Open Access Article
Open Access ArticleEnhancing a clenbuterol immunosensor based on poly(3,4-ethylenedioxythiophene)/multi-walled carbon nanotube performance using response surface methodology†
Nurul Ain A. Talibab,
Faridah Salamc,
Nor Azah Yusofab,
Shahrul Ainliah Alang Ahmad ab,
Mohd Zulkhairi Azidd,
Razali Miradd and
Yusran Sulaiman
ab,
Mohd Zulkhairi Azidd,
Razali Miradd and
Yusran Sulaiman *ab
*ab
aFunctional Devices Laboratory, Institute of Advance Technology, Universiti Putra Malaysia, 43400 Serdang, Selangor, Malaysia
bDepartment of Chemistry, Faculty of Science, Universiti Putra Malaysia, 43400 Serdang, Selangor, Malaysia. E-mail: yusran@upm.edu.my; Fax: +60-389435380; Tel: +60-389466779
cBiodiagnostic-Biosensor Programme, Biotechnology and Nanotechnology Research Centre, Malaysian Agricultural Research and Development Institute, 43400 Serdang, Selangor, Malaysia
dUtilization of Agrobiodiversity Resource Programme, Agrobiodiversity and Environmental Research Centre, Malaysian Agricultural Research and Development Institute, 43400 Serdang, Selangor, Malaysia
First published on 24th April 2018
Abstract
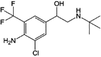
Clenbuterol (CLB) is an illegal antibiotic for livestock, which is misused as a growth promoter drug. In this study, an immunosensor modified with poly(3,4-ethylenedioxythiophene) (PEDOT), multi-walled carbon nanotubes (MWCNT) and anti-clenbuterol antibody (Ab) was developed for the detection of CLB. A screen-printed carbon electrode (SPCE) was modified with PEDOT/MWCNT as a sensor platform before immobilizing Ab for specific CLB binding through a competitive-type immunoassay. Free CLB in the sample solution competed with clenbuterol-horseradish peroxide (CLB–HRP) to bind with Ab. A high current signal was obtained after optimization of the electrochemical immunoassay conditions (pH, incubation temperature, antigen (Ag) incubation time and % blocking) using the response surface methodology/central composite design (RSM/CCD). The developed immunosensor is highly reproducible and sensitive with good storage stability, which are necessary for practical application. In real sample application, this immunosensor produces comparable results with liquid chromatography-mass spectrometry; thus, it is useful for CLB screening and monitoring in real meat samples.
1. Introduction
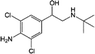
Clenbuterol (CLB) is one of the beta-2-agonist antibiotics that was initially used for treating respiratory problems.1 However, due to its ability to enhance muscle growth,2 this drug has been misused in poultry to produce animals with leaner meat. CLB also has a long half-life, long withdrawal period, slow metabolism in animal bodies, and is stable and will only decompose at high temperature (over 172 °C). Thus, CLB residue can still be detected from the liver of CLB-medicated animals after 56 days, while the effect of CLB as a growth promoter can show up to 70 days after withdrawal.3 The use of CLB in livestock is forbidden by the World Health Organization (WHO) due to its health effects such as muscular tremors, heart throbbing, glaucoma, fever and respiratory problems in human, which are influenced by CLB residue in meat products.4 Therefore, the use of this antibiotic in meat products requires continuous monitoring to avoid pharmacological effects to consumers.In previous studies, a few methods were developed to detect and monitor antibiotics including CLB such as microbiology assay, enzyme-linked immunosorbent assay (ELISA),5 high performance liquid chromatography (HPLC),6,7 liquid chromatography-mass spectrometry (LC-MS),8 gas chromatography-mass spectrometry (GC-MS),9 capillary electrophoresis,10,11 surface plasma resonance12 and electrochemical13 methods. However, even though analytical methods such as HPLC, LC-MS and GS-MS are very sensitive and effective, these methods are relatively expensive, time-consuming and unsuitable for on-site monitoring due to the use of large instruments.
Electrochemical sensors can be developed into various intradisciplinary applications such as food monitoring,14 pharmaceutical15 and pesticide detection,16,17 wastewater screening,18,19 chemical sensors20 and biosensors21 based on the modification of the sensor platform. Many methods have been developed by utilizing various materials such as molecular imprinted polymers,22,23 antibodies (Ab)24,25 and enzymes26,27 to increase the sensitivity, selectivity and effectiveness of electrochemical detection.
A biosensor that utilizes Ab to capture antigen (Ag) is known as an immunosensor.28 Immunosensors based on modified electrode surfaces are fabricated using various materials to enhance their electrochemical properties, which improves the electrode charge transfer, produces a better signal and increases the sensitivity. Carbon nanotubes (CNTs) have good conductivity29 and provide many binding sites for Ab binding due to their high surface area, and thus are suitable for the development immunosensors.30,31 The electrochemical properties of CNT can be enhanced by incorporating conducting polymers such as polyaniline,32 polypyrrole,33 and poly(3,4-ethylenedioxythiophene) (PEDOT).34 Their biocompatibility and ability to improve the efficiency of bio-recognition events have been reported as the advantages of using conducting polymers in the design of immunosensors.35 The fabrication of biosensors utilizing PEDOT as one of the materials successfully produced an amperometric glucose sensor.36 PEDOT has high stability in phosphate buffer solution37 which is very useful for immunoassay and it is also compatible for doping with nanoparticles. A previous study showed that the addition of multi-walled carbon nanotubes (MWCNT) as a dopant into PEDOT improved the electrochemical properties of the composite.38
Recently, electrochemical sensors for CLB monitoring have been developed widely since this method is suitable for on-site monitoring. In addition, the detection process can be performed by semi-skilled personnel due to its relatively simple handling process, and it is relatively less expensive39 in comparison with other analytical detection methods. CLB is a hapten, which is a very small molecule that has little effect on electron transfer. Herein, a CLB immunosensor is developed by fabricating PEDOT/MWCNT/Ab on a screen-printed carbon electrode (SPCE). SPCEs are reported to be more practical for on-site monitoring.40 Additionally, they are inexpensive,41 easily modified and reproducible; hence, they are suitable for mass production. By using SPCE, only small amount of sample and electrolyte are needed for each detection process.
