Open Access Article
Open Access ArticleYixin-Shu facilitated cardiac-like differentiation of mesenchymal stem cells in vitro†
Jingjing Zhang‡
 a,
Feifei Guo‡a,
Hongwei Wua,
Junying Weia,
Minghua Xian
a,
Feifei Guo‡a,
Hongwei Wua,
Junying Weia,
Minghua Xian a,
Fangfang Fanb,
Shihuan Tanga,
Ye Zhaoa,
Yi Zhanga,
Defeng Lia and
Hongjun Yang*a
a,
Fangfang Fanb,
Shihuan Tanga,
Ye Zhaoa,
Yi Zhanga,
Defeng Lia and
Hongjun Yang*a
aInstitute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing 100700, China. E-mail: hongjun0420@vip.sina.com; Fax: +86-10-64013996; Tel: +86-10-64032656
bCollege of Ethnic Medicine, Chengdu University of TCM, Chengdu 610072, China
First published on 12th March 2018
Abstract
Yixin-Shu capsules (YXS) are a Chinese Materia Medica standardized product used for heart disease and their effectiveness has been demonstrated through both clinical and experimental research. However, the mechanism involved has remained unclear. The effect of YXS on the cardiac-like differentiation of mesenchymal stem cells was investigated in this study. The intestinal absorption liquid of YXS was prepared using an in vitro intestinal absorption method and 62 compounds have been identified. A compound-target-function network constructed by a network pharmacology-based approach indicated that these compounds had an effect on cell differentiation. The effects of YXS on cardiac-like differentiation of mesenchymal stem cells was verified by detecting cardiac-specific protein expression such as α-actinin, cardiac troponin-I and desmin through real time-PCR, western blotting and immunofluorescence staining. A network pharmacology analysis indicated that the facilitation of YXS on the cardiac-like differentiation may be through the TGF-β signaling pathway, Wnt signaling pathway and MAPK signaling pathway. The observed improvements on cardiac differentiation may be due to the novel molecular mechanism for YXS that could also benefit developments in cardiac tissue engineering.
Introduction
Myocardial ischemia and myocardial infarction typically result in a loss of a large number of cardiomyocytes and produce local scar tissue due to the limited self-repair abilities, which eventually leads to heart failure.1 It is difficult to recover a sufficient number of cardiomyocytes (heart cells are “non-renewable cells”) to restore normal heart function. The application of stem cell technologies is a promising approach to prevent heart failure and facilitate cardiac remodelling and reserve function.2,3 Mesenchymal stem cells (MSCs) are unique pluripotent stem cells with self-renewal and differentiation abilities, which are able to differentiate into several specific cell types, such as chondrocytes, osteoblasts, adipocytes, and myocytes.4 Additionally, as circulating cells in the body, MSCs can reach damaged sites through spontaneous mobilization.5 Compared with other stem cells, MSCs have many advantages, including ease access from various tissues, high proliferation capacity, multiple differentiation potentials, lack of immune rejection, and inhibition of immune response, which has attracted much attention.4In 1999, Makino was first to demonstrate that MSCs had the ability to differentiate into cardiomyocytes under the stimulation of 5-azacytidine in vitro.6 Animal experiments further demonstrated that transplanted bone marrow mesenchymal stem cells could reach injured sites of the heart, differentiate into cardiomyocytes, and significantly improve cardiac function.7,8 Apart from the cardiomyocyte differentiation, MSCs can also differentiate into endothelial cells and smooth muscle cells to promote angiogenesis, attenuate inflammation,9 improve myocardial ischemia and replace damaged myocardium.10 In addition, MSCs can secrete growth factors such as vascular endothelial growth factor (VEGF) and basic fibroblast growth factor (bFGF), which could facilitate angiogenesis, decrease cardiomyocyte apoptosis, and enhance cardiac compliance.11,12 In the United States, clinical trials confirmed that intracoronary injection of MSCs could improve the quality of life in chronic heart failure patients and reduce mortality.13 Thus, the application of stem cells is a promising approach to prevent heart failure and facilitate cardiac reserve function.
Recent studies indicate that traditional Chinese medicine has a promising effect on stem cell differentiation and cell homing. For example, ginsenoside-Rg1 mediates microenvironment-dependent endothelial differentiation of human mesenchymal stem cells in vitro.14 When combined with bone marrow MSCs transplantation, ginsenoside Rg1 decreased cell apoptosis and facilitated the differentiation of MSCs into neurons and glial cells.15 Astragalus promoted neuron-like differentiation of MSCs.16 Moreover, tanshinone IIA17 alone or combined with astragaloside IV18 enhanced MSCs recruitment to regions of myocardial infarction by activating stromal cell-derived factor-1/CXC chemokine receptor 4 axis. Importantly, these chemical compounds, including ginsenoside Rg1, Astragalus, tanshinone IIA and astragaloside IV, are the main components of Yixin-Shu capsules (Yixin-Shu), a Chinese Materia Medica standardized product approved to treat coronary heart disease,19,20 indicating that YXS capsules may have an effect on stem cell.
