Open Access Article
Open Access ArticleAntioxidant and anti-inflammatory effects of polyphenols extracted from Ilex latifolia Thunb
Tian-Tian Zhang†
 ab,
Ting Hu†c,
Jian-Guo Jiang
ab,
Ting Hu†c,
Jian-Guo Jiang *a,
Jing-Wen Zhaod and
Wei Zhu*d
*a,
Jing-Wen Zhaod and
Wei Zhu*d
aDepartment of Food Science and Technology, South China University of Technology, Guangzhou, 510640, China. E-mail: jgjiang@scut.edu.cn; Fax: +86 20 87113843; Tel: +86 20 87113849
bCollege of Food Science and Engineering, Ocean University of China, Qingdao 266003, China
cSchool of Chemical Engineering & Pharmacy, Wuhan Institute of Technology, Wuhan 430205, China
dThe Second Institute of Clinical Medicine, Guangzhou University of Chinese Medicine, Guangzhou 510120, China. E-mail: zhuwei9201@163.com; Fax: +86 20 39318571; Tel: +86 20 39318571
First published on 14th February 2018
Abstract
To promote the rational and effective application of Ilex latifolia Thunb., a Chinese bitter tea widely consumed as a health beverage, polyphenols were extracted from its leaves and their cellular antioxidant activity (CAA) and anti-inflammatory effect against mouse macrophage RAW 264.7 cells were analyzed. Results showed that the antioxidant capacity of polyphenols was high, and their CAA values in PBS wash and no PBS wash protocols were 6871.42 ± 85.56 and 25161.61 ± 583.55 μmol QE (quercetin equivalents)/100 g phenolic extracts, respectively. In addition, polyphenols from I. latifolia displayed strong inhibition on LPS-induced NO-production in RAW 264.7 cells. Polyphenol treatment inhibited the release of pro-inflammatory cytokines (TNF-α, IL-1β and IL-6) induced by LPS in a dose-dependent manner by ELISA and mRNA expression analysis. Western blot results showed that the anti-inflammatory activity of polyphenols from I. latifolia might be exerted through inhibiting the activation of MAPKs (ERK and JNK) and NF-κB to decrease NO, COX-2 and pro-inflammatory cytokines production. Thus, the polyphenol enriched extracts from I. latifolia are a good source of natural antioxidants with a beneficial effect against inflammation, and they may be applied as a food supplement and/or functional ingredient.
Introduction
Permanent exposure to reactive oxygen species (ROS), including superoxide or hydroxyl radicals, is inevitable and common to all aerobic organisms.1 Oxidative stress is a negative effect produced by free radicals in the body, and is considered to be an important factor leading to aging and disease.2,3 Moreover, oxidative stress is linked to the persistence of chronic inflammatory processes through the activation of inducible transcription factors such as nuclear factor-κB (NF-κB), which is involved in multiple physiological and pathological processes including inflammatory and immune response, cell survival and apoptosis.4Recent important epidemiological studies believed that certain natural foods and plants could be involved in preventing or hindering the development of various diseases.5 Synthetic antioxidants have been considered as endocrine disrupters or even carcinogenic agents, and interest in developing natural nutritional antioxidants has been increasing due to their well-documented impact on human health.6–8 Dietary supplements are becoming attractive alternatives as disease-preventive agents. Among the secondary metabolites of various plant species, polyphenols have been extensively studied because of their antioxidant, anti-cancer, anti-inflammatory and anti-hyperglycemic effects in humans.9
Ilex latifolia Thunb., a kind of herbal tea, is called ku-ding-cha in Chinese and is widely consumed in China and other Southeast Asia countries including Singapore, Malaysia and Vietnam.10 I. latifolia has become more popular in the last few years due to its public acceptance as a functional tea beverage with highly advantageous nutrition and many beneficial functions, such as antioxidant, anti-obesity, anti-diabetic, hepatoprotective, neuroprotective effects and so on.11,12 Recently, more studies have been devoted to the extraction and identification of main chemicals in I. latifolia such as flavonoids, saponins, polysaccharides and alkaloids and their potential health benefits in antioxidant, anti-obesity, neuroprotective, antidiabetic, hepatoprotective and anti-complementary effects.11–13 However, little attention has been paid to the extraction and bioactivities of polyphenols of I. latifolia. Therefore, searching for alternative natural active ingredients such as polyphenols in I. latifolia is necessary for its application on food additives and health products.
On the basis of these reports, polyphenols obtained from I. latifolia were evaluated antioxidant, and anti-inflammatory activities. The aim of this work was to (i) measure the total phenolic content; (ii) determine the cellular antioxidant capacity; (iii) determine the anti-inflammatory activity on NO production in LPS-stimulated macrophage RAW 264.7 cells; (iv) demonstrate possible targeted pathway.
