-MTX through the cell membrane requires about 30 ms of time and 110 pN of force. The “force tracing” technique has actually made important contributions towards tracking the ultra-fast event of membrane transportation.
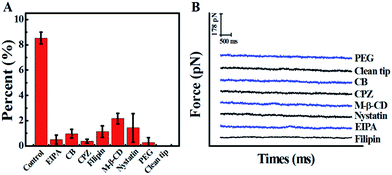
In view of the previous reports, the main method of cellular uptake for the PAMAM dendrimer nanoparticles is endocytosis,31 including clathrin-dependent endocytosis, caveolae-dependent endocytosis, and macropinocytosis pathways.32 Here, the pathway of PAMAM-MTX nano-drug endocytosis was examined using several specific drugs that hinder the different possible delivery mechanisms. Before the inhibition experiment, the cells needed to be pre-incubated with inhibitors for some time to bring the drug into effect. The cytoskeleton induces the internalization of the plasma membrane to participate in the process of endocytosis. Therefore, we destroyed the cytoskeleton to prevent endocytosis. The actin depolymerization drug CB (cytochalasin B, with an ultimate concentration of 2 μg mL−1), which can depolymerize actin and degrade the cytoskeleton,33 was incubated with cells for 20 min at 37 °C. After the actin fibers were depolymerized, the probability of observing a force signal in the “force tracing” curves was reduced to 0.97 ± 0.35%. The results show that actin contributes to the endocytosis of G8-PAMAM-MTX. CPZ (chlorpromazine), which hinders the assembly of grid-coated pits, can be used to evaluate the clathrin-mediated endocytosis pathway.34 After the cells were treated with CPZ (with a final concentration of 10 μg mL−1) for 30 min at 37 °C, the probability of observing a force signal was reduced to 0.37 ± 0.15%. Caveolae-dependent endocytosis requires the participation of cholesterol, therefore the cholesterol-binding drugs nystatin (with a final concentration of 0.03 mM), filipin (with a final concentration of 5 μg mL−1), and M-β-CD (methyl-β-cyclodextrin, 5 mM) were incubated with the Vero cells at 37 °C for 30 min, 30 min, and 10 min respectively. The probability of observing force signals was reduced to 1.4 ± 1.13%, 1.1 ± 0.45%, and 2.16 ± 0.42%, respectively.
Macropinocytosis, as one of the important endocytosis pathways, was examined using EIPA (amiloride, 5-(N-ethyl-N-isopropyl)).35,36 After incubation with EIPA (final concentration 60 μM) for 1 h at 37 °C, the probability of force signal is only 0.5 ± 0.36%. All the results are clearly shown in Fig. 4, which indicates that caveolin-dependent endocytosis, clathrin-dependent endocytosis, and macropinocytosis all participate in the trans-membrane transportation of the PAMAM-MTX nano-drug. For the control experiments, to exclude the effects of the AFM tip and the effects of molecules on the AFM tip, the “force tracing” test was performed using a PEG-modified tip and an unmodified tip, and almost no force signal was detected.
In order to study the trans-membrane dynamic process of the PAMAM related nano-drugs in depth, we also explored the trans-membrane transportation process of G4-PAMAM-MTX synthesized via conjugation. A typical force curve for G4-PAMAM-MTX is shown in Fig. 5A, and the process used is similar to that for the G8-PAMAM-MTX. The duration and force were measured using the “force tracing”curves; the duration ranges from 11.30 to 79.40 ms, with an average value of 39.19 ± 11.28 ms, and the corresponding force distribution is in the range of 20.08–244.35 pN, with an average value of 95.72 ± 42.10 pN, as shown in Fig. 5B and C. Using the same equations and method as for G8-PAMAM-MTX, the average speed during the trans-membrane process was calculated as 0.71 μm s−1. The probability (7.0 ± 1.85%) of observing a force signal is similar to that for G8-PAMAM-MTX. It is noted that the force value for transporting a single G4-PAMAM-MTX molecule is slightly smaller than that for G8-PAMAM-MTX (a box plot graph comparing the force values is shown in ESI Fig. S5†). The result may be affected by the dendrimer particle size (the diameter of G8-PAMAM is 9.7 nm and that of G4-PAMAM is 4.5 nm) and the conjugation of MTX molecules to the G4-PAMAM dendrimer terminals (enlarging the particle size). Aside from the force value difference, the duration for transporting a single G4-PAMAM-MTX nano-drug into the cell is a little bit longer than that for G8-PAMAM-MTX. This probably results from changes in the surface charge and the terminal group of the newly formed G4-PAMAM-MTX nano-drug, which leads to a longer required time for interaction with the cell membrane receptors, since the MTX drug molecule is modified to the surface of the PAMAM dendrimer.