Generally, optimization of the immunoassay can enhance the sensitivity of the sensor. One parameter with the one-time approach is usually applied for this purpose, but this method just describes each parameter individually and is unable to evaluate the interaction between parameters. Therefore, a statistical technique such as the response surface methodology (RSM) can be used instead since it indicates the significance of the studied parameters and developed model for better understanding the optimization process. Currently, limited reports are available on RSM for optimization in biosensors. Mirmoghtadaie and co-workers42 reported the successful optimization of a DNA electrochemical sensor using RSM for parameters such as pH, concentration of DNA probe and deposition potential. Optimization of the effective parameters for the fabrication of modified electrodes and the immobilization of DNA probes (immobilization time and concentration of DNA) using RSM has enhanced the sensitivity of DNA detection.43 The reliability of RSM as an optimization tool was also reported in other literature.44–46
In this work, an electrochemical sensor is designed by utilizing PEDOT, MWCNT and polyclonal anti-clenbuterol antibody (Ab) to produce a high-performance CLB immunosensor. A screen-printed electrode (SPCE) is modified with PEDOT/MWCNT composites through the electrodeposition technique. Ab is immobilized on the modified surface by exploiting the carboxyl group on the MWCNT walls through the EDC/NHSS protocol, while the direct competitive immunoassay format is adapted for the detection of CLB. This work also focuses on the optimization of the immunoassay conditions (pH, incubation temperature, Ag incubation time and % blocking) using the response surface methodology/central composite design (RSM/CCD) approach for the enhancement of the electrochemical detection of CLB. To the best of our knowledge, the optimization of the immunoassay conditions for PEDOT/MWCNT/SPCE using the RSM/CCD approach has not been previously reported. The immunosensing strategy reported here presents a significant electrochemical performance improvement of the sensor platform due to its modification with PEDOT/MWCNT composites. Also, the application of this immunosensor for CLB detection in real samples is validated with LC-MS.
2. Experimental
2.1 Materials and methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio in 0.01 M PBS at pH 7.4. Clenbuterol–horseradish peroxide (CLB–HRP) was diluted in 0.01 M PBS with a 1
1 ratio in 0.01 M PBS at pH 7.4. Clenbuterol–horseradish peroxide (CLB–HRP) was diluted in 0.01 M PBS with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 640 volume ratio, as recommended by the manufacturer (Fitzgerald). The washing buffer was prepared by mixing 0.05% TWEEN 20 (Sigma Aldrich) with 0.01 M PBS at pH 7.4. Dry milk (Blotto, non-fat Santa Cruz Biotechnology) was used as a blocking agent by diluting in 0.01 M PBS at pH 7.4.
640 volume ratio, as recommended by the manufacturer (Fitzgerald). The washing buffer was prepared by mixing 0.05% TWEEN 20 (Sigma Aldrich) with 0.01 M PBS at pH 7.4. Dry milk (Blotto, non-fat Santa Cruz Biotechnology) was used as a blocking agent by diluting in 0.01 M PBS at pH 7.4.2.2 Preparation of clenbuterol immunosensor
MWCNT were treated with an acid mixture of concentrated sulphuric acid (H2SO4) and concentrated nitric acid (HNO3) in a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. The treated MWCNT were further washed with deionized water until pH 7 was obtained, followed by drying in an oven at 60 °C. Before surface modification, the SPCE underwent pre-treatment by cyclic voltammetry (CV) at −1.5 to 0 V for 3 cycles in 1.0 M H2SO4 solution. The PEDOT/MWCNT modified SPCE was prepared by electropolymerization in 0.01 M EDOT in the presence of 0.1 mg mL−1 MWCNT suspension at 1.31 V for 99.1 s38 via chronoamperometry (CA). All electrochemical experiments were performed using a potentiostat (Autolab PGSTATM101) at a scan rate of 0.1 V s−1.
1 ratio. The treated MWCNT were further washed with deionized water until pH 7 was obtained, followed by drying in an oven at 60 °C. Before surface modification, the SPCE underwent pre-treatment by cyclic voltammetry (CV) at −1.5 to 0 V for 3 cycles in 1.0 M H2SO4 solution. The PEDOT/MWCNT modified SPCE was prepared by electropolymerization in 0.01 M EDOT in the presence of 0.1 mg mL−1 MWCNT suspension at 1.31 V for 99.1 s38 via chronoamperometry (CA). All electrochemical experiments were performed using a potentiostat (Autolab PGSTATM101) at a scan rate of 0.1 V s−1.
The fabrication of the CLB immunosensor began with the activation of the carboxyl functional groups in the PEDOT/MWCNT modified electrode surface by dropping 10 μL of crosslinker solution (EDC/NHSS) for 15 min at ambient temperature followed by removal of the unbound crosslinker with washing buffer. The PEDOT/MWCNT/Ab modified electrode was prepared via the incubation of 10 μL of Ab solution on the PEDOT/MWCNT electrode surface for 1 h at 37 °C followed by rinsing with washing buffer.
The detection of CLB commenced by performing a blocking step which included incubation with 20 μL of dry milk for 30 min at 37 °C followed by rinsing with washing buffer. The competitive method was applied by adding 10 μL of CLB standard (0 to 250 ng mL−1) or sample solution and 10 μL of CLB–HRP solution onto the immunosensor during incubation, followed by rinsing with washing buffer. 50 μL of tetramethylbenzidine (TMB) was dropped onto the electrode before the chronoamperogram was recorded using a potentiostat for 300 s. The standard calibration graph was fitted with linear regression and the limit of detection (LOD) was determined.