Materials and methods
Plant materials
I. latifolia cultivated in Hainan Province, China, was purchased from Forest Drugstore Chain Co. (Guangzhou, China) and were authenticated by South China Botanical Garden, Chinese Academy of Sciences, where voucher specimens were kept. Samples were air-dried under shade for one week and ground in a cutting mill to pass through a 50-mesh sieve to obtain fine powder. The dried materials were stored at room temperature in a desiccator until use.Reagents
Dulbecco's modified eagle medium (DMEM), fetal bovine serum (FBS), Hank's balanced salt solution (HBSS) and penicillin/streptomycin were obtained from GIBCO (Grand Island, NY, USA). (3-4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), 2′,7′-dichlorofluorescin diacetate (DCFH-DA), fluorescein disodium salt, 2,2′-azobis(2-amidinopropane)dihydrochloride (ABAP), Folin–Ciocalteu reagent, lipopolysaccharides (LPS), dexamethasone (DXM) and bicinchoninic acid (BCA) kit were purchased from Sigma Aldrich (St. Louis, MO, USA). TRIzol Reagent was purchased from Invitrogen (Carlsbad, CA, USA). The antibodies against phospho-ERK, phospho-JNK, NF-κB and GAPDH were obtained from Cell Signaling Technology (Beverly, MA, USA). The water used in all assays was ultrapure by a Milli-Q water purification system from Millipore. All other reagents used in this study were of analytical grade.Extraction of polyphenols
Ultrasound-assisted extraction was conducted following the method described by Izadiyan et al. (2016) with some modifications.14 Briefly, ten grams of the dried powder were weighed and mixed with 200 mL of 50% (v/v) aqueous ethanol. The mixture was subjected to the continuous ultrasonic treatment in a 60 W (ultrasonic power) ultrasonic bath for 100 min at 30 °C. The samples were then centrifuged at 1500 × g for 10 min. The supernatant was recovered and then evaporated to near dryness under a vacuum at 40 °C. The samples were collected and stored at 4 °C in a refrigerator for further analysis.Determination of total phenolic content and phenolic composition
The total phenolic content (TPC) was determined using the Folin–Ciocalteu method with some modifications.15,16 Two hundred micro liters of phenolic extract was transferred to a 10 mL of volumetric flask and then mixed with 0.5 mL of undiluted Folin–Ciocalteu reagent. After 1 min, 1.5 mL of 20% (w/v) Na2CO3 was added and the volume was made up to 10 mL with water. The samples were incubated at 40 °C for 2 h. The absorbance of the sample and standard were measured at 760 nm, using a UV-Vis spectrophotometer. The total polyphenolic content was calculated on the basis of the standard curve for gallic acid and expressed as mg of gallic acid equivalents (GAE) per g dry material plant (mg GAE/g plant material). The calibration curves (five data points) were linear with R2 = 0.999.The phenolic composition was determined by the LC-MS (liquid chromatography-mass spectrometry) analysis according to our published article.17
Cell culture
Human hepato cellular carcinoma HepG-2 cell lines and RAW 264.7 murine macrophages from the cell bank of the Chinese Academy of Sciences (CAS, Shanghai, China) were cultured in plastic dishes containing DMEM supplemented with 10% FBS, penicillin/streptomycin in a 5% CO2 humidified atmosphere at 37 °C.Cytotoxicity
The cell viability was measured using MTT assay.18–21 Briefly, exponential growth phase HepG-2 cells and RAW 264.7 cells were seeded at a density of 6 × 103 cells per well in 96-well culture plates and treated with the various concentrations of polyphenols for 24 h. After incubation, the media solution was removed, and the cells were washed twice with phosphate-buffered saline (PBS). One hundred micro liters of media containing 0.1% MTT (0.5 mg mL−1) was added to each well and the cells were incubated for additional 4 h at 37 °C. Then the medium in each well were removed and replaced with 150 μL of dimethyl sulfoxide to dissolve the formazan crystals. The optical density was measured using an ELISA plate reader at 570 nm. Control cells were cultured with fresh medium only. The optical density of formazan generated by control cells was taken as 100% survival. Cell viability (%) = polyphenols (OD570)/control (OD570) × 100%.Cellular antioxidant activity (CAA)
The CAA of phenolic extracts was determined using the method of Wolfe & Liu (2007).22 Briefly, exponential growth phase HepG-2 cells with a density of 6 × 104 cells per mL were seeded in 96-well culture plates (100 μL per well) and incubated for 24 h. After incubation, the media solution was removed, and the cells were washed twice with PBS. Micro plates were treated in triplicate for 1 h with 100 μL of medium containing phenolic extracts plus 50 μM DCFH-DA. Certain wells were washed with 100 μL of PBS (PBS wash protocol) and certain wells were not washed (no PBS wash protocol). Then 600 μM ABAP in 100 μL of Hanks' Balanced Salt Solution (HBSS) was added to the cells, and the 96-well micro plate was placed in a Fluoroskan Ascent fluorescent plate reader (Soft Max systems, Molecular Devices, US) at 37 °C. Emission at 538 nm was measured after excitation at 485 nm every 5 min for 1 h. PBS wash protocol means that cells are pretreated with polyphenols before ABAP was added, and no PBS wash means that cells are co-treated with polyphenols and ABAP. Each plate included triplicate control and blank wells. Control wells contained cells treated with DCFH-DA and ABAP. Blank wells contained cells treated with DCFH-DA and HBSS. After the subtraction of the blank and the initial fluorescence values, the area under the fluorescence versus time curve was calculated to determine the CAA value at various concentrations of polyphenols using the following equation:23| CAA (units) = 1 − (∫SA/∫CA) |
The EC50 (median effective dose) of the polyphenols was calculated from the median effect plot of log(fa/fu) versus log(dose), where fa is the fraction affected by the treatment (CAA unit) and fu is the fraction unaffected by the treatment (1 – CAA unit). Results were expressed as μmol quercetin equivalents (QE) per 100 g of phenolic extracts.