Yixin-Shu capsules (YXS), developed from a famous old formula of Sheng-Mai-San that was recorded in a traditional herbal text written by Yi-Xue-Qi-Yuan from the Jin dynasty (AD 1127-1279), are composed of seven herbal medicines, including Radix Ginseng, Radix Ophiopogonis, Fructus Schisandrae Chinensis, Salvia miltiorrhiza, Astragalus membranaceus, Fructus Crataegi and Ligusticum Wallichii. As a well-known traditional Chinese recipe, Yixin-Shu in the formula of pills, tablets, capsules, granule preparations was used for chest pain, chest tightness, palpitation, shortness of breath and cyanosis, which is typical symptom of heart disease in modern medicine.21 Clinically, obvious therapeutic effect of YXS has been achieved when used for angina pectoris and coronary heart disease.22–24 However, the mechanism of YXS used for heart disease remained unclear. Recent studies indicated that YXS reduced myocardial ischemia/reperfusion injury through inhibition of oxidative stress and mitochondrial-mediated apoptosis20 and also inhibited myocardial dysfunction in a pluripotent stem cell (iPS)-derived cardiomyocytes.24 In this study, YXS intestinal absorption liquid was prepared by intestinal absorption in vitro. The intestinal absorption liquids were prepared through mimicking natural absorption process of medicine with valgus sausage sac and the ingredients that were absorbed through the intestines may be the real effective ingredients.24 A compound-target-function network was then constructed by a network pharmacology-based strategy. Finally, the observed function of the YXS intestinal absorption liquid was further verified with additional experiments. The possible mechanism of YXS on cardiac-like differentiation was analysed by a network pharmacology approach.
Materials and methods
Animals
Male Sprague-Dawley rats were purchased from the Experimental Animal Center of Peking University Health Science Center, Beijing, China [certificate no. SCXK (Jing) 2009-0017]. This study was approved by the Laboratory Animal Management Committee of Chinese Academy of Medical Science's Administrative Panel on Laboratory Animal Care and implemented according to the organizational guidelines and ethics of the Committee of Chinese Academy of Medical Sciences (February 1, 2016).Preparation and quality control of YXS capsules
Yixin-Shu capsule was purchased from Guizhou Xinbang Pharmaceutical Co., Ltd (Guiyang, China) (drug approval number: Z52020038, batch number: 20140810). Panax ginseng (200 g) was crushed into fine powder. Schisandra chinensis (133 g) and Salviae miltiorrhiza Radix (267 g) was extracted with 85% ethanol for two times with the first for 3 hours and the second time for 1.5 hours, then combined the filtered solution and then harvested the ethanol and concentrate it to relative density of 1.25 to 1.30 at 80 °C. Astragalus membranaceous (200 g), Crataegus pinnatifida (200 g), Ophiopogon japonicas (200 g) and Ligusticum chuanxiong Hort (133 g) were extracted with water for two times with the first for 2.5 h and the second for 1.5 h. After that combined the filtered solution and concentrated to relative density of 1.10–1.15 at 80 °C, then mixed with equal amount of 85% ethanol. The solution was filtered and concentrated before the addition of the extract of Schisandra chinensis and Salvia miltiorrhiza Radix, the powder of Panax ginseng and starch. The solution was mixed well, dried and then smashed into fine powder to make 1000 capsules.21 As for the quality control of YXS capsule, total content of ginsenoside Re and Rg1 were not less than 0.4 mg per capsule and salvianolic acid B was not less than 1.0 mg per capsule when determined by HPLC according to “Chinese Pharmacopoeia”. The quality control data of Yixin-Shu capsules was provided as ESI.†Preparation of YXS intestinal absorption liquid
The YXS solution (5% g ml−1, w/v) was prepared by dissolving YXS capsule powder into ethanol (1000 ml, 95% v/v) which was then heated until boiling for 2 hours under reflux before being filtered. The filtrate was rotary evaporated to dryness at 65 °C. The YXS extraction solution (16%, w/v) was obtained by adding Tyrode buffer solution (NaCl 8.00 g, KCl 0.28 g, MgCl2 0.10 g, CaCl2 0.20 g, NaHCO3 1.00 g, NaH2PO4 0.05 g, glucose 1.00 g, pH 7.4). Adult male Sprague-Dawley rats (220 ± 10 g) were maintained under fasting conditions for 12 h before the experiment. Intestines were harvested and cut into four 14 cm segments in Tyrode buffer solution at 0 °C. Each segment was ligated to form a sac at one end after being turned inside-out. The sac was filled with Tyrode buffer and then exchanged with YXS solution (16%, w/v), which was maintained at 37 °C and continuously injected with O2/CO2 (95%/5%). After 2 h, the sacs containing absorbed materials were filtered with a 0.22 μm microfiltrate membrane and then stored at −20 °C for further experiments. The original concentration of the YXS intestinal absorption liquid was 2 mg ml−1 (crude drug).Prediction of pharmacological effects based on a network pharmacology strategy
