DOI:
10.1039/C7RA13581E
(Paper)
RSC Adv., 2018,
8, 15069-15077
Natural deep eutectic solvents as new green solvents to extract anthraquinones from Rheum palmatum L.
Received
22nd December 2017
, Accepted 16th April 2018
First published on 20th April 2018
Abstract
Natural deep eutectic solvents (NADESs) are efficient in extracting natural products. However, traditional organic solvents are toxic in the extraction of anthraquinones from Rheum palmatum L. To solve this problem, we applied natural deep eutectic solvent ultrasound-assisted extraction in this study for the extraction of total anthraquinones from R. palmatum L. Principal component analysis revealed that the selected NADES which consisted of lactic acid, glucose and water (LGH), was highly efficient in extracting anthraquinones from R. palmatum L. The ratio of lactic acid/glucose and the addition of water in LGH were investigated via a single-factor experiment. With a lactic acid/glucose ratio of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (mol/mol), and 10% of water (v/v), LGH had a high extraction yield to anthraquinones. Optimized by response surface methodology (RSM), the optimized extraction conditions of extraction time, extraction temperature and solvent-to-solid ratio of 1.5 h, 82 °C and 26 mL g−1, respectively. Under optimum conditions, the extraction yields of aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones were 2.60 ± 0.01, 5.78 ± 0.02, 2.21 ± 0.02, 5.87 ± 0.02, 8.81 ± 0.01 and 25.27 ± 0.07 mg g−1, respectively. The enrichment and separation of five anthraquinones in LGH extraction solution were efficiently achieved using DM130 macroporous resin, with purities of 90.98%, 96.67%, 92.37%, 95.80% and 91.61% as indicated by HPLC, and recovery yields of 84.08%, 79.51%, 84.96%, 81.83% and 78.35%, respectively. LGH was environmentally friendly and highly efficient in extracting anthraquinones from R. palmatum L., and NADESs showed potential for the extraction of effective components from natural products.
1 (mol/mol), and 10% of water (v/v), LGH had a high extraction yield to anthraquinones. Optimized by response surface methodology (RSM), the optimized extraction conditions of extraction time, extraction temperature and solvent-to-solid ratio of 1.5 h, 82 °C and 26 mL g−1, respectively. Under optimum conditions, the extraction yields of aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones were 2.60 ± 0.01, 5.78 ± 0.02, 2.21 ± 0.02, 5.87 ± 0.02, 8.81 ± 0.01 and 25.27 ± 0.07 mg g−1, respectively. The enrichment and separation of five anthraquinones in LGH extraction solution were efficiently achieved using DM130 macroporous resin, with purities of 90.98%, 96.67%, 92.37%, 95.80% and 91.61% as indicated by HPLC, and recovery yields of 84.08%, 79.51%, 84.96%, 81.83% and 78.35%, respectively. LGH was environmentally friendly and highly efficient in extracting anthraquinones from R. palmatum L., and NADESs showed potential for the extraction of effective components from natural products.
1. Introduction
Rheum palmatum L. (Da Huang), as a well-known traditional Chinese medicine, has been used for medical purposes by the Chinese for thousands of years.1 This plant is described in The Divine Farmer's Herb-Root Classic, which was likely compiled about 2700 years ago.2 R. palmatum L. is grown widely, especially in temperate and subtropical regions in Europe and Asia. It is utilized not only for traditional formulations and modern herbal medications, but also for pies and desserts; for instance, it is infused with strawberry juice to make a strawberry rhubarb pie.3 R. palmatum L. preparations are traditionally applied to treat various diseases and adverse conditions, such as high fever, intense sweating, constipation and abdominal pain.4 The roots and stems of R. palmatum L. are rich in anthraquinones, whose major components are aloe-emodin, rhein, emodin, chrysophanol and physcion (Fig. 1A), which have been extensively investigated because of their excellent bioactivities. Anthraquinones have anti-inflammatory, antitumorigenic, antiviral activities, anti-obesity and anticoagulant properties.5–9 They have also been commonly used in pharmaceutical, functional foods, and natural yellow dyes.10,11 As such, an efficient green extraction method should be developed to obtain anthraquinones from R. palmatum L.
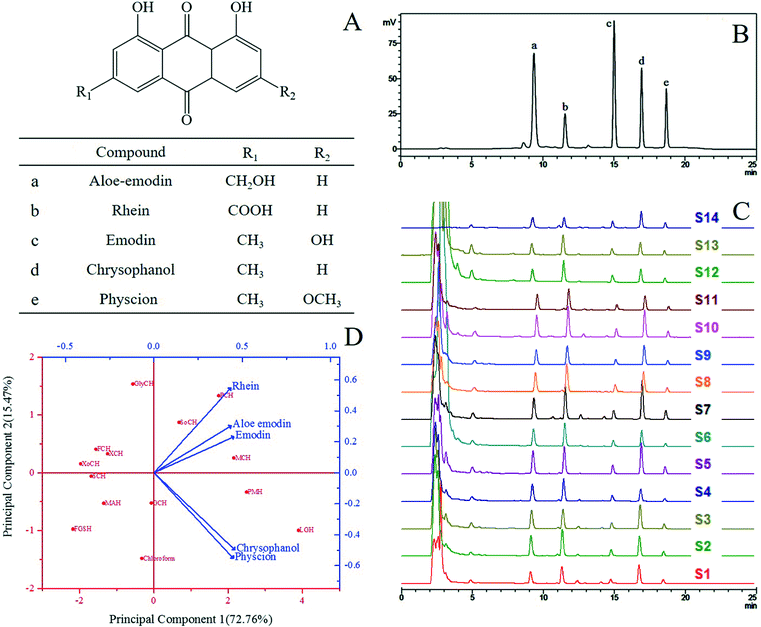 |
| | Fig. 1 (A) The chemical structures of five anthraquinones in R. palmatum L.; (B) the HPLC chromatograms of the standards; (C) the HPLC chromatograms of the extract samples of R. palmatum L. in different solvent systems in Table 1 (S1: FGSH, S2: SoCH, S3: GCH, S4: GlyCH, S5: PMH, S6: FCH, S7: LGH, S8: PCH, S9: MAH, S10: MCH, S11: XoCH, S12: SCH, S13: XCH, S14: CHCl3); (D) biplot of the loading plot (blue) and the score plot (red) through principal component analysis of the extracts from R. palmatum L. with NADES and chloroform. | |
Green techniques have been explored with the development of the green chemistry. For example, rhubarb is a good source of anthraquinones, and extraction is usually carried out with organic hazardous solvents, such as methanol, chloroform and ethyl acetate, which possess disadvantages, including toxicity, presence of solvent traces in the final product, and disposal problems. Anthraquinones can also be extracted by using supercritical fluid (SCF) extraction method. Nevertheless, SCFs are quite costly because of energy consumption for compression, thereby making the whole process inconvenient and uneconomic. Ionic liquids (ILs) contain organic cations and inorganic anions with unique properties, such as low melting temperature, high thermal stability, wide liquid phase range, non-flammability and low vapor pressure, but imidazolium-based ILs entail high cost and exhibit toxicity.12–14 Therefore, the use of green and economic solvents instead of hazardous solvents is one of the most important considerations in green technology.15–17 In 2011, Robert Verpoorte and his co-workers18 postulated a new type of solvents, called natural deep eutectic solvents (NADESs), which are composed of the primary metabolites of living cells. NADESs generally consist of biological components, such as sugars, organic acid and choline chloride, which are abundant in our daily life. NADESs are also excellent alternative media to organic hazardous solvents, SFs, ILs and conventional deep eutectic solvents,19 which are costly, difficult to prepare, and hazardous. Similar to ILs and conventional deep eutectic solvents, NADESs possess many advantages, such as low cost, easy preparation, adjustable viscosity, biodegradability, pharmaceutically acceptable toxicity and sustainability.4,20–22
Anthraquinones are conventionally extracted from plants through maceration extraction, heat reflux extraction, Soxhlet extraction and alkali extraction with organic solvents.23,24 Extraction methods can substantially affect the quality and concentration of targeted compounds in extract production. Ultrasound-assisted extraction (UAE) requires less amount of solvent and occurs faster while providing equivalent or higher extraction yields of target compounds than conventional extractions do.25 Ultrasonic waves change the physical and chemical properties of plant materials. The use of ultrasound can accelerate extraction by increasing the mass transfer. Cavity collapse leads to cell disruption through the formation of micro high-speed jets because of asymmetrical bubble collapse in the vicinity of a solid surface. The effect of cavitation facilitates the release of extractable compounds and improves the mass transport by disrupting plant cell walls, thereby allowing solvents to penetrate plant matrices.26–28 Therefore total anthraquinones may be rapidly extracted from R. palmatum L. via environmentally friendly NADES-UAE.