Nitric oxide assay
Nitric oxide assay was determined using the Griess reagent system.20,24 RAW 264.7 cells were seeded at a density of 2.5 × 105 cells per well in 24-well plates and then incubated with or without LPS in the absence or presence of various concentrations of polyphenols for 24 h. The final concentration of LPS was 1 μg mL−1. After LPS-stimulation for 24 h, cell culture medium was collected to measure the amount of NO using Griess reagent, which was made by mixing equal volumes of 1% sulphanilamide in 5% phosphoric acid and 0.1% N-1-naphtylethylenediamine dihydrochloride in distilled water. In brief, one hundred microlitres of Griess reagent were added to an equal volume of cell culture medium in a 96-well plate and incubated for 10 min at room temperature, and then the absorbance was measured on a microplate reader at 540 nm using serial dilutions of sodium nitrite as a standard.25,26 Fresh culture medium was used as the blank in all experiments and dexamethasone was used as a positive drug in this assay. NO inhibition rate = [LPS (OD540) − sample (OD540)]/[LPS (OD540) − blank (OD540)] × 100%.Enzyme-linked immunosorbent assay (ELISA)
Tumour necrosis factor-α (TNF-α), interleukin-1β (IL-1β) and IL-6 levels in cell culture medium were determined by ELISA following the manufacturer's instructions. RAW 264.7 cells were seeded at 2.5 × 105 cells per well into 24-well plates. Cells were treated with or without LPS (1 μg mL−1) in the absence or presence of various concentrations of phenolic extracts for 24 h. After LPS-stimulation for 24 h, the supernatant was collected to test the levels of TNF-α, IL-1β and IL-6 using ELISA kits specific for mouse (R&D systems) according to the manufacturer's instructions.Quantitative real-time PCR analysis
For the analysis of mRNA expression by quantitative real-time PCR, RAW 264.7 cells were harvested after incubation with or without LPS (1 μg mL−1) in the absence or presence of different concentrations (100, 200, 300 μg mL−1) of polyphenols for 12 h. Total cellular RNA from RAW 264.7 cells was isolated using TRIzol Reagent kit following the manufacturer's instructions. RNA was reversely transcribed into cDNA using an RT-PCR kit with an oligo-adaptor primer. Quantification of mRNA levels was performed by “IQ5 Real-Time PCR Detection System” using the Light Cycler instrument with the Fast Start DNA Master SYBR Green I kit. The nucleotide sequences of the primers used were as follows: TNF-α (forward, 5′-GGG GAT TAT GGC TCA GGG TC-3′, reverse, 5′-CGA GGC TCC AGT GAA TTC GG-3′), iNOS (forward, 5′-CGG CAA ACA TGA CTT CAG GC-3′, reverse, 5′-GCA CAT CAA AGC GGC CAT AG-3′), IL-1β (forward, 5′-CCA TGG AAT CCG TGT CTT CCT-3′, reverse, 5′-GTC TTG GCC GAG GAC TAA GG-3′), IL-6 (forward, 5′-GTA CTC CAG AAG ACC AGA GG-3′, reverse, 5′-TGC TGG TGA CAA CCA CGG CC-3′), COX-2 (forward, 5′-CAG CAA ATC CTT GCT GTT CC-3′, reverse, 5′-TGG GCA AAG AAT GCA AAC ATC-3′) and GAPDH (forward, 5′-CAC TCA CGG CAA ATT CAA CGG CAC-3′, reverse, 5′-GAC TCC ACG ACA TAC TCA GCA-3′). Each sample was tested three times and a mean value was used for the determination of messenger RNA levels. GAPDH was used to normalize gene expression levels for further analysis. Following the normalization, the group data were calculated by formula 2−ΔΔCt and expressed as mean (SD) and differences in transcript levels between groups were analyzed by SPSS 17.0.27,28Western blotting