Structural information (*.inchi) of 62 identified compounds (including 75 possible chemical structures) for YXS were obtained based on metabolomics profiling analysis.24 The structural data (*.inchi) is in the ESI.† Target predictions of the YXS chemical compounds were executed by BATMAN-TCM, which is a web service for discovering TCM therapeutic mechanisms.25 In BATMAN-TCM, the target prediction method ranks potential drug–target interactions based on their similarity to known drug–target interactions that have been previously published and validated. Three types of validation methods, including “leave-one-interaction-out” cross-validation, “leave-one-drug-out” cross-validation and validation of the independent test set, were applied to measure the performance of BATMAN-TCM: in total, 517 potential targets of YXS were predicted with prediction scores larger than 20. Enrichment analysis of potential targets was based on the hyper-geometric cumulative distribution test, and the multiple testing corrections were based on the Benjamini–Hochberg correction method. The“compound-target-function” network consists of YXS compounds, potential targets, related function and interaction of drug–target and target-function. KEGG signaling pathways related to cell differentiation and migration were investigated using pathway enrichment analysis tool of DAVID. Only significantly enriched pathways were selected (p-value ≤ 0.05 by hyper-geometric cumulative distribution test).Cell culture, MTT assay, F-actin staining
MSCs were harvested from the bone marrow of Sprague-Dawley rats (male, 4 week old). Briefly, bone marrow aspirate was harvested by flushing the rat femurs and tibias before centrifugation and subsequent culturing in LG-DMEM medium containing 10% fetal bovine serum (FBS) at 37 °C in a 5% CO2 atmosphere. The non-adherent cells were removed and the culture media was replaced every 3 days. When 70–80% confluence was reached, adherent cells were trypsinized and further expanded. The MSCs at passage 3-5 were used for the following experiments.3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT, M2128; Sigma) assay was used to search for the optimum YXS concentration. Briefly, five thousand MSCs were seeded into 96-well culture plates. After culturing for 24 and 48 h, 100 ml of MTT solution was added and the cells were incubated for 3 h before dissolved by dimethyl sulfoxide (DMSO) to detect signal at 570 nm using a microplate reader (Molecular Devices, USA). Cell proliferation was also evaluated with MTT after 1, 3 and 8 days. There were five groups including the DMEM group (culture medium), control group (intestinal absorption liquid without YXS), the 5-Aza group (10 μM 5-aza for 24 h before being replaced by DMEM medium), the YXS group (62.5 μg ml−1 YXS intestinal absorption liquid), and the YXS + 5-Aza group (10 μM 5-aza for 24 h before treatment with 62.5 μg ml−1 YXS). Four replicates were used for each experiment. As for the F-actin staining, 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 MSCs were seeded into 6-well culture plates and the induction media was replaced every 3 days. After 3 and 28 days in culture, the samples were fixed with 4% paraformaldehyde for 30 min before being permeabilized in 0.1% Triton X-100. Rhodamine phalloidin (PHDR1, cytoskeleton) was applied for 30 min, followed by nuclear counter staining with 4,6-diamidino-2-phenylindole (DAPI; Sigma, USA) for 10 min. The stained samples were observed under the CLSM (LSM510; Zeiss).
000 MSCs were seeded into 6-well culture plates and the induction media was replaced every 3 days. After 3 and 28 days in culture, the samples were fixed with 4% paraformaldehyde for 30 min before being permeabilized in 0.1% Triton X-100. Rhodamine phalloidin (PHDR1, cytoskeleton) was applied for 30 min, followed by nuclear counter staining with 4,6-diamidino-2-phenylindole (DAPI; Sigma, USA) for 10 min. The stained samples were observed under the CLSM (LSM510; Zeiss).
Western blotting, real-time PCR and immunofluorescent staining
As for the western blotting, real-time PCR and immunofluorescent staining experiment, 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 MSCs were seeded into 6-well culture plates and the induction media was replaced every 3 days. After 28 days in culture, the samples were lysed in RIPA lysis buffer (R0020; Solarbio) with fresh protease inhibitor 0.1% phenylmethanesulfonyl fluoride (PMSF; Solarbio). The total cell lysate was boiled after mixing with 4% sodium dodecyl sulfonate (SDS) loading buffer (P1015; Solarbio, People's Republic of China). Protein concentration was determined using the BCA Protein Quantification Kit (ab102536). Equal amounts of protein (30 μg for each slot) were added to the SDS-PAGE gels to perform western blotting according to the manufacturers' protocol. Antibodies used for this experiment are as follows: cTnI (ab47003, Abcam), and α-actinin (A7811, Sigma). Horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibodies were loaded before detected by the ChemiDoc XRS + Molecular Imager (XRS: X-ray Spectrometer; Bio-Rad, USA) and then quantified using the Quantity One image software (Bio-Rad).
000 MSCs were seeded into 6-well culture plates and the induction media was replaced every 3 days. After 28 days in culture, the samples were lysed in RIPA lysis buffer (R0020; Solarbio) with fresh protease inhibitor 0.1% phenylmethanesulfonyl fluoride (PMSF; Solarbio). The total cell lysate was boiled after mixing with 4% sodium dodecyl sulfonate (SDS) loading buffer (P1015; Solarbio, People's Republic of China). Protein concentration was determined using the BCA Protein Quantification Kit (ab102536). Equal amounts of protein (30 μg for each slot) were added to the SDS-PAGE gels to perform western blotting according to the manufacturers' protocol. Antibodies used for this experiment are as follows: cTnI (ab47003, Abcam), and α-actinin (A7811, Sigma). Horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibodies were loaded before detected by the ChemiDoc XRS + Molecular Imager (XRS: X-ray Spectrometer; Bio-Rad, USA) and then quantified using the Quantity One image software (Bio-Rad).