In this study, we aimed to develop a fast and green NADES-UAE method for the extraction of total anthraquinones from R. palmatum L. The investigated technique shows excellent extraction for aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones at a low cost. To the best of our knowledge, this study is the first to use NADES for the extraction of target anthraquinones from R. palmatum L. The effect of different extraction parameters, namely, water content of NADES, extraction temperature, extraction time and ratio of NADES to rhubarb powders, on the extraction properties were analyzed through response surface methodology (RSM). Three levels, three variables, and Box–Behnken experimental design (BBD) were employed to obtain the optimal conditions for the extraction of functional components from R. palmatum L. Macroporous resin was used to separate five anthraquinones rapidly and directly.
2. Experimental
2.1. Materials and reagents
R. palmatum L. (Maerkang County, Tibetan Qiang Autonomous Prefecture of Ngawa, Sichuan Province, China) was dried to a constant weight and pulverized with a disintegrator. The pulverized material was passed through a 40-mesh nylon sieve and stored in a sealed plastic bag before use.
The chemical reference substances of chrysophanol, rhein, emodin, aloe-emodin and physcion were purchased from the Chengdu Purechem-standard Co., Ltd. (Chengdu, China), and their purities were above 98%. Each reference substance was accurately weighed and dissolved in methanol to make stock solutions for the preparation of standard solutions at concentrations within the calibration range through successive dilution with methanol. HPLC grade methanol was purchased from Thermo Fisher Scientific Inc. NADES components, namely, malic acid, choline chloride, glycerol, β-alanine, proline, fructose, xylose, glucose, sucrose, 1,2-propanediol, lactic acid, sorbitol and xylitol, and other chemicals, such as chloroform, sulfuric acid, and analytical grade ethanol, were bought from Chengdu kelong chemical reagent factory (Sichuan, China). DM130 macroporous resin was procured from Tianjin Juhao Resin Technology Co., Ltd. (Tianjin, China).
2.2. HPLC analysis
Chrysophanol, rhein, emodin, aloe-emodin and physcion were simultaneously quantitated using a Shimadzu LC-20 series liquid chromatography system equipped with an LC-20AB pump, a DGU-20A degasser, a SPD-20A UV detector, and a SIL-20A autoinjector. The five anthraquinones were chromatographically separated on an Agilent Eclipse XDB-C18 column (4.6 mm × 250 mm, 5 μm) protected with a guard column. The mobile phase consisted of 0.1% phosphoric acid aqueous solution (A) and methanol (B), and the following gradient elution program was used for separation: 0–15 min, 70–100% B; 15–20 min, 100–70% B; and 20–25 min, 70% B. The flow rate was 1.0 mL min−1, the injection volume was 10 μL, the column temperature was 30 °C, and the detection wavelength was set at 264 nm. After re-equilibration was performed for 5 min, the column was ready for a new injection. All of the samples were filtered through 0.22 μm microporous membranes prior to HPLC analysis. The original HPLC chromatograms of the standards and the target compounds are shown in Fig. 1B and C.
2.3. NADES preparation
In this work, all NADES were prepared according to Young Hae Choi's previously reported method.20 Different component mixtures were heated to 80 °C in a water bath for 30–60 min with magnetic stirring until a homogeneous liquid was formed. Thirteen different NADES systems were prepared (Table 1).
Table 1 Different systems of natural deep eutectic solvents20a
| System abbreviation |
Component |
Mole ratio |
| Component 1 |
Component 2 |
Component 3 |
Component 4 |
| —: not added. |
| FGSH |
Fructose |
Choline chloride |
Water |
Glucose |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
| SoCH |
Sorbitol |
Choline chloride |
Water |
— |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 6 |
| GCH |
Glucose |
Choline chloride |
Water |
— |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
| GlyCH |
Glycerol |
Choline chloride |
Water |
— |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
| PMH |
Proline |
Malic acid |
Water |
— |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
| FCH |
Fructose |
Choline chloride |
Water |
— |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
| LGH |
Lactic acid |
Glucose |
Water |
— |
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
| PCH |
1,2-Propanediol |
Choline chloride |
Water |
— |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
| MAH |
Malic acid |
β-Alanine |
Water |
— |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
| MCH |
Malic acid |
Choline chloride |
Water |
— |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| XoCH |
Xylitol |
Choline chloride |
Water |
— |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
| SCH |
Sucrose |
Choline chloride |
Water |
— |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4 |
| XCH |
Xylose |
Choline chloride |
Water |
— |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
2.4. UAE with different solvents
The pulverized R. palmatum L. (1.00 g) was placed in a beaker flask and acidified with 10% H2SO4. Afterward, 20 mL of the different NADESs was added, and the flask was placed in an ultrasonic cleaner and extracted for 10 min at 40 °C. The mixture was stirred and heated at 75 °C for 1 h in a water bath. The radiation power was set at 600 W. After extraction was completed, the solutions were collected and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm min−1 for 15 min. The supernatant (5 mL) was removed and diluted five times. Finally, the diluted solutions were filtered through a 0.22 μm nylon membrane and analysed through HPLC. These experiments were repeated thrice.