The culture medium was discarded and the remaining cells were harvested at the indicated conditions for western blot analysis as described previously.29,30 The cells were washed twice by ice-cold PBS and lysed with RIPA cell lysis buffer (50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1% Nonidet P-40, and 0.1% sodium dodecyl sulphate, SDS) supplemented with 5 μg mL−1 each of leupeptin and aprotinin for 30 min at 4 °C. The cell debris was removed by micro-centrifugation and the protein concentration was determined using a BCA protein assay kit according to the manufacturer's instruction. Equal amount of cellular protein were separated by 12% sodium dodecyl sulfate-polyacryl amide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene difluoride (PVDF) membranes, which were activated in methanol. The membranes were blocked using 5% skim milk for 1 h at room temperature and sequentially incubated overnight at 4 °C in primary antibodies against phospho-ERK, phospho-JNK, NF-κB and GAPDH. After washing with Tween 20/Tris–buffered saline (TTBS), the membranes were incubated with horseradish-peroxidase-conjugated secondary antibody for 2 h at room temperature, and then followed by ECL detection. GAPDH was used to normalize for protein loading and the intensity of the bands was determined using densitometric analysis.Statistical analysis
The results were expressed as mean values ± standard deviations of the three independent experiments. Statistical significance of differences between the mean values were calculated using one-way analysis of variance (ANOVA) followed by the Tukey post hoc test (P < 0.05). All analyses were carried out using SPSS 11.5 software.Results and discussion
The contents of polyphenols and phenolic composition
The content of total polyphenols was 187.86 mg GAE/g plant material using the Folin–Ciocalteu method. In addition, LC-MS analysis of polyphenols from I. latifolia revealed the presence of (1) quinic acid, (2) 3-caffeoylquinic acid, (3) 5-caffeoylquinic acid, (4) shikimic acid, (5) 4-caffeoylquinic acid, (6) rutin, (7) hyperoside, (8) 3,4-di-caffeoylquinic acid, (9) 3,5-di-caffeoylquinic acid and (10) 4,5-di-caffeoylquinic acid as shown in our previous published paper.17Cellular antioxidant activity
The cellular antioxidant activity of polyphenols from I. latifolia was measured using CAA assay. CAA assay is more physiologically relevant to biological systems and reflects the absorption, metabolism, and distribution of antioxidants at the cellular level.31 Initially, the cell viability experiment of HepG-2 cells was performed at 100–300 μg mL−1 concentration and there was no significant cytotoxicity (cell viability ≥ 96%) of polyphenols at the concentration up to 300 μg mL−1 (shown in Table 1). In this assay, PBS wash and no PBS wash protocols were both used to manage the cells after one hour of incubation. The EC50 and CAA values of polyphenols from I. latifolia in PBS wash and no PBS wash protocols are presented in Table 2. In the PBS wash protocol, EC50 and CAA values of polyphenols were 251.94 ± 6.18 μg mL−1 and 6871.42 ± 85.56 μmol QE/100 g phenolic extracts, respectively. Those compounds that are only loosely associated with the cell membrane can be removed by PBS wash protocols.32 In the no PBS wash protocol, EC50 and CAA values of polyphenols were 28.85 ± 0.68 μg mL−1 and 25161.61 ± 583.55 μmol QE/100 g phenolic extracts, respectively. EC50 is calculated from the linear regression of the median effect curve, and then converted into CAA values. The CAA and EC50 values are negatively correlated, and the lower level of EC50 means higher CAA activity.| Concentration (μg mL−1) | Cell viabilitya (%) | |
|---|---|---|
| HepG-2 cells | RAW 264.7 cells | |
| a All data were expressed as relative cell viability (%) compared with untreated group which cell viability was considered as 100%. Values are expressed as mean ± SD (n = 6). | ||
| 100 | 100.42 ± 1.98 | 100.10 ± 2.45 |
| 200 | 99.69 ± 2.15 | 98.11 ± 3.29 |
| 300 | 96.28 ± 2.68 | 97.39 ± 2.56 |
| EC50 (μg mL−1) | CAA (μmol QE/100 g phenolic extracts) | |
|---|---|---|
| PBS wash | 251.94 ± 6.18 | 6871.42 ± 85.56 |
| No PBS wash | 28.85 ± 0.68 | 25161.61 ± 583.55 |
Effect of polyphenols on LPS-induced NO production in macrophages
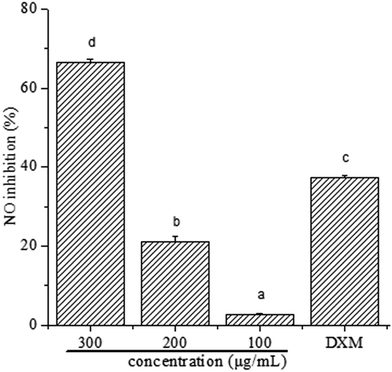