2.3 Determination of potential applied
Different applied potentials in the range of −0.6 to 0.6 V were used for the detection of various concentrations of standard CLB solutions (0 to 100 ng mL−1) using CA for 100 s. The same preparation procedure as that for clenbuterol immunosensor was applied here.2.4 Optimization of the electrochemical immunoassay condition
Optimization of the Ab concentration was conducted by comparing the standard calibration curves at various concentrations of Ab (10.0, 1.0 and 0.1 mg mL−1). Meanwhile, the other electrochemical immunoassay parameters such as pH (A), incubation temperature (B), Ag incubation time (C) and % blocking (D) were determined by applying RSM/CCD. The study range of each parameter was decided based on previous studies, which were pH 6.4 to 8.4,47 28 °C to 46 °C incubation temperature,13 10 to 70 min Ag incubation time48 and 0 to 0.10% blocking. The experiment was designed based on these ranges and the output of this model was the current response.A total of 26 experiment runs (Table S1†) were suggested including 16 factorial points, 8 axial points and 2 center points using the statistical package (Design Expert 6.0, Stat Ease Inc., MN, USA). A mathematical model was generated based on the current response data from the performed experiments to fit the general quadratic model equation (eqn (1)):
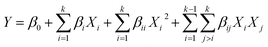 | (1) |
| Y = β0 + β1X1 + β2X2 + β3X3 + β4X4 + β11X12 + β22X22 + β33X32 + β44X42 + β12X1X2 + β13X1X3 + β14X1X4 + β23X2X3 + β24X2X4 + β34X3X4 | (2) |
The significance of the developed model was evaluated based on prob < F value and F-value analyzed by the analysis of variance (ANOVA), while the quality of the model was determined based on regression (R2), adjusted R2, predicted R2, lack of fit and adequate precision.
2.5 Characterization
The optimized immunosensor was characterized in a mixture solution of 5 mM K3[Fe(CN)6], 5 mM K4[Fe(CN)6], and 0.1 M KCl via CV (0.2 V to 0.6 V potential range) and electrochemical impedance spectroscopy (EIS) within the frequency range of 1 Hz to 10 MHz using a sinusoidal current of 0.005 V amplitude at open circuit potential to evaluate its electrochemical behavior. Field emission scanning electron microscopy (FESEM, JEOL JSM-7600F) was performed to study its surface morphology. Confirmation of the Ab immobilization was determined by performing a bicinchoninic acid (BCA) protein assay.49 100 μL of the substrate containing reagent A and reagent B in a 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio was added on each electrode surface for each fabrication stage. The electrodes were incubated for 30 min at 37 °C before the solutions were transferred to a 96-well plate followed by absorbance reading at 560 nm using a microplate reader (VersaMax, Molecular Devices). The protein standard curve was obtained based on bovine serum albumin (BSA) protein. A stock solution of BSA (5 mg mL−1) was prepared in PBS and diluted to a series of standard solutions (0 to 4.0 mg mL−1). 10 μL per well of each solution was inserted into the 96-well plate followed by 100 μL of the substrate. The procedure was continued with incubation for 30 min at 37 °C followed by absorbance reading at 560 nm.
1 ratio was added on each electrode surface for each fabrication stage. The electrodes were incubated for 30 min at 37 °C before the solutions were transferred to a 96-well plate followed by absorbance reading at 560 nm using a microplate reader (VersaMax, Molecular Devices). The protein standard curve was obtained based on bovine serum albumin (BSA) protein. A stock solution of BSA (5 mg mL−1) was prepared in PBS and diluted to a series of standard solutions (0 to 4.0 mg mL−1). 10 μL per well of each solution was inserted into the 96-well plate followed by 100 μL of the substrate. The procedure was continued with incubation for 30 min at 37 °C followed by absorbance reading at 560 nm.
2.6 Sample preparation for analysis of clenbuterol in meat samples
Fresh beef samples were minced using a homogenizer and weighed (0.1 g) into centrifugal tubes. The samples were spiked with 1 mL of CLB standard (50 and 100 ng mL−1) followed by the addition of 19 mL acetonitrile (ACN) and DI water. The samples were shaken using a shaker for 5 min before they were centrifuged at 3900 rpm for 5 min at 4 °C. The supernatants were transferred into centrifuge tubes containing 0.5 g LiChroprep® RP-18 (25–40 μm, Merck), followed by the addition of 10 mL saturated hexane (saturated with ACN). The samples were shaken for another 1 min before centrifugation at 3900 rpm for 5 min at 4 °C. The upper layers containing hexane were removed. Only 5.0 mL of the aliquots was transferred into centrifuge tubes and evaporated to 1.0 mL. The extracts were filtered with a 0.45-micron nylon membrane syringe filter into vials for analysis via LCMS. Meanwhile, for analysis with the PEDOT/MWCNT/Ab/SPCE immunosensors, the extracts were evaporated until they were fully dried before dissolving them in buffer solution.3. Results and discussion
3.1 Immunosensing strategy
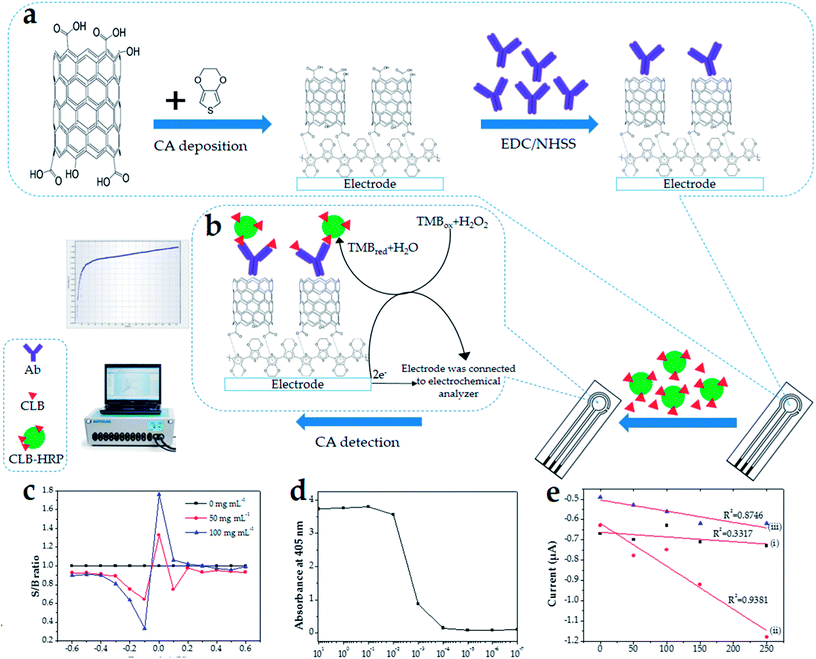