000 rpm min−1 for 15 min. The supernatant (5 mL) was removed and diluted five times. Finally, the diluted solutions were filtered through a 0.22 μm nylon membrane and analysed through HPLC. These experiments were repeated thrice.
2.5. Experimental design and statistical analysis
The parameters of NADES-UAE were examined systematically to obtain appropriate extraction conditions for the total anthraquinones in R. palmatum L. The mole ratios of lactic acid to glucose (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 4
1, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 5
1, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 6
1, 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 7
1 and 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and the added proportions of water (0%, 10%, 20%, 30% and 40%) in LGH were investigated via a single-factor experiment. BBD was used to determine the most appropriate combination of extraction time (A), extraction temperature (B) and solvent-to-solid ratio (C). The extraction yields of aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones were fitted with the following second-order polynomial model:
1) and the added proportions of water (0%, 10%, 20%, 30% and 40%) in LGH were investigated via a single-factor experiment. BBD was used to determine the most appropriate combination of extraction time (A), extraction temperature (B) and solvent-to-solid ratio (C). The extraction yields of aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones were fitted with the following second-order polynomial model:| |
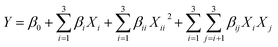 | (1) |
where Y is the response function; Xi and Xj are the independent variables influencing Y; and β0, βi, βii and βij are the regression coefficients of intercept, linear, quadratic and interaction terms, respectively.
All above experiments were performed in triplicate. The data of BBD were subjected to multiple regression to obtain the second order polynomial model using Design-Expert 10. Analysis of variance (ANOVA) was carried out to evaluate the model obtained by BBD.
2.6. Enrichment and separation of anthraquinones from R. palmatum L. by using NADESs
The anthraquinones in the NADES extraction solution were directly enriched and separated in a column (15 mm × 500 mm) we-packed with 10 g (dry weight) of DM130 macroporous resin. The bed volume (BV) of resin was 40 mL. The NADES extraction solution (30 mL) obtained under the optimized extraction conditions was flowed through the column packed with DM130 macroporous resin at a flow rate of 1 BV h−1. The macroporous resin column was washed with an appropriate amount of deionized water several times until Molisch reaction occurred to detect the eluate without sugar. Afterward, 90% aqueous ethanol (v/v) was eluted at a flow rate of 1.5 BV h−1. The eluate was collected and analysed through HPLC until it was nearly colourless. The solution was concentrated and dried with a rotary evaporator. The recoveries of the five target anthraquinones were calculated. These experiments were carried out in triplicate.
3. Results and discussion
3.1. Extractability of NADES and chloroform for the total anthraquinones in R. palmatum L.
Thirteen NADES systems (Fig. 1) were tested. However, the viscosity of NADESs is generally high, thereby hindering mass transport from plants to solutions. As such, 20% (v/v) water in NADES solutions was utilized in this study to extract the five target anthraquinones. Chloroform was chosen as a reference extraction solvent of the five target anthraquinones.29,30 The results are summarized in Table 2 and Fig. 1C. The extraction yields of the target compounds with the tested NADES and chloroform differed. Principal component analysis (PCA) was conducted to visualize these differences (Fig. 1D). Two principal components were obtained, suggesting that the date varied by 72.76% and 15.47%. Each blue arrowhead on the biplot represented the contribution of each variable (five target anthraquinones) to the score, while each red point on the biplot corresponded to the tested solvents. In Fig. 1D, principal component 1 correlated well with the yields of chrysophanol and physcion with the loadings of 0.458 and 0.450, respectively, and principal component 2 mainly correlated with the yield of rhein with the loading of 0.548. PCA confirmed the similarity between LGH and PMH, and these solvents were efficient for the extraction of chrysophanol and physcion. LGH was located in the rightmost position of principal component 1, indicating its higher extraction efficiencies for chrysophanol and physcion than for the other target compounds. GlyCH was placed in the uppermost position of principal component 2, indicating its higher extraction efficiency for rhein than for the other target compounds. However, overall, in the biplot of PCA, the five target anthraquinones clustered around LGH, indicating that this solvent was more suitable than other solvents for the simultaneous extraction of these compounds. Principal component 1 possessed more similarity than principal component 2 did. As such, LGH was selected for the extraction of anthraquinones from R. palmatum L.
Table 2 Extraction yields of five anthraquinones from R. palmatum L. by UAE with NADES and chloroform
| NADES |
The extraction yield (mean ± SD)/(mg g−1) |
| Aloe-emodin |
Rhein |
Emodin |
Chrysophanol |
Physcion |
Total anthraquinones |
| FGSH |
1.34 ± 0.02 |
2.15 ± 0.04 |
0.79 ± 0.01 |
2.81 ± 0.03 |
2.31 ± 0.03 |
9.40 ± 0.13 |
| SoCH |
2.45 ± 0.03 |
3.54 ± 0.05 |
1.19 ± 0.03 |
2.97 ± 0.04 |
2.69 ± 0.03 |
12.84 ± 0.18 |
| GCH |
2.15 ± 0.03 |
2.51 ± 0.04 |
1.11 ± 0.03 |
3.52 ± 0.03 |
2.84 ± 0.02 |
12.13 ± 0.13 |
| GlyCH |
2.12 ± 0.03 |
3.27 ± 0.04 |
1.24 ± 0.02 |
2.06 ± 0.03 |
1.67 ± 0.02 |
10.36 ± 0.12 |
| PMH |
2.56 ± 0.02 |
3.64 ± 0.02 |
1.37 ± 0.02 |
4.47 ± 0.04 |
4.10 ± 0.05 |
16.14 ± 0.14 |
| FCH |
1.72 ± 0.03 |
2.80 ± 0.04 |
0.95 ± 0.02 |
2.35 ± 0.03 |
1.84 ± 0.03 |
9.66 ± 0.12 |
| LGH |
2.37 ± 0.01 |
4.11 ± 0.03 |
1.55 ± 0.02 |
5.68 ± 0.03 |
4.99 ± 0.02 |
18.70 ± 0.10 |
| PCH |
2.50 ± 0.02 |
4.01 ± 0.03 |
1.48 ± 0.02 |
3.14 ± 0.02 |
2.92 ± 0.02 |
14.05 ± 0.11 |
| MAH |
1.55 ± 0.02 |
2.57 ± 0.02 |
0.93 ± 0.02 |
2.86 ± 0.03 |
2.51 ± 0.02 |
10.42 ± 0.09 |
| MCH |
2.34 ± 0.01 |
3.82 ± 0.02 |
1.46 ± 0.02 |
4.04 ± 0.03 |
3.59 ± 0.02 |
15.25 ± 0.09 |
| XoCH |
1.70 ± 0.02 |
2.53 ± 0.02 |
0.86 ± 0.02 |
2.03 ± 0.03 |
1.99 ± 0.02 |
9.11 ± 0.10 |
| SCH |
1.25 ± 0.01 |
2.48 ± 0.01 |
1.16 ± 0.02 |
1.87 ± 0.02 |
2.43 ± 0.03 |
9.19 ± 0.09 |
| XCH |
1.15 ± 0.02 |
2.92 ± 0.03 |
1.29 ± 0.02 |
1.86 ± 0.01 |
2.46 ± 0.02 |
9.68 ± 0.09 |
| CHCl3 |
1.79 ± 0.05 |
1.89 ± 0.02 |
1.20 ± 0.03 |
3.40 ± 0.04 |
3.51 ± 0.05 |
11.79 ± 0.08 |
3.2. Effect of the lactic acid/glucose ratio
The NADES systems with different lactic acid/glucose ratios were evaluated for the extraction of the five target anthraquinones. In Fig. 2A, the extraction yields of these anthraquinones changed significantly when the lactic acid/glucose ratio increased from 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 7
1 to 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (mol/mol). The amounts of the extracted target anthraquinones increased as the lactic acid/glucose ratio increased from 2
1 (mol/mol). The amounts of the extracted target anthraquinones increased as the lactic acid/glucose ratio increased from 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 5
1 to 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (mol/mol). Conversely, the extraction yields of the five target anthraquinones continuously decreased as the lactic acid/glucose ratio increased from 5
1 (mol/mol). Conversely, the extraction yields of the five target anthraquinones continuously decreased as the lactic acid/glucose ratio increased from 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 7
1 to 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. On the basis of these results, we chose a lactic acid/glucose ratio of 5
1. On the basis of these results, we chose a lactic acid/glucose ratio of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (mol/mol) for the following tests.