Samples were fixed in 4% paraformaldehyde at room temperature for 30 min before permeabilization with 0.5% Triton X-100 for 15 min. After blocking with 1% bovine serum albumin containing 10% donkey serum for 1 h, the samples were incubated with various primary antibodies at 4 °C overnight. After thorough washing, secondary antibodies conjugated with Alexa Fluor 555 or Alexa Fluor 488 was incubated for 1 h. Nuclei were stained with DAPI (10 μg ml−1, w/v). Images were captured with an Olympus IX81-FV1000 microscope. Antibodies used in this study are as follows: cTnI (ab47003, Abcam), α-actinin (A7811, sigma), desmin (sc-14026, Santa Cruz) and Cx43 (sc-9059, Santa Cruz). Images were captured using a confocal microscope.
Trizol (15596-026; Invitrogen) was used to extract RNA from induced cells at day 14 and day 28 following the manufacturer's instructions. cDNA was synthesized with 500 ng RNA according to the instructions of the iScriptTM cDNA synthesis kit (Bio-Rad). The PCR experiment was performed using the Applied Biosystem 7500 Real-Time PCR System (Applied Biosystem) with Power SYBR Green PCR Master Mix (Applied Biosystem, USA) according to standard protocols. The target genes of α-actinin, cTnI and Cx43 were normalized by the reference gene β-actin. The sequences of the primers were listed in Table 1.
| Target primer | Sequence forward and reverse (from 5′ to 3′) |
|---|---|
| β-Actin | TCATGAAGTGTGACGTTGACATCCGT |
| CCTAGAAGCATTTGCGGTGCACGATG | |
| cTnI | ATGACCTGCGTGGCAAGTTTAA |
| TTCTCAATGTCCTCCTTCTTCACC | |
| α-Actinin | CCGAGATCTCACCGACTACC |
| TCCAGAGCGACATAGCACAG | |
| Cx43 | GGTGGGCACAGACACGAATAT |
| CTCAACAACCTGGCTGCGAAA |
Statistical analysis
SPSS V17.0 was used to analyze the data [one-way analysis of variance (ANOVA), LSD (least-significant difference, p < 0.05)]. The data are expressed as the mean ± standard deviation with significant p values, *p < 0.05.Results
The targets of YXS enriched in cell differentiation
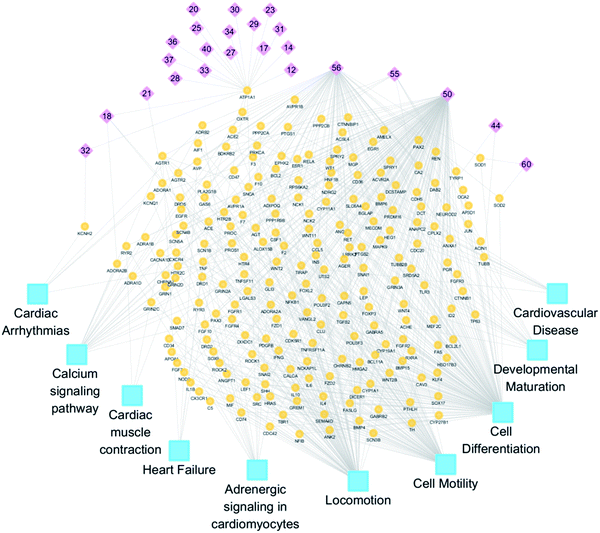
The intestinal absorption liquid of YXS was collected by turning rat intestines inside-out at 37 °C for 2 hours, which allowed for absorption by small intestine villi and intestinal barrier penetration. And 62 compounds have been identified in intestinal absorption liquid of YXS (ESI Fig. S1 and Table S1†).24 There were 517 potential targets that were predicted to bind with the 62 compounds found in the YXS intestinal absorption liquid by BATMAN-TCM. Functional enrichment of potential targets was used to discover the biological processes related to YXS. The enrichment results showed that potential targets of YXS tended to participate in biological processes related to cell movement and differentiation and pathways related to heart failure (e.g., cardiac arrhythmias, calcium signaling pathway, cardiac muscle contraction, and adrenergic signaling in cardiomyocytes; Fig. 1). As shown in Table 2, the potential targets of the identified 62 compounds were obviously enriched in gene ontology (GO) such as cell differentiation (GO: 0030154) (p < 0.01).| Function term | Adjusted P-value | Mapped gene | Enrich ratio |
|---|---|---|---|
| Developmental maturation (GO:0021700) | 3.8 × 10−7 | 19 | 4.2 |
| Cell differentiation (GO:0030154) | 1.94 × 10−26 | 173 | 2.3 |
| Locomotion (GO:0033058) | 1.6 × 10−19 | 98 | 2.7 |
| Cell motility (GO:0048870) | 4.58 × 10−21 | 81 | 3.3 |
| Cardiovascular disease (TTD) | 3.59 × 10−2 | 13 | 2.3 |
| Heart failure (TTD) | 1.89 × 10−2 | 8 | 3.4 |
| Cardiac arrhythmias (TTD) | 4.95 × 10−5 | 7 | 7.9 |
| Calcium signaling pathway (HAS:04020) | 1.13 × 10−11 | 40 | 3.6 |
| Cardiac muscle contraction (HAS:04260) | 3.63 × 10−5 | 17 | 3.6 |
| Adrenergic signaling in cardiomyocytes (HAS:04261) | 2.03 × 10−3 | 21 | 2.3 |
Cell viability, proliferation and morphology
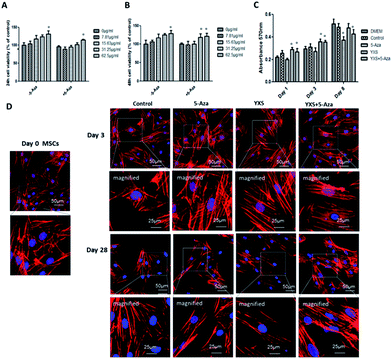