Macrophages are the important immune cells in mammals and produce great amount of NO after stimulated by LPS, which results in inflammation.33,34 To further study the activity of polyphenols against inflammation, treatment of LPS-induced RAW 264.7 macrophages with polyphenols was performed to determine its dosage-effect against NO production. Initially, the effect of polyphenols on the cell viability of RAW 264.7 cells was measured by a MTT assay and the data were shown as relative cell viability referred to control (equal to 100%). Results showed that polyphenols did not cause toxicity to the cells (cell viability ≥ 97%), implementing that the NO suppressive action of polyphenols at the highest concentration was not due to the cytotoxicity effect (Table 1).To determine whether polyphenols from I. latifolia can regulate inflammatory mediators in LPS-induced RAW 264.7 macrophages, NO production was determined in the culture medium using Griess reagent. The results (Fig. 1) were reported as NO inhibition rate means ± standard error measured from at least three independent experiments. Fig. 1 shows a concentration-dependent NO release inhibition rates of polyphenols, where NO release inhibition rates of polyphenols were 66.53 ± 0.87, 21.17 ± 1.42, 2.71 ± 0.29%, corresponding to the concentration of 300, 200 and 100 μg mL−1. Interestingly, NO release inhibition rate of polyphenols at the concentration of 300 μg mL−1 was found to exceed that of dexamethasone at the concentration of 50 μg mL−1, where the NO release inhibition rate is 37.30 ± 0.52%. Many previous literatures showed that phenolic extracts had good anti-inflammatory activity. After the treatment of polyphenols from Hibiscus sabdariffa leaves, the reduction of LPS-induced NO production dose-dependently in RAW 264.7 cells indicates the phenolic extracts' potential anti-inflammatory activity.35 Polyphenols extracted from chokeberry (Aronia melanocarpa (Michx.) Elliot) exhibited anti-inflammatory activity.1 Thus, it is possible to demonstrate that polyphenols from I. latifolia may be a promising candidate for anti-inflammation treatment. Previous research has reported that polyphenols from I. latifolia possesses anti-inflammatory activity, however there are few literature reports on its anti-inflammatory mechanism.12
Effects of polyphenols on pro-inflammatory cytokines (TNF-α, IL-1β, IL-6) production by ELISA
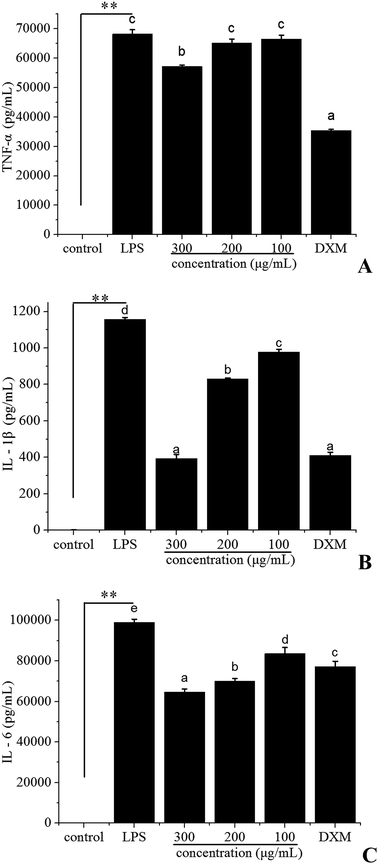
Macrophages play an important role in both the specific and non-specific immune responses, and they are a major source of various cytokines involved in inflammation, hematopoiesis and other homeostatic processes.36 Activation by inflammatory stimuli could produce many inflammatory mediators including NO and pro-inflammatory cytokines such as TNF-α, IL-1β and IL-6.37 However, if out of control, the inflammatory mediators could be involved in the pathogenesis of many inflammatory disorders. Therefore, the inhibition of the release of these inflammatory mediators is a crucial target in the treatment of inflammatory diseases.In order to test the possibility that polyphenols from I. latifolia might modulate the inflammatory response, the pro-inflammatory cytokines (TNF-α, IL-1β, IL-6) were evaluated by ELISA. The concentration of dexamethasone (DXM) was 50 μg mL−1. To reach detectable ranges of released pro-inflammatory cytokines in cultured media, LPS treatment was extended up to 24 h. After stimulation by LPS for 24 h, the production of TNF-α (Fig. 2A), IL-1β (Fig. 2B), IL-6 (Fig. 2C) was markedly increased. Polyphenols treatment (at the concentration of 300, 200, 100 μg mL−1) inhibited the release of pro-inflammatory cytokines induced by LPS in a dose-dependent manner (Fig. 2). Especially, the IL-1β level after treatment with the polyphenols at the concentration of 300 μg mL−1 was 394.01 ± 22.17 pg mL−1, which was very close to that of dexamethasone at the concentration of 50 μg mL−1 (Fig. 2B). Previous studies have demonstrated that polyphenols possess anti-inflammatory activity due to the suppression of the release of pro-inflammatory mediators.38 The above results suggested that polyphenols from I. latifolia suppressed the initial phase of the LPS-induced inflammatory response.