Based on the high temporal–spatial resolution technique of “force tracing”, the dynamic trans-membrane process of a single PAMAM-MTX nano-drug obtained by either encapsulation (G8-PAMAM-MTX) or covalent conjugation (G4-PAMAM-MTX) was tracked in real-time. Compared to G4-PAMAM-MTX, the G8-PAMAM-MTX nano-drug needed a slightly larger force to accomplish trans-membrane transportation behaviour; however, the G4-PAMAM-MTX obtained by chemical conjugation needed a longer time due to the terminal group interaction with the cell surface. The further studies indicated that the PAMAM nano-drug delivery depended on caveolin-dependent endocytosis, clathrin-dependent endocytosis, and macropinocytosis. This technique provides a new way to screen nano-drugs and the loading of nano-drugs.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key R&D Program of China (No. 2017YFA0505300 to H. W.), National Natural Science Foundation of China (no. 31330082, 21773017 and 21673023 to Y. S., no. 21525314, 21703231 and 21721003 to H. W., and no. 21503213 to M. J.), and Jilin Provincial Science Research Foundation of China (no. 20160520133JH to Y. S. and no. 2017010119633JC to M. J.).Notes and references
- L. Albertazzi, M. Serresi, A. Albanese and F. Beltram, Mol. Pharm., 2010, 7, 680–688 CrossRef CAS PubMed.
- F. Assa, H. Jafarizadeh, H. Ajamein, H. Vaghari, N. Anarjan, O. Ahmadi and A. Berenjian, Crit. Rev. Biotechnol., 2017, 37, 492–509 CrossRef CAS PubMed.
- S. Y. Qin, A. Q. Zhang, S. X. Cheng, L. Rong and X. Z. Zhang, Biomaterials, 2017, 112, 234–247 CrossRef CAS PubMed.
- P. Silva, J. Bernegossi, M. Lima Gonçalez, M. Rillo Sato, C. Queico Fujimura Leite, F. Pavan and M. Chorilli, J. Biomed. Nanotechnol., 2016, 12, 241 CrossRef PubMed.
- Q. Wang, H. Cheng, H. Peng, H. Zhou, P. Y. Li and R. Langer, Adv. Drug Delivery Rev., 2015, 91, 125–140 CrossRef CAS PubMed.
- Y. Yuan and B. Liu, Chem. Sci., 2017, 8, 2537–2546 RSC.
- W. D. Tian and Y. Q. Ma, Chem. Soc. Rev., 2013, 42, 705–727 RSC.
- R. Esfand and D. A. Tomalia, Drug Discovery Today, 2001, 6, 427–436 CrossRef CAS PubMed.
- C. C. Lee, J. A. MacKay, J. M. J. Frechet and F. C. Szoka, Nat. Biotechnol., 2005, 23, 1517–1526 CrossRef CAS PubMed.
- A. Alvarez, C. Guzmán, S. Rivas, L. A. Godinez, A. Saccà, A. Carbone, E. Passalacqua, L. G. Arriaga and J. Ledesma-García, Int. J. Hydrogen Energy, 2014, 39, 16686–16693 CrossRef CAS.
- H. Kulhari, D. Pooja, S. Shrivastava, M. Kuncha, V. G. M. Naidu, V. Bansal, R. Sistla and D. J. Adams, Sci. Rep., 2016, 6, 23179 CrossRef CAS PubMed.
- J. Zhu and X. Shi, J. Mater. Chem. B, 2013, 1, 4199–4211 RSC.
- J. F. Jansen, D. B. D. B. Em and E. W. Meijer, Science, 1994, 266, 1226 CAS.
- V. Jain, P. Maiti and P. Bharatam, J. Chem. Phys., 2016, 145, 1211 CrossRef PubMed.
- Y. Gao, Z. Li, X. Xie, C. Wang, J. You, F. Mo, B. Jin, J. Chen, J. Shao, H. Chen and L. Jia, Eur. J. Pharm. Sci., 2015, 70, 55–63 CrossRef CAS PubMed.