1 (mol/mol) for the following tests.
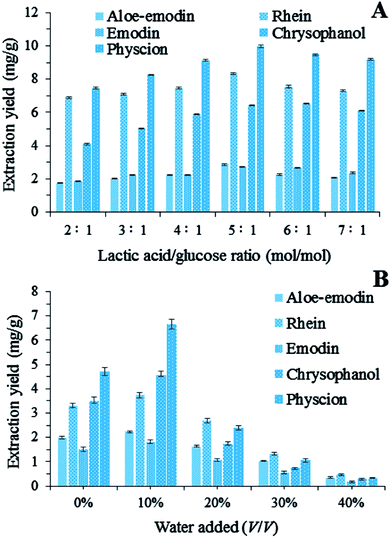 |
| | Fig. 2 (A) Extraction yields of five anthraquinones by using NADESs with different lactic acid/glucose ratios; (B) extraction yields of total anthraquinones by using LGH (lactic acid/glucose ratios = 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, molar ratio) diluted with different percentages of water. 1, molar ratio) diluted with different percentages of water. | |
3.3. Effect of the addition of water to LGH
The addition of a small amount of water to NADESs can increase the solubilizing capacity, but the optimum water content depends on a particular compound.20 in our study, extractions were carried out with different percentages of water added to LGH (from 0% to 40%, v/v). The addition of water to LGH could cause a decrease in solvent viscosity, which is beneficial to the mass transport from plant matrices to solutions. Fig. 2B shows that the extraction yields of the five anthraquinones from R. palmatum L. changed significantly as the content of water in LGH increased. For instance, the addition of 10% water to LGH markedly improved the extraction process. By contrast, 10–40% water in LGH led to a decrease in the amounts of the five target anthraquinones probably because high water contents decreased the interactions between LGH and anthraquinones and increased the polarity of the solvent mixture. Anthraquinones are also insoluble in water. Therefore, the addition of 10% (v/v) water in LGH was selected for the subsequent BBD experiments.
3.4. Optimization of NADES-UAE by using BBD
The NADES-UAE conditions (extraction time, extraction temperature and solvent-to-solid ratio) were further optimized with BBD. (Table 3) and analysed using Design Expert 10 for second-order polynomial regression analysis and ANOVA. The mathematical regression models of aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones are shown below in terms of the coded levels:| | |
YAloe-emodin = 2.40 − 0.049A + 0.028B − 0.061C − 0.038AB + 0.085AC + 0.0025BC − 0.12A2 − 0.17B2 − 0.12C2
| (2) |
| | |
YRhein = 5.49 − 0.022A − 0.11B + 0.29C + 0.012AB + 0.19AC − 0.070BC − 0.34A2 − 0.50B2 − 0.68C2
| (3) |
| | |
YEmodin = 2.19 + 0.053A + 0.051B + 0.049C − 0.12AB + 0.040AC + 0.0075BC − 0.067A2 − 0.17B2 − 0.17C2
| (4) |
| | |
YChrysophanol = 6.07 + 0.00125A + 0.11B + 0.061C − 0.18AB + 0.0601C + 0.012BC − 0.45A2 − 0.45B2 − 0.56C2
| (5) |
| | |
YPhyscion = 9.08 + 0.11A + 0.38B − 0.025C + 0.100AB − 0.48AC + 0.43 − 0.85A2 − 0.79B2 − 0.45C2
| (6) |
| | |
YTatal anthraquinones = 25.23 + 0.28A + 0.58B + 0.36C − 0.35AB − 0.21AC + 0.38BC − 1.95A2 − 2.12B2 − 1.95C2
| (7) |
where Y is the extraction yield of the target compounds aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones (mg g−1), and A, B and C are the extraction time, extraction temperature and solvent-to-solid ratio, respectively. ANOVA was performed to evaluate the optimal conditions of the NADES-UAE and the relationship between the response and the variables. Table 4 shows the ANOVA results of the quadratic models of aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones. The model F-values of more than 9.25 of emodin, chrysophanol, physcion and total anthraquinones implied that these models were significant. “Lack of fit F-value” of less than 3.0 indicated that the “lack of fit” of four quadratic models were not significant. The regression analysis of the data revealed that the coefficients of determination (R2) of emodin, chrysophanol, physcion and total anthraquinones were more than 0.92, suggesting that these quadratic models could be used to describe the response of the experiment pertaining to the three target anthraquinones and the total anthraquinones. Moreover, no abnormality was obtained from the diagnoses of residuals. Thus, the developed quadratic models were significant. “P-values > F” less than 0.05 indicated the model terms were significant. In Table 4, in most of the six quadratic models, the linear terms of A, B and C and the quadratic terms of B2 and C2 were significant. The effects of extraction temperature (B) and solvent-to-solid ratio (C) were more intense than that of extraction time (A).