In this study, SPCE underwent modification with PEDOT/MWCNT before being further developed into an immunosensor (Fig. 1a). Our strategy was to directly electrodeposit the PEDOT/MWCNT composite instead of using the common covalent attachment of CNT to the electrode surface.50 To avoid entrapment of Ab into the polymer composite,51 only PEDOT and MWCNT were electrodeposited on the electrode. The Ab was covalently linked to the functionalized MWCNT in the next steps using the EDC/NHSS protocol.30 The MWCNT used for this modification contains abundant hydroxyl and carboxyl groups due to the functionalization with the acid mixture. The existence of carboxyl groups allows Ab to bind to the modified electrode surface through the EDC/NHSS crosslinker. The high surface area of MWCNT provide numerous binding sites for Ab to bind to the electrode surface, thus increasing the sensitivity of this immunosensor. Dry milk was used as a blocking agent to avoid unspecific binding on the PEDOT/MWCNT/Ab/SPCE surface. The blocking agent covered the electrode area that was not bound with Ab to avoid background noise from interrupting the current signal or giving any false signal due to unbound protein. Therefore, a washing step was included after each immobilization process to remove the unbound protein.The CLB immunosensor developed in this study is based on the direct competitive detection method (Fig. 1b). Generally, CLB as a hapten has a very little effect on electron transfer. Label enzymes such as HRP are commonly used in immunoassay studies since their enzyme activity can be determined by fluorescence and electrochemical methods. To overcome the abovementioned problem, CLB conjugated with HRP (CLB–HRP) was utilized to amplify the effect of electron transfer, which produces a measurable signal. In this study, the signal in the form of current value was measured based on the reduction of TMB catalyzed by HRP. The reduction of TMB is due to the catalyzation of CLB–HRP on the modified electrode surface resulting in a measurable electrochemical current signal. Sine CLB–HRP was competing with free CLB in the sample solution, the signal current produced was variable depending on the amount of CLB–HRP successfully bound to Ab on the modified electrode surface. As the concentration of free CLB in the sample increased, fewer CLB–HRP managed to bind with Ab immobilized on the electrode surface, and vice versa.
3.2 Determination of set potential
Since this study utilized CA to generate a signal for the detection of CLB, the optimum applied potential was investigated. The signal current over background current (S/B) ratio using step amperometry from −0.6 to 0.6 V for 0 to 100 ng mL−1 CLB concentration was calculated and the graph was plotted (Fig. 1c) to observe the pattern of the current signal. Even though various concentrations of CLB produced different S/B ratios, the highest values were obtained at the same applied potential. These results show that −0.1 V is the best potential for this system due to its largest S/B ratio among the potentials measured at different CLB concentrations. Thus, −0.1 V was chosen as the applied potential for further measurements.3.3 Optimization of the immunoassay conditions
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 based on Fig. 1d, whereby this high Ab dilution indicates the high Ab activity towards the target (CLB).53 Three concentrations of Ab (0.01, 0.1 and 1.0 mg mL−1) were shortlisted based on the result of the ELISA titer (Fig. 1d). The standard CLB calibration curve for each Ab concentration was plotted (Fig. 1e) and the efficiency of the immunosensor was evaluated based on the R2 value of the calibration curve. An ideal calibration curve has an R2 value closer to 1.000. Based on Fig. 1e, the immunosensor developed using 0.1 mg mL−1 Ab has a better R2 (0.9381) than 0.01 mg mL−1 Ab (R2 = 0.8746). As the concentration of Ab increases, there are more chances for the formation of Ab–Ag binding, thus increasing the immunosensor efficiency. However, when the concentration of Ab was increased to 1.0 mg mL−1, the R2 significantly decreased to 0.3317. This phenomenon occurred because saturation of the binding sites was achieved. A further increase in the Ab concentration to more than 0.1 mg mL−1 does not increase the immunosensor efficiency, thus the R2 does not improve. Consequently, the concentration of 0.1 mg mL−1 Ab was chosen as the optimum Ab concentration.
000 based on Fig. 1d, whereby this high Ab dilution indicates the high Ab activity towards the target (CLB).53 Three concentrations of Ab (0.01, 0.1 and 1.0 mg mL−1) were shortlisted based on the result of the ELISA titer (Fig. 1d). The standard CLB calibration curve for each Ab concentration was plotted (Fig. 1e) and the efficiency of the immunosensor was evaluated based on the R2 value of the calibration curve. An ideal calibration curve has an R2 value closer to 1.000. Based on Fig. 1e, the immunosensor developed using 0.1 mg mL−1 Ab has a better R2 (0.9381) than 0.01 mg mL−1 Ab (R2 = 0.8746). As the concentration of Ab increases, there are more chances for the formation of Ab–Ag binding, thus increasing the immunosensor efficiency. However, when the concentration of Ab was increased to 1.0 mg mL−1, the R2 significantly decreased to 0.3317. This phenomenon occurred because saturation of the binding sites was achieved. A further increase in the Ab concentration to more than 0.1 mg mL−1 does not increase the immunosensor efficiency, thus the R2 does not improve. Consequently, the concentration of 0.1 mg mL−1 Ab was chosen as the optimum Ab concentration.3.4 Optimization of pH, incubation temperature, Ag incubation time and % blocking
| Current = 0.146 + 0.002A + 0.052B − 0.052C − 0.019D − 0.037A2 + 0.059B2 − 0.146C2 − 0.039D2 + 0.012AB − 0.020AC − 0.004AD − 0.007BC − 0.003BD + 0.008CD | (3) |
Four independent variables, i.e. pH, incubation time, Ag incubation time and % blocking, are represented by coded values (X1 = A, X2 = B, X3 = C, and X4 = D, respectively). In this study, B, C, D, B2, C2 and AC were indicated as significant model terms due to their prob > F values of less than 0.0500. Factor pH (model term A) was identified as an insignificant term, indicating the pH range of 6.40 to 8.40 does not affect the current response.