Effect of polyphenols on the involved mRNA expression
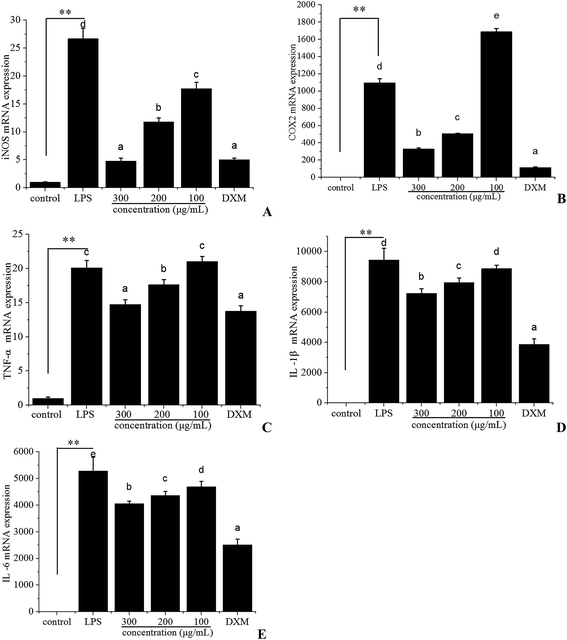
Nitric oxide production is a characteristic feature of activated macrophages. In macrophages, NO is primarily released by inducible nitric oxide synthase (iNOS), which is the enzyme produced by external stimulation, and high levels of this free radical could cause damage to a target tissue during an infection.39,40 Prostaglandin E2 (PGE2), an inflammatory mediator, exerts a crucial role during inflammation. The synthesis of PGE2 is dependent on cyclooxygenase-2 (COX-2), which is a prostaglandin-endoperoxide synthase enzyme.35 It is involved in inflammatory diseases.Because iNOS and COX-2 are the crucial enzymes for the production of NO and PGE2, respectively, we examined the mRNA expression level of iNOS and COX-2 in LPS-stimulated RAW 264.7 cells using quantitative real-time PCR to investigate the anti-inflammatory effect of polyphenols from I. latifolia. When RAW 264.7 cells were treated with LPS for 12 h, the iNOS (Fig. 3A) and COX-2 (Fig. 3B) mRNA expression were markedly increased; however, the mRNA expression of iNOS and COX-2 were significantly suppressed in a dose-dependent manner in LPS-stimulated RAW 264.7 cells treated with polyphenols from I. latifolia at a concentration of 300, 200 and 100 μg mL−1. Interestingly, Fig. 3A showed that polyphenols significantly inhibited the mRNA expression of iNOS at the concentration of 300 μg mL−1, which was very close to that of dexamethasone at the concentration of 50 μg mL−1, supporting that polyphenols from I. latifolia suppressed NO production by decreasing iNOS expression. Additionally, when RAW 264.7 cells were treated with polyphenols from I. latifolia at a concentration of 300 μg mL−1, COX-2 production was decreased by about 70%. Moreover, the effects of polyphenols from I. latifolia on the mRNA expression of pro-inflammatory cytokines including TNF-α (Fig. 3C), IL-1β (Fig. 3D), IL-6 (Fig. 3E) were further investigated. The mRNA expression of pro-inflammatory cytokines was up-regulated after LPS stimulation and was significantly down-regulated by polyphenols treatment in a concentration dependent manner. These results were consistent with the above outcomes obtained by ELISA. It was confirmed that polyphenols from I. latifolia exhibited strong anti-inflammatory effect.
Effects of polyphenols on the MAPKs pathway and NF-κB signal pathway in LPS-stimulated RAW 264.7 macrophages
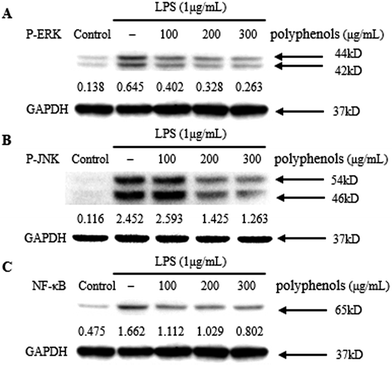
MAPKs play an important role in the signal transduction pathways that appear to be critical in inflammatory processes.20 JNK signaling regulates the expression of iNOS, whereas ERK signaling up-regulate iNOS expression and the production of pro-inflammatory cytokines including TNF-α and IL-6 in LPS-stimulated macrophages.41 Therefore, the effect of polyphenols from I. latifolia on the LPS-induced MAPK phosphorylation was determined to investigate whether the MAPK pathways are involved in the inhibition of inflammatory mediators by polyphenols. As shown in Fig. 4A and B, LPS treatment for 1 h significantly induced the phosphorylation of ERK and JNK compared to un-stimulated cells. Three hours of pre-treatment with polyphenols from I. latifolia (100–300 μg mL−1) significantly suppressed the LPS-induced phosphorylation of ERK and JNK in a concentration dependent manner. The experimental results are in accordance with published reports. Our previous report has shown that phosphorylation of MAPKs (ERK and JNK) was significantly inhibited by flavonoid glycosides from Rubus chingii Hu fruits in LPS-stimulated RAW 264.7 macrophages.20 These data indicate that the inhibition of inflammatory mediators by polyphenols from I. latifolia occurs probably through the inhibitions of the ERK and JNK activation.As NF-κB regulates the gene expression of pro-inflammatory cytokines and COX-2, it is a good target for treating inflammatory diseases. Moreover, many anti-inflammatory ingredients exert their effects by suppressing the NF-κB signaling pathway.41 Therefore, the effect of polyphenols (100–300 μg mL−1) on the NF-κB pathway activated by LPS in the macrophage cell line RAW 264.7 was investigated. RAW 264.7 cells were pre-incubated for 3 h with the polyphenols from I. latifolia and then stimulated with LPS (1 μg mL−1) for another 1 h. Although LPS treatment markedly promoted NF-κB activation in RAW 264.7 macrophages, the activation of NF-κB was inhibited by polyphenols treatment in a dose-dependent manner (Fig. 4C). These data suggested that polyphenols from I. latifolia might decrease NO, COX-2 and pro-inflammatory cytokines production via inhibitions of MAPKs and NF-κB activation. Interestingly, our previous study also showed that the polyphenols from I. latifolia exerted anti-atherosclerotic activity through suppressing NF-κB activation and phosphorylation of ERK1/2 in oxidized low-density lipoprotein (ox-LDL)-induced macrophages, indicating the specific role of the I. lactifolia polyphenols in the regulation of these two pathways.
Conclusion
In conclusion, polyphenols from I. latifolia have high antioxidant and anti-inflammatory activities. The CAA values of polyphenols in PBS wash and no PBS wash protocols were 6871.42 ± 85.56 and 25161.61 ± 583.55 μmol QE/100 g phenolic extracts, respectively. Moreover, the polyphenols from I. latifolia showed strong inhibition on LPS-induced NO-production in RAW 264.7 cells. The polyphenols could decrease the production of NO, COX-2 and pro-inflammatory cytokines via inhibitions of MAPKs (ERK and JNK) and NF-κB activation. Thus, the polyphenols enriched extracts from I. latifolia are good source of natural antioxidants with a beneficial effect against inflammation, and they may be applied as food supplement and/or functional ingredient. However, further research is needed to evaluate their anti-inflammatory activity in vivo.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This project was supported by the National Natural Foundation of China (31301453), Scientific Research Projects of the State Administration of traditional Chinese medicine (JDZX2015205); Guangdong Science and Technology Project (2013B090700015); Guangdong Provincial Science and Technology Department Project (2015B020211013)References
- K. Appel, P. Meiser, E. Millán, J. A. Collado, T. Rose, C. C. Gras, R. Carle and E. Muñoz, Fitoterapia, 2015, 105, 73–82 CrossRef CAS PubMed.
- C. M. Bergamini, S. Gambetti, A. Dondi and C. Cervellati, Curr. Pharm. Des., 2004, 10, 1611–1626 CrossRef CAS PubMed.
- W. Liao, Z. Ning, L. Ma, X. Yin, Q. Wei, E. Yuan, J. Yang and J. Ren, Rejuvenation Res., 2014, 17, 422–429 CrossRef CAS PubMed.
- B. Hoesel and J. A. Schmid, Mol. Cancer, 2013, 12, 1 CrossRef PubMed.
- T. T. Zhang, C. L. Lu and J. G. Jiang, J. Funct. Foods, 2015, 18, 423–431 CrossRef CAS.
- I. Kowalska, D. Jedrejek, L. Ciesla, L. Pecio, M. Masullo, S. Piacente, W. Oleszek and A. Stochmal, J. Agric. Food Chem., 2013, 61, 4417–4423 CrossRef CAS PubMed.
- A. Panusa, A. Zuorro, R. Lavecchia, G. Marrosu and R. Petrucci, J. Agric. Food Chem., 2013, 61, 4162–4168 CrossRef CAS PubMed.
- R. Ilahy, G. Piro, I. Tlili, A. Riahi, R. Sihem, I. Ouerghi, C. Hdider and M. S. Lenucci, Food Funct., 2016, 7, 574–583 CAS.
- K. Ogawa, S. Hirose, S. Nagaoka and E. Yanase, J. Agric. Food Chem., 2016, 64, 204–209 CrossRef CAS PubMed.
- L. Liu, Y. Sun, T. Laura, X. Liang, H. Ye and X. Zeng, Food Chem., 2009, 112, 35–41 CrossRef CAS.
- T. Hu, X. W. He, J. G. Jiang and X. L. Xu, Food Funct., 2014, 5, 876–881 CAS.
- T. Hu, X. W. He and J.-G. Jiang, J. Agric. Food Chem., 2014, 62, 8608–8615 CrossRef CAS PubMed.
- J. Zheng, L. Tang, X.-D. Xian, S. X. Zhou, H. M. Shi, Y. Jiang, Y.-Q. Gu, G. Liu and P.-F. Tu, Planta Med., 2009, 75, 1410–1414 CrossRef CAS PubMed.
- P. Izadiyan and B. Hemmateenejad, Food Chem., 2016, 190, 864–870 CrossRef CAS PubMed.
- G. Picariello, V. De Vito, P. Ferranti, M. Paolucci and M. G. Volpe, J. Food Compos. Anal., 2016, 45, 50–57 CrossRef CAS.
- E. A. Abourashed, C. L. A. Roberson and N. Elsharkawy, J. Diet. Suppl., 2016, 13, 171–184 CrossRef CAS PubMed.
- T. T. Zhang, C. Y. Zheng, T. Hu, J. G. Jiang, J. W. Zhao and W. Zhu, MedChemComm, 2018 10.1039/c7md00477j.