Table 3 Experimental data and observed response values with different combinations of extraction time (A, min), extraction temperature (B, °C) and solvent-to-solid ratio (C, mL g−1) used in BBD
| Run |
Factor |
Extraction yield/(mg g−1) |
| A |
B |
C |
Aloe-emodin |
Rhein |
Emodin |
Chrysophanol |
Physcion |
Total anthraquinones |
| 1 |
1.5 |
80 |
25 |
2.27 |
5.55 |
2.19 |
6.07 |
9.18 |
25.26 |
| 2 |
2.0 |
70 |
25 |
2.17 |
4.14 |
2.07 |
5.42 |
7.37 |
21.17 |
| 3 |
1.5 |
80 |
25 |
2.27 |
5.48 |
2.19 |
6.04 |
9.18 |
25.16 |
| 4 |
1.0 |
70 |
25 |
1.99 |
5.40 |
1.70 |
4.83 |
6.81 |
19.93 |
| 5 |
1.5 |
70 |
20 |
2.12 |
4.16 |
1.83 |
4.79 |
7.77 |
20.67 |
| 6 |
1.0 |
80 |
20 |
2.49 |
3.68 |
1.85 |
5.19 |
7.61 |
20.42 |
| 7 |
2.0 |
80 |
20 |
2.02 |
4.44 |
1.84 |
4.85 |
8.26 |
21.41 |
| 8 |
2.0 |
90 |
25 |
2.16 |
3.92 |
1.98 |
5.14 |
8.27 |
21.68 |
| 9 |
1.0 |
90 |
25 |
2.13 |
5.13 |
2.07 |
5.28 |
7.31 |
21.84 |
| 10 |
1.0 |
80 |
30 |
2.14 |
4.11 |
1.98 |
5.14 |
8.27 |
21.64 |
| 11 |
2.0 |
80 |
30 |
2.01 |
5.64 |
2.13 |
5.04 |
6.99 |
21.81 |
| 12 |
1.5 |
70 |
30 |
2.05 |
4.63 |
1.80 |
4.94 |
7.12 |
20.54 |
| 13 |
1.5 |
80 |
25 |
2.31 |
5.36 |
2.16 |
6.17 |
9.27 |
25.27 |
| 14 |
1.5 |
80 |
25 |
2.70 |
5.78 |
2.22 |
5.87 |
8.81 |
25.38 |
| 15 |
1.5 |
80 |
25 |
2.45 |
5.27 |
2.20 |
6.19 |
8.97 |
25.08 |
| 16 |
1.5 |
80 |
30 |
2.10 |
4.30 |
1.88 |
5.34 |
8.78 |
22.40 |
| 17 |
1.5 |
80 |
25 |
2.16 |
4.11 |
1.88 |
5.14 |
7.72 |
21.01 |
Table 4 ANOVA of the quadratic model of the extraction yields of aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones
| Source |
Aloe-emodin |
Rhein |
Emodin |
Chrysophanol |
Physcion |
Total anthraquinones |
| F-value |
Pa > F |
F-value |
Pa > F |
F-value |
Pa > F |
F-value |
Pa > F |
F-value |
Pa > F |
F-value |
Pa > F |
| Probability values (P-values). |
| Model |
1.25 |
0.394 |
1.21 |
0.412 |
10.36 |
0.003 |
13.52 |
0.001 |
9.25 |
0.004 |
645.15 |
0.001 |
| A |
0.60 |
0.465 |
0.0091 |
0.927 |
4.94 |
0.062 |
0.0004 |
0.984 |
0.81 |
0.398 |
58.16 |
0.001 |
| B |
0.19 |
0.676 |
0.21 |
0.658 |
4.89 |
0.063 |
3.50 |
0.104 |
9.28 |
0.019 |
247.17 |
0.001 |
| C |
0.94 |
0.364 |
1.48 |
0.264 |
4.43 |
0.073 |
0.99 |
0.353 |
0.041 |
0.845 |
96.12 |
0.001 |
| AB |
0.18 |
0.687 |
0.0014 |
0.971 |
11.84 |
0.011 |
4.40 |
0.074 |
0.33 |
0.585 |
45.39 |
0.001 |
| AC |
0.91 |
0.373 |
0.33 |
0.581 |
1.43 |
0.270 |
0.48 |
0.513 |
7.63 |
0.028 |
15.54 |
0.006 |
| BC |
0.0008 |
0.978 |
0.044 |
0.840 |
0.081 |
0.784 |
0.021 |
0.890 |
5.99 |
0.044 |
53.51 |
0.001 |
| A2 |
1.75 |
0.228 |
1.08 |
0.334 |
4.39 |
0.074 |
28.05 |
0.001 |
25.13 |
0.002 |
1490.66 |
0.001 |
| B2 |
3.93 |
0.088 |
2.41 |
0.164 |
26.84 |
0.001 |
28.36 |
0.001 |
21.45 |
0.002 |
1753.33 |
0.001 |
| C2 |
1.90 |
0.210 |
4.44 |
0.073 |
28.45 |
0.001 |
44.25 |
0.000 |
6.86 |
0.034 |
1490.66 |
0.001 |
| Lack of fit |
0.88 |
0.522 |
25.71 |
0.005 |
20.84 |
0.007 |
2.99 |
0.159 |
6.74 |
0.048 |
0.59 |
0.654 |
| R2 |
0.6161 |
0.6077 |
0.9301 |
0.9456 |
0.9224 |
0.9988 |
Three-dimensional RSM is used as a tool to visualize the effect of independent variables on dependent ones.31 In our study, to depict the interactive effects of operational variables on responses, we maintained one variable at its middle level and varied the two other variables in the defined ranges. Fig. 3 illustrates the three-dimensional plots of the response surface of the extraction yields of aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones related to the extraction time (A), extraction temperature (B) and solvent-to-solid ratio (C). The yields of the five anthraquinones increased as the extraction temperature and time simultaneously increased. High temperatures could decrease physical adsorption and chemical interactions between plant secondary metabolites and matrices and increase NADES penetration, thereby facilitating the mass transfer of the target anthraquinones from the interior of the plant matrices to NADESs. However, extraction yields slightly decreased when the extraction temperature exceeded 80 °C.