A dose-dependent increase in cell viability was observed when cells were treated with 0–62.5 μg ml−1 YXS (Fig. 2A), and the addition of 5-Aza didn't impair cell viability when compared with the 0 μg ml−1 group, indicating that 62.5 μg ml−1 YXS was the optimal concentration for the following experiments. Cell proliferation was increased by 62.5 μg ml−1 YXS treatments at day 1 and day 3 and showed no effects by day 8 (Fig. 2C). In contrast, the combination of 5-Aza and YXS (62.5 μg ml−1) didn't impair cell proliferation at day 1 and day 3, but was inhibited when compared with the control group at day 8 (Fig. 2C). The arrangement of actin filaments was analyzed at day 3 and day 28 with phalloidin-staining of F-actin, and nucleus-staining with DAPI. Confocal micrographs revealed that MSCs showed fibroblast morphologies and remained unchanged at day 3 (Fig. 2D). However, cells became elongated at day 28 in the YXS, 5-Aza and YXS+5-Aza groups when compared to the multilateral morphology of the control group.YXS facilitated cardiac-like differentiation of MSCs
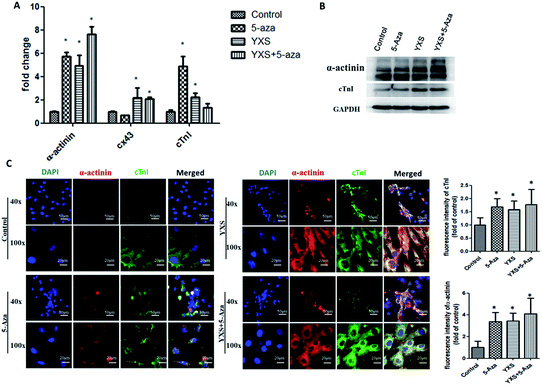
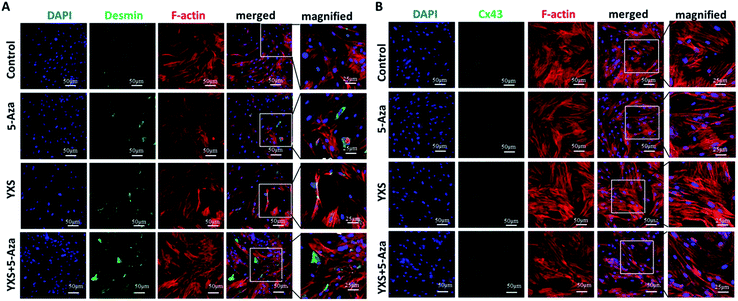
Consistent with previous reports, 5-Aza treatment enhanced cardiac-like differentiation of MSCs as indicated by the increased expression of α-actinin and cardiac troponin-I (cTnI) at both the transcription mRNA and protein levels (Fig. 3). YXS facilitated the gene expression of cardiac-specific proteins such as α-actinin, cardiac troponin-I (cTnI) and Cx43 at day 10 when compared to the control group (Fig. 3A). After 28 days, western blotting experiments confirmed that YXS alone or combination with 5-Aza enhanced the expression of α-actinin and cTnI when compared to the control group (Fig. 3B). As indicated by the immunofluorescent staining results (Fig. 3C and 4), treated cells were positive for cardiac-specific markers, such as α-actinin, cardiac troponin-I (cTnI) and desmin, but negative for Cx43. As shown in Fig. 3C, the fluorescence intensity of α-actinin and cTnI in YXS group was significantly higher than the control group (p < 0.05). However, there was no significance in the fluorescence intensity of α-actinin between YXS and 5-Aza, as well as the fluorescence intensity of cTnI. The combination of 5-Aza and YXS facilitated cardiac-like differentiation of MSCs as indicated by the increased fluorescence intensity of α-actinin and cTnI when compared to the control (p < 0.05), but demonstrated no significance when compared to either 5-Aza or YXS. These data indicated that YXS facilitated the cardiac-like differentiation of MSCs.Prediction analysis of pharmacological mechanism based on network pharmacology
To explain the mechanism of YXS on cardiac-like differentiation, network pharmacology was used to predict the potential signaling pathway involved (as shown in Table 3). Need to notice, MAPK signaling pathway, TGF-β signaling pathway and Wnt signaling pathway, which was found to play an important role in cardiac differentiation and development, were also enriched via pathway enrichment for analyzing the potential targets of YXS intestinal absorption liquid.| Term | Count | % | P-Value | Genes | Fold enrichment |
|---|---|---|---|---|---|
| hsa04010:MAPK signaling pathway | 36 | 5.7507 | 1.57 × 10−4 | FGFR2, MEF2C, FGFR1, FGFR4, HRAS, FGF7, FGFR3, TNF, PDGFB, FASLG, FGF10, NFKB1, TGFB2, AKT1, FOS, CDC42, IL1B, FAS, EGF, CHUK, MAP2K5, PRKCA, EGFR, RELA, CACNA1I, MECOM, PRKCB, RPS6KA2, ARRB1, JUN, CACNA1G, MAPK9, CACNA1C, IKBKB, CACNA1D, CACNA1B | 1.947164 |
| hsa04350:TGF-beta signaling pathway | 14 | 2.2364 | 0.0069 | BMP4, AMHR2, TNF, ROCK1, SMAD7, TGFB2, ACVR2A, ACVR1B, ID2, PPP2CA, IFNG, PPP2CB, BMP6, ACVR1 | 2.298736 |
| hsa04310:Wnt signaling pathway | 18 | 2.8753 | 0.0209 | PRKCA, ROCK2, CAMK2G, VANGL2, FZD1, LEF1, FZD2, PRKCB, CTNNB1, WNT2B, CTNNBIP1, WNT2, WNT4, JUN, CAMK2D, MAPK9, WNT11, SOX17 | 1.799011 |
Discussion