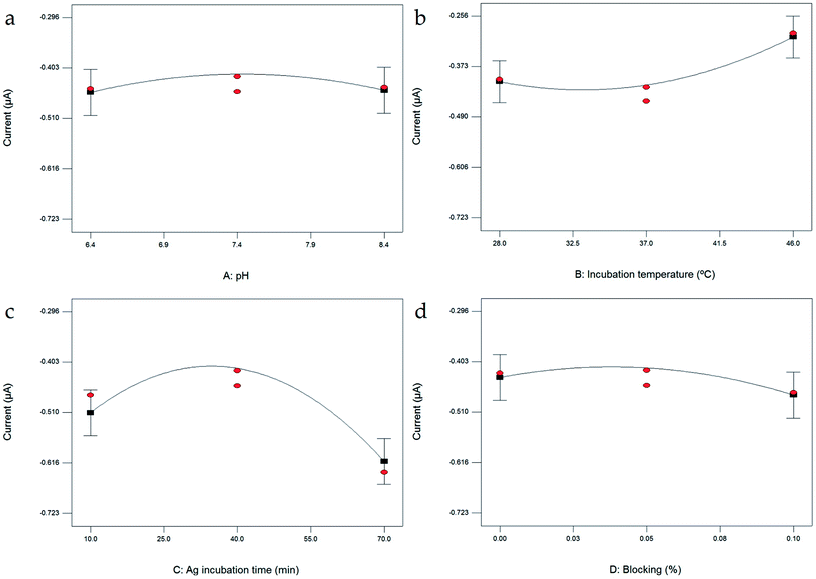 | ||
| Fig. 2 One factor plots of peak current response as a function of (a) pH; (b) incubation temperature; (c) Ag incubation time; and (d) blocking. | ||
In this study, incubation temperature was evaluated from 28 °C to 46 °C (Fig. 2b). The reactivity of Ab and Ag is strongly affected by temperature. A very small current change occurred as the incubation temperature increased from 28 °C to 37 °C, while the current obviously increased as the temperature increased to 46 °C. The highest current reading observed at 46 °C shows that the Ab–Ag interaction formed the best at this temperature. The formation of Ab–Ag bonding also depends on the incubation time; thus, the incubation time of 10 to 70 min. The current increases as the incubation time increases from 10 to 40 min (Fig. 2c), at which point, the highest current response was observed. However, the current reading decreases as the incubation time increases to 70 min, showing that an incubation time longer than 40 min is not effective.
To avoid unspecific binding of any unwanted molecules, dry milk was introduced as a blocking agent. The percentage of blocking was evaluated in the range of 0% to 0.10%. An increase in current was observed as the blocking was increased to 0.05% (Fig. 2d). As the blocking was further increased to 0.10%, the current response decreased. An increase in blocking agent of more than 0.05% not only blocked the empty electrode surface but also caused current interruption. Thus, blocking agent of more than 0.05% is not necessary.
One of the advantages of RSM is its specialty to allow the interaction between factors to be studied. The interaction between factors is presented in Fig. S1.† Individually, the evaluation of pH shows no significant terms based on the graph (Fig. 2a) and statistical analysis (Table S2†). However, based on ANOVA (Table S2†), as the interaction between factors was evaluated in the term AC (representing the interaction between pH and Ag incubation time), the prob > F value is 0.0233. The pH value was affecting the Ag incubation time significantly. Thus, the study of the pH factor cannot be ignored even though individually this factor is considered as not significant.
| Parameter | Fabrication of PEDOT/MWCNT/Ab/SPCE | |
|---|---|---|
| Original | Optimized | |
| pH | 7.4 | 7.78 |
| Ab incubation temperature (°C) | 37 | 46 |
| Ag incubation time (min) | 60 | 33 |
| Blocking agent (%) | 0.05 | 0.03 |
| R2 | 0.9381 | 0.9921 |
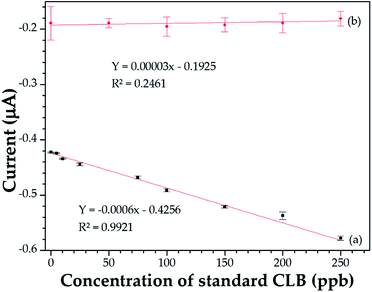 | ||
| Fig. 3 Standard CLB calibration curve after optimization with RSM. (a) Immunosensor modified with PEDOT/MWCNT. (b) Sensor without MWCNT. | ||
| Technique | Detection limit | Linear range | Reference |
|---|---|---|---|
| a NA: not available. | |||
| GC-MS | 2 ng mL−1 | 0.06–8.0 ng mL−1 | 58 |
| Surface-enhanced Raman spectroscopy (SERS) | 0.5 ng mL−1 | 0.5–20 ng mL−1 | 59 |
| Surface-enhanced Raman spectroscopy (SERS) | NA | 1–1000 pg mL−1 | 60 |
| Surface plasmon resonance | 1.26 ng mL−1 | NA | 61 |
| Quartz crystal microbalance sensor | 3.0 ng mL−1 | NA | 55 |
| Fluorometry/FRET | 3.96 ng mL−1 | 200–1800 ng mL−1 | 62 |
| Electrochemical immunosensor | 4.66 ng mL−1 | 0–250 ng mL−1 | This work |
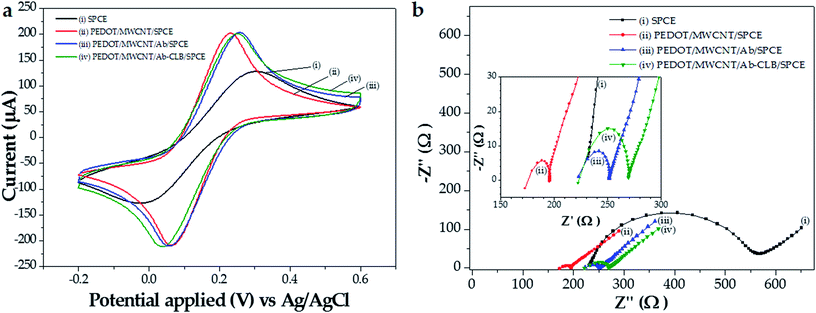
3.5 Characterization
3.6 Analytical performance of the immunosensor
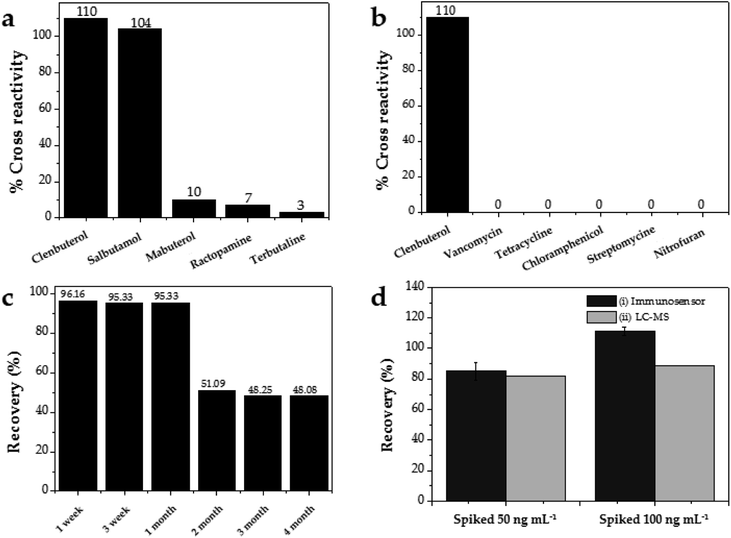
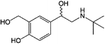
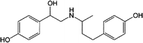
The specificity of this CLB immunosensor was evaluated by detected other antibiotics from the β-agonist group, such as salbutamol, mabuterol, ractopamine and terbutaline, along with other antibiotics from other groups such as vancomycin, tetracycline, chloramphenicol, streptomycin and nitrofuran. The current response of each antibiotic with the same concentration of 100 ng mL−1 was investigated. The inhibition percentages were calculated as 110%, 104%, 10%, 7% and 3% for CLB, salbutamol, mabuterol, ractopamine and terbutaline, respectively (n = 3) (Fig. 6a).Other antibiotics from other families such as vancomycin, tetracycline, chloramphenicol, streptomycin and nitrofuran (n = 3) show 0% cross reactivity (Fig. 6b). Other β-agonists and antibiotics do not affect the CLB detection except salbutamol, which implies that this immunosensor has good selectivity towards CLB. Salbutamol shows a high% cross reactivity because its structure is closer to CLB than other antibiotics (Table 3).
The reproducibility of this immunosensor was determined for n = 10 under the same conditions, resulting in the standard deviation (SD) value of 0.110, indicating excellent reproducibility. The coefficient of variation of this reproducibility test is 1.33%. The coefficient of variation is defined as the ratio of the standard deviation to mean [(SD/mean)100%], where a value a less than 5%, as reported in this study, implies excellent reproducibility. The immunosensor was also stored at 4 °C for one month to study its storage stability. After 1 month of storage, 95.33% (Fig. 6c) of the initial current response was obtained, indicating relatively good storage stability.
3.7 Comparative study
Real sample analyses were performed to detect CLB in 10 beef samples using the developed CLB immunosensor. The same samples were also detected using LC-MS for comparison. Since no CLB was detected, standard solutions of CLB were spiked in the samples to evaluate the applicability of the developed immunosensor. As shown in Fig. 6d, good recoveries with 85% to 111% recovery obtained for both the 50 and 100 ng mL−1 spiked samples, respectively, as detected by the immunosensor. Comparable results were obtained from this immunosensor with that from LC-MS, which proves the reliability of this immunosensor for CLB detection in real samples.4. Conclusions
A PEDOT/MWCNT/Ab/SPCE immunosensor was successfully developed for the detection of CLB. The optimum conditions for the electrochemical immunoassay were determined using RSM/CCD. Incubation time, Ag incubation time and % blocking were determined as significant parameters along with the interaction between pH and Ag incubation time. A linear graph of CLB standard with R2 = 0.9921 was obtained under the optimized electrochemical immunoassay conditions. Modification of SPCE with the PEDOT/MWCNT composite improved the electrochemical properties of the electrode and provided a high surface area for Ab–CLB binding. Consequently, high reproducibility, storage stability and selectivity towards CLB was produced. This method was successfully employed for CLB detection in real meat samples, which produced comparable results with LC-MS, and thus is reliable for CLB screening and monitoring in real applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank Fundamental Research Grant Scheme (01-01-15–1707FR) and Malaysian Agricultural Research and Development Institute (MARDI) for funding this research project.References
- L. A. Cudmore, T. Muurlink, T. Whittem and S. R. Bailey, Res. Vet. Sci., 2013, 94, 682–686 CrossRef CAS PubMed.
- D. S. Huckins and M. F. Lemons, J. Emerg. Med, 2013, 44, 444–449 CrossRef PubMed.
- C. T. Elliott, S. R. H. Crooks, J. G. D. McEvoy, W. J. McCaughey, S. A. Hewitt, D. Patterson and D. Kilpatrick, Vet. Res. Commun., 1993, 17, 459–468 CrossRef CAS PubMed.
- G. Brambilla, T. Cenci, F. Franconi, R. Galarini, A. Macrì, F. Rondoni, M. Strozzi and A. Loizzo, Toxicol. Lett., 2000, 114, 47–53 CrossRef CAS PubMed.
- A. Posyniak, J. Zmudzki and J. Niedzielska, Anal. Chim. Acta, 2003, 483, 61–67 CrossRef CAS.
- B. Liu, H. Yan, F. Qiao and Y. Geng, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2011, 879, 90–94 CrossRef CAS PubMed.
- K. Yan, H. Zhang, W. Hui, H. Zhu, X. Li, F. Zhong, X. e. Tong and C. Chen, J. Food Drug Anal., 2016, 24, 277–283 CrossRef CAS PubMed.
- R. Nicoli, M. Petrou, F. Badoud, J. Dvorak, M. Saugy and N. Baume, J. Chromatogr. A, 2013, 1292, 142–150 CrossRef CAS PubMed.
- L. Amendola, C. Colamonici, F. Rossi and F. Botrè, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2002, 773, 7–16 CrossRef CAS.
- J. Zhou, X. Xu and Y. Wang, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2007, 848, 226–231 CrossRef CAS PubMed.
- H. Wang, H. Yan, M. Shi, F. Wang, K. Mei, Q. Yu, X. Lv and X. Wang, Anim. Feed Sci. Technol., 2015, 209, 257–267 CrossRef CAS.
- S. Suherman, K. Morita and T. Kawaguchi, Appl. Surf. Sci., 2015, 332, 229–236 CrossRef.
- P. Zhan, X.-W. Du, N. Gan, S.-C. Lin, T.-H. Li, Y.-T. Cao and W.-G. Sang, Anim. Feed Sci. Technol., 2013, 41, 828–834 CAS.
- M. Sheikhshoaie, I. Sheikhshoaie and M. Ranjbar, J. Mol. Liq., 2017, 231, 597–601 CrossRef CAS.
- N. K. Bhajanthri, V. K. Arumugam, R. Chokkareddy and G. G. Redhi, J. Mol. Liq., 2016, 222, 370–376 CrossRef CAS.
- X.-P. Wei, Y.-L. Luo, F. Xu, Y.-S. Chen and L.-H. Yang, Mater. Des., 2016, 111, 445–452 CrossRef CAS.
- P. B. Deroco, B. C. Lourencao and O. Fatibello-Filho, Microchem. J., 2017, 133, 188–194 CrossRef CAS.
- P. Devi, G. Kaur, A. Thakur, N. Kaur, A. Grewal and P. Kumar, Talanta, 2017, 170, 49–55 CrossRef CAS PubMed.
- A. Ferancová, M. K. Hattuniemi, A. M. Sesay, J. P. Räty and V. T. Virtanen, J. Hazard. Mater., 2016, 306, 50–57 CrossRef PubMed.
- Y. Zhou, L. Yang, S. Li and Y. Dang, Sens. Actuators, B, 2017, 245, 238–246 CrossRef CAS.
- R. Sha, K. Komori and S. Badhulika, Electrochim. Acta, 2017, 233, 44–51 CrossRef CAS.
- M. Roushani, A. Nezhadali, Z. Jalilian and A. Azadbakht, Mater. Sci. Eng., C, 2017, 71, 1106–1114 CrossRef CAS PubMed.
- Y. Liu, L. Zhu, Y. Hu, X. Peng and J. Du, Food Chem., 2017, 221, 1128–1134 CrossRef CAS PubMed.
- A. J. Zaitouna and R. Y. Lai, Anal. Chim. Acta, 2014, 828, 85–91 CrossRef CAS PubMed.
- S. Campuzano, P. Yánez-Sedeño and J. M. Pingarrón, Electrochim. Acta, 2017, 230, 271–278 CrossRef CAS.
- J. Zhou, X. Qiu, K. Su, G. Xu and P. Wang, Sens. Actuators, B, 2016, 235, 170–178 CrossRef CAS.
- D. Antuña-Jiménez, M. C. Blanco-López, A. J. Miranda-Ordieres and M. J. Lobo-Castañón, Sens. Actuators, B, 2015, 220, 688–694 CrossRef.
- F. Ricci, G. Volpe, L. Micheli and G. Palleschi, Anal. Chim. Acta, 2007, 605, 111–129 CrossRef CAS PubMed.
- W. Duan, G. Chen, C. Chen, R. Sanghvi, A. Iddya, S. Walker, H. Liu, A. Ronen and D. Jassby, J. Membr. Sci., 2017, 531, 160–171 CrossRef CAS.
- X. Yu, B. Munge, V. Patel, G. Jensen, A. Bhirde, J. D. Gong, S. N. Kim, J. Gillespie, J. S. Gutkind, F. Papadimitrakopoulos and J. F. Rusling, J. Am. Chem. Soc., 2006, 128, 11199–11205 CrossRef CAS PubMed.
- A. Juzgado, A. Soldà, A. Ostric, A. Criado, G. Valenti, S. Rapino, G. Conti, G. Fracasso, F. Paolucci and M. Prato, J. Mater. Chem. B, 2017, 5, 6681–6687 RSC.
- M. Eising, C. E. Cava, R. V. Salvatierra, A. J. G. Zarbin and L. S. Roman, Sens. Actuators, B, 2017, 245, 25–33 CrossRef CAS.