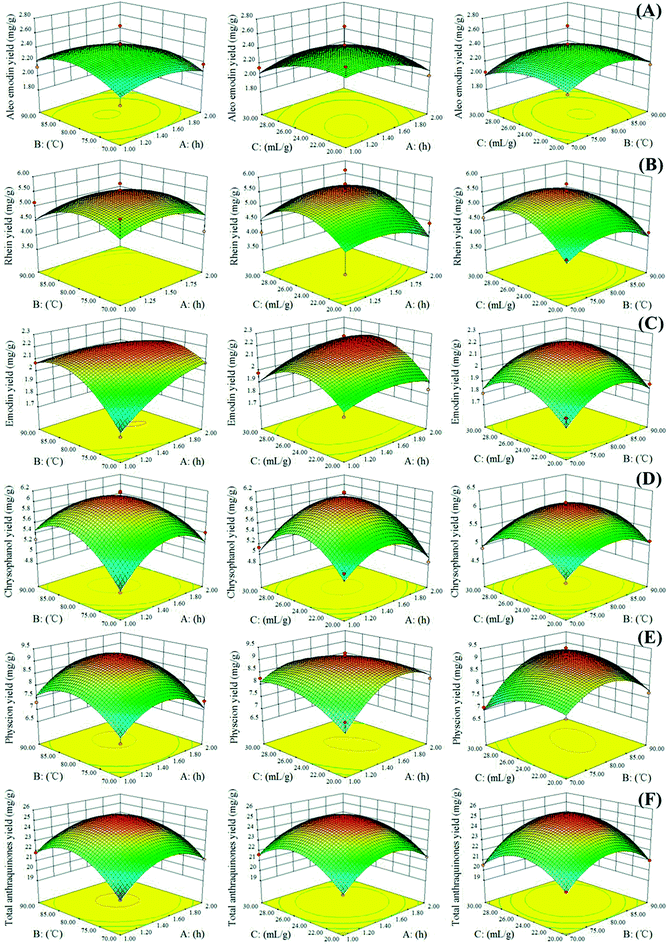 |
| | Fig. 3 Response surface representations for (A) aloe-emodin, (B) rhein, (C) emodin, (D) chrysophanol, (E) physcion and (F) total anthraquinones. A: extraction time; B: extraction temperature; C: solvent-to-solid ratio. | |
In Fig. 3, the extraction yields of aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones significantly increased as the solvent-to-solid ratio increased, especially at high extraction temperature and middle solvent-to-solid ratio. Increasing the solvent-to-solid ratio from 20 to 26 mL g−1 and the extraction temperature from 70 to 80 °C improved the extraction yields of the five anthraquinones. By contrast, their extraction yields decreased when the solvent-to-solid ratio and temperature exceeded 26 mL g−1 and 80 °C, respectively. These results indicated that a long extraction time and a high extraction temperature are unsuitable for the extraction of target anthraquinones.32
3.5. Verification of the models and comparison of our solvent with traditional organic solvents
The optimal conditions for the simultaneous extraction of the total anthraquinones obtained with our models were as follows: extraction time 1.53 h, extraction temperature 81.42 °C and solvent-to-solid ratio 25.51 mL g−1, the predicted extraction yield of the total anthraquinones was 25.30 mg g−1. Considering that the parameters are difficult to operate in actual experiments, we selected the extraction temperature of 82 °C, the solvent-to-solid ratio of 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mL g−1, and the extraction time 1.5 h. To confirm the fitness of the predicted response values, we performed verification experiments under the optimized conditions in three replicates. The observed extraction yields of aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones were 2.60 ± 0.01, 5.78 ± 0.02, 2.21 ± 0.02, 5.87 ± 0.02, 8.81 ± 0.01 and 25.27 ± 0.07 mg g−1, respectively, which were largely consistent with the predicted values. These findings indicated that the established quadratic models were statistically reliable and reasonable. The extraction yields of the five free anthraquinones and the total anthraquinones showed good repeatability.
1 mL g−1, and the extraction time 1.5 h. To confirm the fitness of the predicted response values, we performed verification experiments under the optimized conditions in three replicates. The observed extraction yields of aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones were 2.60 ± 0.01, 5.78 ± 0.02, 2.21 ± 0.02, 5.87 ± 0.02, 8.81 ± 0.01 and 25.27 ± 0.07 mg g−1, respectively, which were largely consistent with the predicted values. These findings indicated that the established quadratic models were statistically reliable and reasonable. The extraction yields of the five free anthraquinones and the total anthraquinones showed good repeatability.
The relative extraction yields of the five anthraquinones and the total anthraquinones were used for comparison (Table 2). The extraction yields of the total anthraquinones obtained with LGH were the highest and even 1.8 times higher than those of anthraquinones traditionally extracted with chloroform (14.04 ± 0.07 mg g−1). Yang's group25 used 84% methanol as a solvent and acquired an average extraction yield of total anthraquinones of 17.55 mg g−1 under optimal conditions. Zhang's group32 utilized 80% ethanol as a solvent, and they obtained the relative extraction yields of emodin, aloe-emodin and rhein of 0.97, 0.54 and 0.89 mg g−1, respectively. Therefore, the cavitation effect of sonication and LGH increased the extraction efficiency. Higher extraction yield and lower toxicity were observed when LGH was used than when traditional organic solvents were applied. NADES should be employed to extract effective components from many kinds of herbal samples.
3.6. Enrichment and separation of the five target anthraquinones from LGH extraction solution
Ethanol or aqueous ethanol is traditionally used as a solvent for the extraction of natural products from rhubarb. To enrich and separate the natural products in ethanol or aqueous ethanol extraction solution, the macroporous resin was used. In this study, the five free anthraquinones were directly enriched and separated from LGH by utilizing DM130 macroporous resin. This resin shows a good adsorption and desorption capacity toward anthraquinones.24 After purification with DM130 macroporous resin was performed, the purities of aloe-emodin, rhein, emodin, chrysophanol and physcion reached 90.98%, 96.67%, 92.37%, 95.80% and 91.61%, respectively, and the recovery yields were 84.08%, 79.51%, 84.96%, 81.83% and 78.35%, respectively. These results indicated that anthraquinones could be efficiently recovered from NADES extracts. DM130 macroporous resin could effectively adsorb the five target anthraquinones, while the polar ingredients of NADES could be eluted and recovered with deionized water. Most of the target anthraquinones could be obtained with 90% ethanol after the polar compounds were eluted with deionized water.
4. Conclusions
NADES-UAE was developed to extract five main anthraquinones, namely, aloe-emodin, rhein, emodin, chrysophanol and physcion, from R. palmatum L. LGH composed of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 lactic acid/glucose and 10% water (v/v) was efficient in extracting the five free anthraquinones. The optimized extraction time, extraction temperature, and solvent-to-solid ratio were 1.5 h, 82 °C, and 26 mL g−1, respectively. Under these conditions, the extraction yield of the total anthraquinones was 25.27 ± 0.07 mg g−1. Therefore, the five anthraquinones in the LGH extraction solution could be convenient and effective. The developed procedure based on NADES-UAE and direct macroporous resin adsorption and desorption could be an alternative to conventional methods involving organic solvents for the extraction and separation of natural products from plants. NADESs possess excellent properties, such as sustainability, biodegradability, pharmaceutically acceptable toxicity and high extractability, and thus show potential as green solvents for the extraction of effective components from plants.
1 lactic acid/glucose and 10% water (v/v) was efficient in extracting the five free anthraquinones. The optimized extraction time, extraction temperature, and solvent-to-solid ratio were 1.5 h, 82 °C, and 26 mL g−1, respectively. Under these conditions, the extraction yield of the total anthraquinones was 25.27 ± 0.07 mg g−1. Therefore, the five anthraquinones in the LGH extraction solution could be convenient and effective. The developed procedure based on NADES-UAE and direct macroporous resin adsorption and desorption could be an alternative to conventional methods involving organic solvents for the extraction and separation of natural products from plants. NADESs possess excellent properties, such as sustainability, biodegradability, pharmaceutically acceptable toxicity and high extractability, and thus show potential as green solvents for the extraction of effective components from plants.
Conflicts of interest
Authors declare no conflicts of interest.
Acknowledgements
This work was financially supported by the Science and Technology Program Project Fund of Sichuan Province (Grant No. 2016SZ0030 and 2016SZ0032).
References
- Y. Liu, L. Li, Y.-Q. Xiao, J.-Q. Yao, P.-Y. Li, D.-R. Yu and Y.-L. Ma, Food Chem., 2016, 192, 531–540 CrossRef CAS PubMed.
- A. D. M. Association, Origin and History of all the Pharmacopeial Vegetable Drugs, Chemicals and Preparations, with Bibliography. Volume I, Vegetable Drugs, ed. John Uri Lloyd, Nabu Press, 1921 Search PubMed.
- B. Neal, Biscuits, Spoonbread, and Sweet Potato Pie, The University of North Carolina Press, 2003 Search PubMed.
- D. Aichner and M. Ganzera, Talanta, 2015, 144, 1239–1244 CrossRef CAS PubMed.
- S.-J. Kim, M.-C. Kim, B.-J. Lee, D.-H. Park, S.-H. Hong and J.-Y. Um, Molecules, 2010, 15, 6436–6451 CrossRef CAS PubMed.
- Y. Gao, X. Chen, L. Fang, F. Liu, R. Cai, C. Peng and Y. Qi, Free Radical Biol. Med., 2014, 72, 104–112 CrossRef CAS PubMed.
- S.-J. Chang, S.-H. Huang, Y.-J. Lin, Y.-Y. Tsou and C.-W. Lin, Arch. Pharmacal Res., 2014, 37, 1117–1123 CrossRef CAS PubMed.
- J. Li, L. Ding, B. Song, X. Xiao, M. Qi, Q. Yang, Q. Yang, X. Tang, Z. Wang and L. Yang, Eur. J. Pharmacol., 2016, 770, 99–109 CrossRef CAS PubMed.
- L. Zhang, J. Chen, D. Jiang and P. Zhang, J. Crit. Care, 2015, 30, 282–289 CrossRef CAS PubMed.
- Y. Zhou, J. Zhang, R.-C. Tang and J. Zhang, Ind. Crops Prod., 2015, 64, 224–232 CrossRef CAS.
- S. A. Khan, Shahid-ul-Islam, M. Shahid, M. I. Khan, M. Yusuf, L. J. Rather, M. A. Khan and F. Mohammad, J. Nat. Fibers, 2015, 12, 243–255 CrossRef CAS.
- I. Lavilla, V. Romero, I. Costas and C. Bendicho, TrAC, Trends Anal. Chem., 2014, 61, 1–10 CrossRef CAS.
- T. Khezeli, A. Daneshfar and R. Sahraei, Talanta, 2016, 150, 577–585 CrossRef CAS PubMed.
- A. Amiri-Rigi and S. Abbasi, Food Chem., 2016, 197, 1002–1007 CrossRef CAS PubMed.
- J. Sherman, B. Chin, P. D. Huibers, R. Garcia-Valls and T. A. Hatton, Environ. Health Perspect., 1998, 106, 253–271 CrossRef CAS PubMed.
- M. Tobiszewski, A. Mechlinska and J. Namiesnik, Chem. Soc. Rev., 2010, 39, 2869–2878 RSC.
- D. Prat, O. Pardigon, H.-W. Flemming, S. Letestu, V. Ducandas, P. Isnard, E. Guntrum, T. Senac, S. Ruisseau, P. Cruciani and P. Hosek, Org. Process Res. Dev., 2013, 17, 1517–1525 CrossRef CAS.
- Y. H. Choi, S. J. Van, Y. Dai, M. Verberne, F. Hollmann, I. W. Arends, G. J. Witkamp and R. Verpoorte, Plant Physiol., 2011, 156, 1701–1705 CrossRef CAS PubMed.
- M. Espino, M. de los Ángeles Fernández, F. J. V. Gomez and M. F. Silva, TrAC, Trends Anal. Chem., 2016, 76, 126–136 CrossRef CAS.
- Y. Dai, J. van Spronsen, G.-J. Witkamp, R. Verpoorte and Y. H. Choi, Anal. Chim. Acta, 2013, 766, 61–68 CrossRef CAS PubMed.
- Y. Dai, G.-J. Witkamp, R. Verpoorte and Y. H. Choi, Anal. Chem., 2013, 85, 6272–6278 CrossRef CAS PubMed.
- Z.-F. Wei, X.-Q. Wang, X. Peng, W. Wang, C.-J. Zhao, Y.-G. Zu and Y.-J. Fu, Ind. Crops Prod., 2015, 63, 175–181 CrossRef CAS.
- L. Tan, D.-d. Geng, F.-z. Hu and Q. Dong, J. Chromatogr. Sci., 2016, 54, 48–57 CAS.
- J. Wang, H. Fang, B. Dong, D. Wang, Y. Li, X. Chen, L. Chen, T. Wei and Q. Wei, Korean J. Physiol. Pharmacol., 2015, 19, 485–489 CrossRef CAS PubMed.
- L.-C. Zhao, J. Liang, W. Li, K.-M. Cheng, X. Xia, X. Deng and G.-L. Yang, Molecules, 2011, 16, 5928–5937 CrossRef CAS PubMed.
- A. Tomsik, B. Pavlic, J. Vladic, M. Ramic, J. Brindza and S. Vidovic, Ultrason. Sonochem., 2016, 29, 502–511 CrossRef CAS PubMed.
- L. Wang and C. L. Weller, Trends Food Sci. Technol., 2006, 17, 300–312 CrossRef CAS.
- M. Vinatoru, Ultrason. Sonochem., 2001, 8, 303–313 CrossRef CAS PubMed.
- M. Ozturk, F. Aydogmus-Ozturk, M. E. Duru and G. Topcu, Food Chem., 2007, 103, 623–630 CrossRef.
- Y. C. Cao and Y. D. Zhao, J. Innovative Opt. Health Sci., 2011, 4, 127–132 CrossRef.
- Y. Kong, Z.-F. Wei, Y.-J. Fu, C.-B. Gu, C.-J. Zhao, X.-H. Yao and T. Efferth, Food Chem., 2011, 128, 596–605 CrossRef CAS.
- L. Wang, D. Li, C. Bao, J. You, Z. Wang, Y. Shi and H. Zhang, Ultrason. Sonochem., 2008, 15, 738–746 CrossRef CAS PubMed.
|
| This journal is © The Royal Society of Chemistry 2018 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article a,
P. Wua,
Y. B. Liac,
T. C. Liua,
L. Zhang*a and
Y. H. Zhoub
a,
P. Wua,
Y. B. Liac,
T. C. Liua,
L. Zhang*a and
Y. H. Zhoub
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (mol/mol), and 10% of water (v/v), LGH had a high extraction yield to anthraquinones. Optimized by response surface methodology (RSM), the optimized extraction conditions of extraction time, extraction temperature and solvent-to-solid ratio of 1.5 h, 82 °C and 26 mL g−1, respectively. Under optimum conditions, the extraction yields of aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones were 2.60 ± 0.01, 5.78 ± 0.02, 2.21 ± 0.02, 5.87 ± 0.02, 8.81 ± 0.01 and 25.27 ± 0.07 mg g−1, respectively. The enrichment and separation of five anthraquinones in LGH extraction solution were efficiently achieved using DM130 macroporous resin, with purities of 90.98%, 96.67%, 92.37%, 95.80% and 91.61% as indicated by HPLC, and recovery yields of 84.08%, 79.51%, 84.96%, 81.83% and 78.35%, respectively. LGH was environmentally friendly and highly efficient in extracting anthraquinones from R. palmatum L., and NADESs showed potential for the extraction of effective components from natural products.
1 (mol/mol), and 10% of water (v/v), LGH had a high extraction yield to anthraquinones. Optimized by response surface methodology (RSM), the optimized extraction conditions of extraction time, extraction temperature and solvent-to-solid ratio of 1.5 h, 82 °C and 26 mL g−1, respectively. Under optimum conditions, the extraction yields of aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones were 2.60 ± 0.01, 5.78 ± 0.02, 2.21 ± 0.02, 5.87 ± 0.02, 8.81 ± 0.01 and 25.27 ± 0.07 mg g−1, respectively. The enrichment and separation of five anthraquinones in LGH extraction solution were efficiently achieved using DM130 macroporous resin, with purities of 90.98%, 96.67%, 92.37%, 95.80% and 91.61% as indicated by HPLC, and recovery yields of 84.08%, 79.51%, 84.96%, 81.83% and 78.35%, respectively. LGH was environmentally friendly and highly efficient in extracting anthraquinones from R. palmatum L., and NADESs showed potential for the extraction of effective components from natural products.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm min−1 for 15 min. The supernatant (5 mL) was removed and diluted five times. Finally, the diluted solutions were filtered through a 0.22 μm nylon membrane and analysed through HPLC. These experiments were repeated thrice.
000 rpm min−1 for 15 min. The supernatant (5 mL) was removed and diluted five times. Finally, the diluted solutions were filtered through a 0.22 μm nylon membrane and analysed through HPLC. These experiments were repeated thrice.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 4
1, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 5
1, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 6
1, 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 7
1 and 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and the added proportions of water (0%, 10%, 20%, 30% and 40%) in LGH were investigated via a single-factor experiment. BBD was used to determine the most appropriate combination of extraction time (A), extraction temperature (B) and solvent-to-solid ratio (C). The extraction yields of aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones were fitted with the following second-order polynomial model:
1) and the added proportions of water (0%, 10%, 20%, 30% and 40%) in LGH were investigated via a single-factor experiment. BBD was used to determine the most appropriate combination of extraction time (A), extraction temperature (B) and solvent-to-solid ratio (C). The extraction yields of aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones were fitted with the following second-order polynomial model: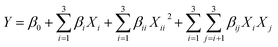
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 7
1 to 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (mol/mol). The amounts of the extracted target anthraquinones increased as the lactic acid/glucose ratio increased from 2
1 (mol/mol). The amounts of the extracted target anthraquinones increased as the lactic acid/glucose ratio increased from 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 5
1 to 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (mol/mol). Conversely, the extraction yields of the five target anthraquinones continuously decreased as the lactic acid/glucose ratio increased from 5
1 (mol/mol). Conversely, the extraction yields of the five target anthraquinones continuously decreased as the lactic acid/glucose ratio increased from 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 7
1 to 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. On the basis of these results, we chose a lactic acid/glucose ratio of 5
1. On the basis of these results, we chose a lactic acid/glucose ratio of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (mol/mol) for the following tests.
1 (mol/mol) for the following tests.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mL g−1, and the extraction time 1.5 h. To confirm the fitness of the predicted response values, we performed verification experiments under the optimized conditions in three replicates. The observed extraction yields of aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones were 2.60 ± 0.01, 5.78 ± 0.02, 2.21 ± 0.02, 5.87 ± 0.02, 8.81 ± 0.01 and 25.27 ± 0.07 mg g−1, respectively, which were largely consistent with the predicted values. These findings indicated that the established quadratic models were statistically reliable and reasonable. The extraction yields of the five free anthraquinones and the total anthraquinones showed good repeatability.
1 mL g−1, and the extraction time 1.5 h. To confirm the fitness of the predicted response values, we performed verification experiments under the optimized conditions in three replicates. The observed extraction yields of aloe-emodin, rhein, emodin, chrysophanol, physcion and total anthraquinones were 2.60 ± 0.01, 5.78 ± 0.02, 2.21 ± 0.02, 5.87 ± 0.02, 8.81 ± 0.01 and 25.27 ± 0.07 mg g−1, respectively, which were largely consistent with the predicted values. These findings indicated that the established quadratic models were statistically reliable and reasonable. The extraction yields of the five free anthraquinones and the total anthraquinones showed good repeatability.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 lactic acid/glucose and 10% water (v/v) was efficient in extracting the five free anthraquinones. The optimized extraction time, extraction temperature, and solvent-to-solid ratio were 1.5 h, 82 °C, and 26 mL g−1, respectively. Under these conditions, the extraction yield of the total anthraquinones was 25.27 ± 0.07 mg g−1. Therefore, the five anthraquinones in the LGH extraction solution could be convenient and effective. The developed procedure based on NADES-UAE and direct macroporous resin adsorption and desorption could be an alternative to conventional methods involving organic solvents for the extraction and separation of natural products from plants. NADESs possess excellent properties, such as sustainability, biodegradability, pharmaceutically acceptable toxicity and high extractability, and thus show potential as green solvents for the extraction of effective components from plants.
1 lactic acid/glucose and 10% water (v/v) was efficient in extracting the five free anthraquinones. The optimized extraction time, extraction temperature, and solvent-to-solid ratio were 1.5 h, 82 °C, and 26 mL g−1, respectively. Under these conditions, the extraction yield of the total anthraquinones was 25.27 ± 0.07 mg g−1. Therefore, the five anthraquinones in the LGH extraction solution could be convenient and effective. The developed procedure based on NADES-UAE and direct macroporous resin adsorption and desorption could be an alternative to conventional methods involving organic solvents for the extraction and separation of natural products from plants. NADESs possess excellent properties, such as sustainability, biodegradability, pharmaceutically acceptable toxicity and high extractability, and thus show potential as green solvents for the extraction of effective components from plants.