- T. T. Zhang, X. L. Xu, M. H. Jiang and J. G. Jiang, Food Funct., 2013, 4, 1581–1585 CAS.
- T. T. Zhang, L. Yang and J. G. Jiang, Food Funct., 2015, 6, 2205–2214 CAS.
- T. T. Zhang, M. Wang, L. Yang, J. G. Jiang, J. W. Zhao and W. Zhu, J. Funct. Foods, 2015, 18, 235–243 CrossRef CAS.
- T. T. Zhang, L. Yang and J. G. Jiang, J. Funct. Foods, 2015, 19, 575–583 CrossRef CAS.
- K. L. Wolfe and R. H. Liu, J. Agric. Food Chem., 2007, 55, 8896–8907 CrossRef CAS PubMed.
- W. Liao, Z. Ning, L. Chen, Q. Wei, E. Yuan, J. Yang and J. Ren, J. Agric. Food Chem., 2014, 62, 8648–8654 CrossRef CAS PubMed.
- T. T. Zhang, C. L. Lu, J. G. Jiang, M. Wang, D. M. Wang and W. Zhu, Carbohydr. Polym., 2015, 130, 307–315 CrossRef CAS PubMed.
- D. Gunawardena, N. Karunaweera, S. Lee, F. van Der Kooy, D. G. Harman, R. Raju, L. Bennett, E. Gyengesi, N. J. Sucher and G. Münch, Food Funct., 2015, 6, 910–919 CAS.
- S. Hooshmand, A. Kumar, J. Y. Zhang, S. A. Johnson, S. C. Chai and B. H. Arjmandi, Food Funct., 2015, 6, 1719–1725 CAS.
- T. T. Zhang, L. Yang and J. G. Jiang, Food Funct., 2015, 6, 2588–2597 CAS.
- A. Filipek, M. E. Czerwińska, A. K. Kiss, M. Wrzosek and M. Naruszewicz, Phytomedicine, 2015, 22, 1255–1261 CrossRef CAS PubMed.
- K. Srisook, E. Srisook, W. Nachaiyo, M. Chan-In, J. Thongbai, K. Wongyoo, S. Chawsuanthong, K. Wannasri, S. Intasuwan and K. Watcharanawee, J. Ethnopharmacol., 2015, 165, 94–102 CrossRef CAS PubMed.
- N. M. Elsherbiny, S. Ahmad, M. Naime, A. M. Elsherbini, S. Fulzele, M. M. Al-Gayyar, L. A. Eissa, M. M. El-Shishtawy and G. I. Liou, Life Sci., 2013, 93, 78–88 CrossRef CAS PubMed.
- Y. Chen, G. Chen, X. Fu and R. H. Liu, J. Agric. Food Chem., 2014, 63, 169–176 CrossRef PubMed.
- Y. Chen, G. Wang, H. Wang, C. Cheng, G. Zang, X. Guo and R. H. Liu, PLoS One, 2014, 9, e108140 Search PubMed.
- S. Ahn, M. H. Siddiqi, H. Y. Noh, Y. J. Kim, Y. J. Kim, C. G. Jin and D. C. Yang, Sci. Bull., 2015, 60, 773–784 CrossRef CAS.
- N. K. Im, Y. S. Jung, J. H. Choi, M. H. Yu and G. S. Jeong, Nat. Prod. Sci., 2014, 20, 51–57 CAS.
- J. Zhen, T. S. Villani, Y. Guo, Y. Qi, K. Chin, M.-H. Pan, C. T. Ho, J. E. Simon and Q. Wu, Food Chem., 2016, 190, 673–680 CrossRef CAS PubMed.
- M. S. Lee, M. S. Kwon, J. W. Choi, T. Shin, H. K. No, J. S. Choi, D. S. Byun, J.-I. Kim and H.-R. Kim, J. Agric. Food Chem., 2012, 60, 9120–9129 CrossRef CAS PubMed.
- B. B. Aggarwal, S. C. Gupta and B. Sung, Br. J. Pharmacol., 2013, 169, 1672–1692 CrossRef CAS PubMed.
- G. Costa, J. P. Ferreira, C. Vitorino, M. E. Pina, J. J. Sousa, I. V. Figueiredo and M. T. Batista, J. Ethnopharmacol., 2016, 178, 222–228 CrossRef CAS PubMed.
- R. Sarkar, R. Roy and A. N. Chaudhuri, Int. J. Curr. Microbiol. Appl. Sci., 2015, 4 Search PubMed.
- Y. Park, S. A. Yoo, W.-u. Kim, C.-S. Cho and C.-H. Yoon, J. Immunol., 2015, 194, 48.22 Search PubMed.
- S. Hirai, S. Horii, Y. Matsuzaki, S. Ono, Y. Shimmura, K. Sato and Y. Egashira, Life Sci., 2014, 117, 1–6 CrossRef CAS PubMed.
Footnote |
| † These authors contributed to the work equally and should be regarded as co-first authors. |
| This journal is © The Royal Society of Chemistry 2018 |