- S. Askarian, K. Abnous, S. Ayatollahi, S. A. Farzad, R. K. Oskuee and M. Ramezani, Carbohydr. Polym., 2017, 157, 929–937 CrossRef CAS PubMed.
- Y. Shan, X. Hao, X. Shang, M. Cai, J. Jiang, Z. Tang and H. Wang, Chem. Commun., 2011, 47, 3377–3379 RSC.
- Y. Shan, S. Ma, L. Nie, X. Shang, X. Hao, Z. Tang and H. Wang, Chem. Commun., 2011, 47, 8091–8093 RSC.
- X. Shang, Y. Shan, Y. Pan, M. Cai, J. Jiang and H. Wang, Chem. Commun., 2013, 49, 8163–8165 RSC.
- Y. Pan, F. Zhang, L. Zhang, S. Liu, M. Cai, Y. Shan, X. Wang, H. Wang and H. Wang, Adv. Sci., 2017, 4, 1600489 CrossRef PubMed.
- Y. Pan, S. Wang, Y. Shan, D. Zhang, J. Gao, M. Zhang, S. Liu, M. Cai, H. Xu, G. Li, Q. Qin and H. Wang, Small, 2015, 11, 2782–2788 CrossRef CAS PubMed.
- J. F. Kukowska-Latallo, K. A. Candido, Z. Cao, S. S. Nigavekar, I. Majoros, T. Thomas, L. Balogh, M. K. Khan and J. Baker, Cancer Res., 2005, 65, 5317 CrossRef CAS PubMed.
- S. Natali and J. Mijovic, Macromolecules, 2010, 43, 3011–3017 CrossRef CAS.
- G. Pan, Y. Lemmouchi, E. O. Akala and O. Bakare, J. Bioact. Compat. Polym., 2005, 20, 113–128 CrossRef CAS.
- P. Singh, U. Gupta, A. Asthana and N. K. Jain, Bioconjugate Chem., 2008, 19, 2239–2252 CrossRef CAS PubMed.
- B. Ding, Y. Tian, Y. Pan, Y. Shan, M. Cai, H. Xu, Y. Sun and H. Wang, Nanoscale, 2015, 7, 7545–7549 RSC.
- Y. Pan, S. Wang, Y. Shan, D. Zhang, J. Gao, M. Zhang, S. Liu, M. Cai, H. Xu, G. Li, Q. Qin and H. Wang, Small, 2015, 11, 2782–2788 CrossRef CAS PubMed.
- B. Yang, H. Xu, S. Wang, M. Cai, Y. Shi, G. Yang, H. Wang and Y. Shan, Nanoscale, 2016, 8, 18027–18031 RSC.
- K. Welsher and H. Yang, Nat. Nanotechnol., 2014, 9, 198–203 CrossRef CAS PubMed.
- F. Kienberger, V. P. Pastushenko, G. Kada, H. J. Gruber, C. Riener, H. Schindler and P. Hinterdorfer, Single Mol., 2000, 1, 123–128 CrossRef CAS.
- L. Albertazzi, M. Serresi, A. Albanese and F. Beltram, Mol. Pharm., 2010, 7, 680–688 CrossRef CAS PubMed.
- S. Zhang, H. Gao and G. Bao, ACS Nano, 2015, 9, 8655–8671 CrossRef CAS PubMed.
- S. MacLean-Fletcher and T. D. Pollard, Cell, 1980, 20, 329–341 CrossRef CAS PubMed.
- H. Xu, X. Hao, S. Wang, Z. Wang, M. Cai, J. Jiang, Q. Qin, M. Zhang and H. Wang, Sci. Rep., 2015, 5, 11753 CrossRef PubMed.
- I. Nakase, M. Niwa, T. Takeuchi, K. Sonomura, N. Kawabata, Y. Koike, M. Takehashi, S. Tanaka, K. Ueda, J. Simpson, A. Jones, Y. Sugiura and S. Futaki, Mol. Ther., 2004, 10, 1011–1022 CrossRef CAS PubMed.
- J. Lim and P. A. Gleeson, Immunol. Cell Biol., 2011, 89, 836–843 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details of experiments and UV-Vis spectra (Fig. S1), 1H-NMR spectroscopy (Fig. S2), the chemical structure of PAMAM and MTX (Fig. S3), topography of AFM tip located on living cells (Fig. S4), and a box plot graph of endocytosis forces (Fig. S5). See DOI: 10.1039/c8ra00134k |
| This journal is © The Royal Society of Chemistry 2018 |