The limited self-repair ability of cardiomyocytes restricts heart repair and causes the formation of local scars in myocardial ischemia and myocardial infarction, which eventually leads to heart failure.1 The application of stem cell technologies is a promising approach to enhance heart repair by supplementing the number of cardiomyocytes.2,4 In this study, YXS intestinal absorption liquid was prepared using an in vitro intestinal absorption method. A compound-target-function network constructed by a network pharmacology-based strategy indicated that these 62 compounds had an effect on cell differentiation. Considering the use of YXS to treat coronary heart disease, the cardiac-like differentiation of MSCs was further verified which demonstrates that YXS facilitates cardiac-like differentiation of MSCs. Finally the potential signaling pathway of YXS on cardiac-like differentiation was analysed by a network pharmacology approach.In our study, YXS facilitated cardiac-like differentiation of MSCs in vitro. YXS, which contains seven herbs including Ginseng, Ophiopogon japonicus, Schisandra, Astragalus, Salvia, Chuanxiong, and Hawthorn, is a Chinese Materia Medica standardized product widely used in the treatment of heart disease.19,20 The pharmacological mechanism of YXS remained in the reduction of lactic dehydrogenase (LDH), serum creatine kinase (CK), aspartate aminotransferase (AST) and malondialdehyde (MDA), which improves blood viscosity, decreases myocardial infarction size, and reduces cell apoptosis as well as oxidative stress injury.20 In this study, we first reported that the intestinal absorption liquid of YXS could facilitate the differentiation of MSCs into cardiac-like cells, as evidenced by the enhancement of the cardiac-specific markers α-actinin, cTnI and desmin. Compared with traditional Chinese medicine monomers and single herbs, prescription YXS has been used to treat coronary heart disease and its safety and effectiveness have been fully validated. Previously, treatment of MSCs with DNA demethylating agent 5-Aza results in cardiomyocyte induction.6 Our result was in agreement with the observation that 5-Aza enhanced cardiac-like differentiation as evidenced by high gene expression and protein levels of cardiac-specific genes and proteins such as cTnI, α-actinin, and desmin. In addition, the removal of methylation from a transcriptionally inactive myogenic-determinant locus was the key mechanism employed by 5-Aza to induce the myogenic differentiation.26 However, 5-Aza induction was accompanied by significant toxicity, indicating serious problems in clinical safety that restrict its application.27 Thus, compared with 5-Aza induction, YXS can facilitate a cardiac-like differentiation in a safe manner.
Based on the network pharmacology analysis, MAPK signaling pathway, TGF-β signaling pathway and WNT signaling pathway was the potential signaling pathway involved in cardiac-like differentiation of YXS in our study (Table 3). The Wnt signaling pathway played a vital role on cardiac cardiogenesiss with early Wnt signaling promoting and later Wnt signaling inhibiting heart development.28 Need to notice, Wnt11 can enhance cardiomyocyte development through inhibition of canonical Wnt signaling29 and Wnt11 also promoted cardiac-like differentiation of MSCs.30 Inhibition of MAPK resulted in enhanced cardiac differentiation, for example, a small molecule SB203580, which was a p38 MAPK inhibitor, promoted cardiac differentiation of human embryonic stem cells.31 Recent study indicated that cardiac specific gene expression can be enhanced by TGF-β in c-kit + bone marrow cells, suggesting TGF-β signaling pathway was an key regulators in cardiomyogenic differentiation.32 Especially, TGF-β1 treatment promoted functional cardiomyocytes differentiation from cardiomyocyte progenitor cells in a high efficiency33 and TGF-β facilitated the immature cardiomyocytes differentiation from bone marrow stem cells.34
In this study, YXS intestinal absorption liquid was prepared using an in vitro intestinal absorption method and 62 compounds had been identified. The compound-target-function network constructed by a network pharmacology-based strategy implied that these 62 compounds had an effect on cell differentiation. The cardiac-like differentiation of MSCs was further verified by real time-PCR, immunofluorescence staining and western blotting experiments. Based on the network pharmacology analysis, MAPK signaling pathway, TGF-β signaling pathway and Wnt signaling pathway may play an important role in facilitating cardiac-like differentiation. This research offers a novel molecular mechanism supporting the application of YXS in heart disease treatments, and facilitates progress in cardiac tissue engineering.