- Y. Li, D. Ye, W. Liu, B. Shi, R. Guo, H. Pei and J. Xie, J. Colloid Interface Sci., 2017, 493, 241–248 CrossRef CAS PubMed.
- G. Xu, B. Li, X. T. Cui, L. Ling and X. Luo, Sens. Actuators, B, 2013, 188, 405–410 CrossRef CAS.
- L. Wilson, J. Vam Wyk, C. Rassie, N. Ross, C. Sunday, H. R. Makelane, M. Bilibana, T. Waryo, S. Mapolie, P. G. Baker and E. I. Iwuoha, Int. J. Electrochem. Sci., 2015, 10, 3207–3222 CAS.
- A. Kros, S. W. F. M. van Hövell, N. A. J. M. Sommerdijk and R. J. M. Nolte, Adv. Mater., 2001, 13, 1555–1557 CrossRef CAS.
- K. Liu, J. Zhang, Q. Liu and H. Huang, Electrochim. Acta, 2013, 114, 448–454 CrossRef CAS.
- N. A. A. Talib, F. Salam, N. A. Yusof, A. S. Alang Ahmad and Y. Sulaiman, RSC Adv., 2017, 7, 11101–11110 RSC.
- L. Wang, R. Yang, J. Chen, J. Li, L. Qu and P. d. B. Harrington, Food Chem., 2014, 164, 113–118 CrossRef CAS PubMed.
- F. Lopes, J. G. Pacheco, P. Rebelo and C. Delerue-Matos, Sens. Actuators, B, 2017, 243, 745–752 CrossRef CAS.
- Z. Taleat, A. Khoshroo and M. Mazloum-Ardakani, Microchim. Acta, 2014, 181, 865–891 CrossRef CAS.
- L. Mirmoghtadaie, A. A. Ensafi, M. Kadivar and P. Norouzi, Mater. Sci. Eng., C, 2013, 33, 1753–1758 CrossRef CAS PubMed.
- J. I. A. Rashid, N. A. Yusof, J. Abdullah, U. Hashim and R. Hajian, J. Mater. Sci., 2015, 51, 1083–1097 CrossRef.
- Z. Urkut, P. Kara, Y. Goksungur and M. Ozsoz, Electroanalysis, 2011, 23, 2668–2676 CrossRef CAS.
- M. D. Gouda, M. S. Thakur and N. G. Karanth, World J. Microbiol. Biotechnol., 2001, 17, 595–600 CrossRef CAS.
- A. Khoobi, S. M. Ghoreishi, M. Behpour and S. Masoum, Anal. Chem., 2014, 86, 8967–8973 CrossRef CAS PubMed.
- Y. Dou, Z. Jiang, W. Deng, J. Su, S. Chen, H. Song, A. Aldalbahi, X. Zuo, S. Song, J. Shi and C. Fan, J. Electroanal. Chem., 2016, 781, 339–344 CrossRef CAS.
- Y. Lai, J. Bai, X. Shi, Y. Zeng, Y. Xian, J. Hou and L. Jin, Talanta, 2013, 107, 176–182 CrossRef CAS PubMed.
- J. M. Walker, in The Protein Protocols Handbook, ed. J. M. Walker, Humana Press, Totowa, NJ, 2002, vol. 11, pp. 11–14, DOI:10.1385/1-59259-169-8.
- F. Patolsky, Y. Weizmann and I. Willner, Angew. Chem., Int. Ed., 2004, 43, 2113–2117 CrossRef CAS PubMed.
- S. Cosnier and M. Holzinger, Chem. Soc. Rev., 2011, 40, 2146 RSC.
- B. H. Shaz, in Transfusion Medicine and Hemostasis, Elsevier, San Diego, 2nd edn, 2013, pp. 307–315, DOI:10.1016/B978-0-12-397164-7.00048-3.
- M. Johnson, Anal. Chem., 2012, 2, 156 Search PubMed.
- T. Lundstedt, E. Seifert, L. Abramo, B. Thelin, Å. Nyström, J. Pettersen and R. Bergman, Chemom. Intell. Lab. Syst., 1998, 42, 3–40 CrossRef CAS.
- F. Feng, J. Zheng, P. Qin, T. Han and D. Zhao, Talanta, 2017, 167, 94–102 CrossRef CAS PubMed.
- J. Kong, L. Jiang, X. Su, J. Qin, Y. Du and B. Lin, Lab Chip, 2009, 9, 1541 RSC.
- X. Yang, F. Wu, D.-Z. Chen and H.-W. Lin, Sens. Actuators, B, 2014, 192, 529–535 CrossRef CAS.
- S. Yang, X. Liu, Y. Xing, D. Zhang, S. Wang, X. Wang, Y. Xu, M. Wu, Z. He and J. Zhao, J. Chromatogr. Sci., 2012, 00, 1–10 Search PubMed.
- J. Cheng, X.-O. Su, S. Wang and Y. Zhao, Sci. Rep., 2016, 6, 32637 Search PubMed.
- M. Yu, Y. Hu and J. Liu, New J. Chem., 2017, 41, 10407–10414 RSC.
- J. Hu, R. Chen, S. Wang, T. Wang, Y. Zhao, J. Li, X. Hu, H. Liang, J. Zhu, X. Sun, L. Ma and M. Jiang, PLoS One, 2015, 10, e0122005 Search PubMed.
- J. Xu, Y. Li, J. Guo, F. Shen, Y. Luo and C. Sun, Food Control, 2014, 46, 67–74 CrossRef CAS.
- P. Yan, Q. Tang, A. Deng and J. Li, Sens. Actuators, B, 2014, 191, 508–515 CrossRef CAS.
- V. Nandakumar, J. T. La Belle, J. Reed, M. Shah, D. Cochran, L. Joshi and T. L. Alford, Biosens. Bioelectron., 2008, 24, 1045–1048 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra00109j |
| This journal is © The Royal Society of Chemistry 2018 |