Author contributions
Hongjun Yang and Jingjing Zhang conceived and designed the experiments. Jingjing Zhang, Hongwei Wu, Junying Wei, Minghua Xian and Fangfang Fan performed the experiments. Jingjing Zhang, and Feifei Guo wrote the manuscript. Jingjing Zhang and Feifei Guo performed the analysis. Shihuan Tang, Ye Zhao, Defeng Li, and Yi Zhang gave useful suggestions and revised the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors acknowledge support from National Natural Science Foundation of China (81603422, 81330086 and 81703951), the National Science and Technology Major Project (2014ZX09201021-009), National Program on Key Basic Research Project (2015CB554406), and National Key Technology R&D Program of China (No 2011BAI07B08).References
- D. H. Kwon, R. Hachamovitch, A. Adeniyi, B. Nutter, Z. B. Popovic, B. L. Wilkoff, M. Y. Desai, S. D. Flamm and T. Marwick, Heart, 2014, 100, 206–213 CrossRef PubMed.
- A. R. Williams, K. E. Hatzistergos, B. Addicott, F. McCall, D. Arvalho, V. Suncion, A. R. Morales, J. D. Silva, M. A. Sussman, A. W. Heldman and J. M. Hare, Circulation, 2013, 127, 213–223 CrossRef PubMed.
- B. Z. Cai, G. Wang, N. Chen, Y. J. Liu, K. Yin, C. P. Ning, X. D. Li, F. Yang, N. Wang, Y. Wang, Z. W. Pan and Y. J. Lua, J. Cell. Mol. Med., 2014, 18, 1407–1416 CrossRef CAS PubMed.
- M. F. Pittenger, A. M. Mackay, S. C. Beck, R. K. Jaiswal, R. Douglas, J. D. Mosca, M. A. Moorman, D. W. Simonetti, S. Craig and D. R. Marshak, Science, 1999, 284, 143–147 CrossRef CAS PubMed.
- L. Li, S. Z. Wu, P. J. Li, L. H. Zhuo, Y. H. Gao and Y. L. Xu, Cell Biochem. Biophys., 2015, 73, 749–757 CrossRef CAS PubMed.
- S. Makino, K. Fukuda, S. Miyoshi, F. Konishi, H. Kodama, J. Pan, M. Sano, T. Takahashi, S. Hori, H. Abe, J. Hata, A. Umezawa and S. Ogawa, J. Clin. Invest., 1999, 103, 697–705 CrossRef CAS PubMed.
- C. Toma, M. F. Pittenger, K. S. Cahill, B. J. Byrne and P. D. Kessler, Circulation, 2002, 105(1), 93–98 CrossRef PubMed.
- S. Tomita, R. K. Li, R. D. Weisel, D. A. G. Mickle, E. J. Kim, T. Sakai and Z. Q. Jia, Circulation, 1999, 100, II247–II256 CrossRef CAS PubMed.
- A. Martire, F. B. Bedada, S. Uchida, J. Pöling, M. Krüger, H. Warnecke, M. Richter, T. Kubin, S. Herold and T. Braun, Basic Res. Cardiol., 2016, 111, 54 CrossRef PubMed.
- A. R. Williams, B. Trachtenberg, D. L. Velazquez, I. McNiece, P. Altman, D. Rouy, A. M. Mendizabal, P. M. Pattany, G. A. Lopera, J. Fishman, J. P. Zambrano, A. W. Heldman and J. M. Hare, Circ. Res., 2011, 108, 792–796 CrossRef CAS PubMed.
- A. Singh, A. Singh and D. Sen, Cell. Physiol. Biochem., 2015, 35, 1219–1229 CrossRef PubMed.
- W. D. Dai, S. L. Hale and R. A. Kloner, Regener. Med., 2007, 2, 63–68 CrossRef PubMed.
- B. E. Strauer, M. Yousef and C. M. Schannwell, Eur. J. Heart Failure, 2010, 12, 721–729 CrossRef PubMed.
- W. He, W. K. Wu, Y. L. Wu, X. H. Yang, Q. X. Lin and W. H. Yu, J. Asian Nat. Prod. Res., 2011, 13, 1–11 CrossRef PubMed.
- C. Bao, Y. Wang, H. Min, M. Zhang, X. Du, R. Han and X. Liu, Cell. Physiol. Biochem., 2015, 37, 901–910 CrossRef CAS PubMed.
- J. F. Zhong, H. Cao, Z. H. Chen, F. Zhou and X. L. Tan, Neurosci. Lett., 2013, 553, 29–34 CrossRef CAS PubMed.
- Y. H. Tong, W. Xu, H. C. Han, Y. Chen, J. Yang, H. X. Qiao, D. S. Hong, Y. J. Wu and C. X. Zhou, Phytomedicine, 2011, 18, 443–450 CrossRef CAS PubMed.
- J. X. Huan, W. T. B. Song, Z. R. Wang, F. L. Jing, M. J. Chen, Y. Y. Nan, H. Yi and W. Wang, Protoplasma, 2013, 250, 521–530 CrossRef PubMed.
- Z. J. Li, Z. Y. Zhang, Y. Y. Li, S. Arnovitz, P. Chen, H. Huang, X. Jiang, G. M. Hong, R. B. Kunjamma, H. M. Ren, C. J. He, C. Z. Wang, A. G. Elkahloun, P. J. M. Valk, K. Döhner, M. B. Neilly, L. Bullinger, R. Delwel, B. Löwenberg, P. P. Liu, R. Morgan, J. D. Rowley, C. S. Yuan and J. J. Chen, Blood, 2013, 121, 1422–1431 CrossRef CAS PubMed.
- Y. C. Zhao, L. W. Xu, Z. Q. Qiao, L. C. Gao, S. Ding, X. Y. Ying, Y. Y. Su, N. Lin, B. He and J. Pu, Sci. Rep., 2016, 6, 23025 CrossRef CAS PubMed.
- C. P. Committee, China Pharmacopoeia, Chemical Industry Press, 2015, vol. 1, pp. 1397–1402 Search PubMed.
- H. Cheng, L. Zhai and X. Li, Chin. J. Pract. Chin. Mod. Med., 2004, 1621–1622 Search PubMed.
- Y. D. Zhang, R. T. Hou, L. J. Tang, H. M. Wang, J. L. Zhang and Y. T. Kang, Chin. J. Integr. Med., 2012, 10, 1414–1415 Search PubMed.
- M. Y. Zhang, H. W. Wu, F. F. Guo, Y. Y. Yu, J. Y. Wei, Y. Geng, S. F. Wang, S. Y. Li and H. J. Yang, Mol. BioSyst., 2017, 13, 1469–1480 RSC.
- Z. Y. Liu, F. F. Guo, Y. Wang, C. Li, X. L. Zhang, H. L. Li, L. H. Diao, J. Y. Gu, W. Wang, D. Li and F. C. He, Sci. Rep., 2016, 6, 21146 CrossRef CAS PubMed.
- S. F. Konieczny and C.P. Emerson Jr, Cell, 1984, 38, 791–800 CrossRef CAS PubMed.
- A. Bel, E. Messas, O. Agbulut, P. Richard, J. L. Samuel, P. Bruneval, A. A. Hagège and P. Menasché, Circulation, 2003, 108, II247–II252 CrossRef PubMed.
- X. J. Lian, C. Hsiao, G. Wilson, K. X. Zhu, L. B. Hazeltine, S. M. Azarin, K. K. Raval, J. H. Zhang, T. J. Kamp and S. P. Palecek, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E1848–E1857 CrossRef CAS PubMed.
- M. Abdul-Ghani, D. Dufort, R. Stiles, Y. D. Repentigny, R. Kothary and L. A. Megeney, Mol. Biol. Cell, 2011, 31, 163–178 CrossRef CAS PubMed.
- Z. S. He, H. X. Li, S. Zuo, Z. Pasha, Y. G. Wang, Y. T. Yang, W. P. Jiang, M. Ashraf and M. F. Xu, Stem Cells Dev., 2011, 20, 1771–1778 CrossRef CAS PubMed.
- R. Graichen, X. Q. Xu, S. R. Braam, T. Balakrishnan, S. Norfiza, S. Sieh, S. Y. Soo, S. C. Tham, C. Mummery, A. Colman, R. Zweigerdt and B. P. Davidson, Differentiation, 2008, 76, 357–370 CrossRef CAS PubMed.
- T. S. Li, M. Hayashi, H. Ito, A. Furutani, T. Murata, M. Matsuzaki and K. Hamano, Circulation, 2005, 111, 2438–2445 CrossRef CAS PubMed.
- M. J. Goumans, T. P. Boer, A. M. Smits, L. W. Laake, P. Vliet, C. H. G. Metz, T. H. Korfage, K. P. Kats, R. Hochstenbach, G. Pasterkamp, M. C. Verhaar, M. A. G. Heyden, D. Kleijn, C. L. Mummery, T. A. B. Veen, J. P. G. Sluijter and P. A. Doevendans, Stem Cell Res., 2007, 1, 138–149 CrossRef CAS PubMed.
- T. S. Li, T. Komota, M. Ohshima, S. L. Qin, M. K. K. Ueda and K. Hamano, Biochem. Biophys. Res. Commun., 2008, 266, 1074–1080 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra13326j |
| ‡ These authors contribute equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |