Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Mixed-solvent liquid exfoliated MoS2 NPs as peroxidase mimetics for colorimetric detection of H2O2 and glucose†
Yuehua Zhao‡
ab,
Yu Huang‡ *a,
Jiangling Wuc,
Xiaoli Zhand,
Yuanyang Xieab,
Dongyun Tanga,
Haiyan Cao
*a,
Jiangling Wuc,
Xiaoli Zhand,
Yuanyang Xieab,
Dongyun Tanga,
Haiyan Cao *e and
Wen Yun*f
*e and
Wen Yun*f
aChongqing Institute of Green and Intelligent Technology, Chinese Academy of Sciences, Chongqing, 400714, China. E-mail: huangyu@cigit.ac.cn
bUniversity of Chinese Academy of Science, Beijing, 100049, China
cDepartment of Clinical Laboratory, University-Town Hospital of Chongqing Medical University, Chongqing, 401331, China
dChongqing Traditional Chinese Medicine Hospital, Chongqing, 400021, China
eSchool of Chemistry and Chemical Engineering, Yangtze Normal University, 408100, Chongqing, China. E-mail: 513923170@qq.com
fChongqing Key Laboratory of Catalysis and Functional Organic Molecules, College of Environment and Resources, Chongqing Technology and Business University, Chongqing, 400067, China. E-mail: 44863542@qq.com
First published on 15th February 2018
Abstract
Ultra-small molybdenum disulfide nanoparticles (MoS2 NPs) were prepared by a facile liquid exfoliation method with ethanol/water as the solvent. The produced MoS2 NPs were of high purity due to the easily removable ethanol/water solution. The prepared MoS2 NPs exhibited an intrinsic peroxidase-like activity in analogy to that of horseradish peroxidase (HRP). A custom-made spectrometer was employed to investigate the peroxidase-like activity of MoS2 NPs in the presence of H2O2 and glucose. The change in absorption detected from MoS2 NPs is proportional to the amount of target. The calibration curve of H2O2 and glucose shows a good relationship between the concentration of target and the change in the absorption of MoS2 NPs. The limit of detection of H2O2 and glucose achieved by this method could approach 1.25 μM and 7 μM respectively. This method has been applied for the detection of glucose in serum from humans. Therefore, these produced MoS2 NPs offer an alternative high-efficiency and economic way to detect diabetes.
Introduction
The natural enzyme, horseradish peroxidase (HRP), is widely used for the detection of H2O2 due to its catalysis capability. The addition of a chromogenic substrate, such as 3,3′,5,5′-tetramethylbenzidine (TMB), is able to produce a color change in the presence of H2O2.1,2 Furthermore, glucose in serum could be oxidized to produce H2O2 under the catalysis of glucose oxidase (GOx). This produced H2O2 then reacts with HRP, which could be used to diagnose diabetes.3–5 However, high costs of preparation, purification and easy inactivation of HRP motivate people to seek alternative mimic enzymes as complementary peroxidase enzyme to sense glucose for clinical diagnosis.6–8 Many nanomaterials exhibit good peroxidase properties, such as Fe3O4,9 Co3O4,10 and Au NPs,11 Prussian blue,12 carbon nanodots,13 etc. Comparing to HRP, these peroxidase mimetics have advantages of lower cost, flexibility in design, and good chemical stability.Molybdenum disulfide (MoS2) is a typical layered transition metal dichalcogenides (TMDs) formed by a stack of S–Mo–S through weak van der Waals force.14,15 MoS2 has been regarded as one of the most promising materials due to its unique structure and electronic properties.16,17 An intrinsic peroxidase-like activity possessed by MoS2 nanomaterials has been densely studied in the past few years.18–23 However, most of these MoS2 nanomaterials were synthesized by hydrothermal method,19–21 which required high temperature and pressure. A few MoS2 nanomaterials were prepared by liquid exfoliation,18,22,23 but the organic solvent and surfactant were difficult to remove and the residual could affect the peroxidase performance. Zhou et al. reported that an alternative way to synthesis MoS2 nanosheet,24 which could avoid the drawbacks inhered from the above-mentioned methods. Jia et al. modified the method introduced by Zhou and successfully fabricated MoS2 nanoplates.25
Herein, a mixed-solvent liquid exfoliation method on the basis of Jia's has been introduced to synthesis MoS2 NPs. A custom made spectrometer has been established to systematically investigate the catalysis performance of MoS2 NPs to get the optimized parameters. The as-prepared MoS2 NPs exhibits good catalysis capability and the detection limit of H2O2 and glucose performed by this method could approach 1.25 μM and 7 μM respectively. A test of serum has been conducted by using this method and the results are comparable to that from commercial glucometer. This indicates the practicality of this prepared MoS2 NPs and its relative test method could be used as an alternative way for the diagnosis of diabetes. However, there is still much room for improvement, compared with these using high-precision glucometer. More fundamental insights could be provided to improve this flexible and economic method for clinical diagnosis.
Experimental section
Chemicals and materials
H2O2, sodium acetate and acetic acid were obtained from Chongqing Chemical Reagent Company (China). Horseradish peroxidase (HRP, >150 U mg−1), glucose, GOx, lactose, sucrose and fructose were obtained from Shanghai Sangon Biotech Co., Ltd (China). 3,3′,5,5′-Tetramethylbenzidine (TMB) were purchased from Merck & Co., Inc (USA). MoS2 powder (≥98%, 2 μm in size) were purchased from Tianjin Chemical Reagent Factory Kaida chemical plant. All chemicals and reagents were of analytical grade and used as received without further purification, and ultrapure water was used throughout the work. Ultrapure water was prepared in the lab using a water treatment device. Serum was kindly donated from the Department of Clinical Laboratory at the University-Town Hospital of Chongqing Medical University (Shapingba District, Chongqing Municipality).Apparatus
High-resolution transmission electron microscopy (HRTEM) images were carried out with an FEI Tecnai G2 F30 transmission electron microscope (USA). X-Ray photoelectron spectroscopy (XPS) data were recorded in a Thermo Fisher ESCALAB 250xi (England) using ALKα radiation (1486.6 eV). Binding energies were calculated with respect to C (1s) at 284.8 eV. Binding energies were measured with a precision of ±0.05 eV. X-Ray diffraction (XRD) spectra were obtained through a X′-Pert3 Powder X-ray diffractometer (PANalytical, Netherlands). Zeta potential was measured by Zetasizer Nano-ZS (Malvern, UK). The atomic force microscopy (AFM) imagines were performed on a Multi-mode Nanoscope III3d scanning probe microscopy system (Burker, USA). The pH of the solutions was detected by a PHS-3D pH meter (Shanghai Precision Scientific Instruments Co., Ltd., China).The custom made thermal controlled spectrometer is shown in Fig. 1. Drying oven is used to control the temperature of the cuvette's surrounding environment. Unpolarized light from a tungsten halogen lamp (HL-2000-HP, Ocean Optics) illuminates the cuvette and the transmitted light is collected by spectrometer (HR4000, Ocean Optics). The illumination and absorption of cuvette is conducted through fibers, which dramatically enhance the flexibility of this setup. The spectral information was recorded and analyzed by a program written in C++ and Matlab.
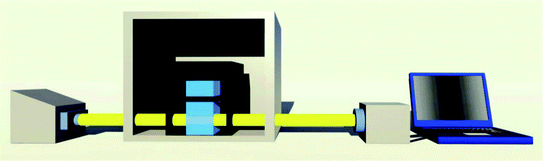 | ||
| Fig. 1 Schematic diagram of this custom made thermal controlled spectrometer. From left to right: tungsten halogen lamp, drying oven, cuvette, spectrometer and computer. | ||
Preparation of MoS2 NPs
The MoS2 NPs were prepared according to the mixed-solvent liquid exfoliation method reported previously.25 Briefly, 300 mg MoS2 powder was added into a 250 mL beaker, 100 mL of ethanol/water with ethanol volume fraction of 45% was added as dispersion solvent. The sealed beaker containing the above mixture was ultrasonicated for 24 h. In order to remove the aggregates, the obtained dark green suspension was centrifuged at 6000 rpm for 20 min three times. After that, collected supernatant was heated at 70 °C in a drying oven to remove the ethanol and water successively. Then the product was resolved in water and centrifuged for 20 min twice at 6000 rpm. At last, the supernatant was further purified by a 0.22 μm Millipore membrane filter. The final product was stored at 4 °C until required for further use.Detection of H2O2 and glucose
The detection of H2O2 was performed as follows: 0.3 mL of TMB solution (0.8 mM in ethanol) performed as peroxidase substrate and 0.3 mL H2O2 with different concentrations were added into 2.1 mL of acetate buffer solution (0.2 M, pH = 3.5) in a 4 mL cuvette. Then, 0.3 mL of MoS2 NPs solution (100 μg mL−1) was injected to the above mixture immediately, and the mixture was incubated at 30 °C for 20 min. The absorption of the mixture was recorded at 656 nm, which is a typical absorption wavelength of oxidized TMB (oxTMB).The detection of glucose was realized as follows: 0.03 mL of 5.0 mg mL−1 glucose oxidase (GOx) and 0.27 mL of glucose with different concentrations in 10 mM acetate buffer solution (pH 5.5) were incubated at 37 °C for 30 min to produce H2O2. The other procedure of detection was the same as that of H2O2.
In selectivity experiment, the target was replaced by sucrose (1.35 mM), lactose (1.35 mM) and fructose (1.35 mM) respectively during the experiment while the detection procedure was the same as that of glucose.
For the detection of glucose in human serum, the proteins in serum samples were separated by modified precipitation process according to the literature.18 Firstly, diluting 0.3 mL of serum with 0.2 mL water, then adding 0.5 mL Ba(OH)2 (0.11 M) and 0.5 mL ZnSO4 (0.0765 M) to above mixture. After centrifugation at speed of 4000 rpm for 15 min, 0.5 mL of the supernatant was extracted and diluted with 1.5 mL of acetate buffer solution (10 mM, pH 5.5). The other detection process was the same as the above mentioned glucose detection procedure.
Results and discussion
Characterization of MoS2 NPs
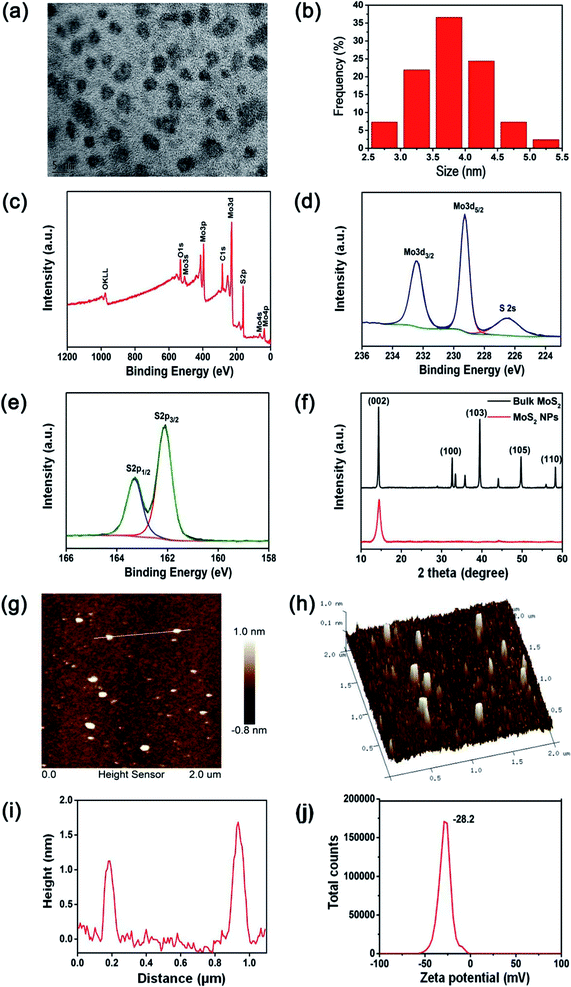
MoS2 NPs were obtained from the bulk MoS2 by the simple mixed-solvent liquid exfoliation method. The high resolution transmission electron microscopy (HRTEM) image of the resulting MoS2 NPs shown in Fig. 2(a) indicates that these NPs are spherical and well-dispersed, with an average diameter of 3.7 nm (Fig. 2(b)). The XPS spectrum shown in Fig. 2(c) demonstrates the Mo and S elemental peaks. And the high-resolution XPS spectra were collected to analyze the chemical states of Mo and S in MoS2 NPs. The peaks at 232.4 and 229.3 eV observed by the high-resolution spectrum of Mo (Fig. 2(d)) are attributed to Mo4+ 3d3/2 and Mo4+ 3d5/2, respectively. In addition, the peaks and 163.3 and 162.1 eV shown in the high-resolution spectrum of S (Fig. 2(e)) are corresponding to the S 2p1/2 and S 2p3/2 orbits of divalent sulfide ions (S2−).22,26 XRD was used to further investigate the crystal structure of the prepared MoS2 NPs and bulk MoS2 crystals. As shown in Fig. 2(f), both bulk MoS2 crystals and MoS2 NPs are mainly identified as 2H-MoS2, which has a dominant peak at 14.3°, representing the (002) plane (JCPDS 37-1492). In addition, the broadening (002) peak and the absence of most other peaks of MoS2 NPs provide a direct evidence of a decrease in the particle size.22,23 The thickness of the MoS2 NPs is around 1–2 nm, as shown in the atomic force microscopy (AFM) image and height profile of MoS2 NPs (Fig. 2(g)–(i)). The zeta potential of the MoS2 NPs was measured to be −28.2 mV (Fig. 2(j)). This indicates that MoS2 NPs exhibit a great colloidal stability in aqueous media.Peroxidase-like activities of MoS2 NPs
The peroxidase-like activity of MoS2 NPs was investigated in terms of the catalytic oxidation of the peroxidase substrate TMB in the presence of H2O2 under acidic conditions (pH = 4). According to previous literatures,18,20,26 the reaction could be characterized as the following process:Total: 2H+ + TMB + H2O2 → oxTMB2+ + 2H2O
(a) H2O2 → 2OH
(b) TMB + 2OH → oxTMB2+ + 2OH−
(c) 2H+ + 2OH− → 2H2O
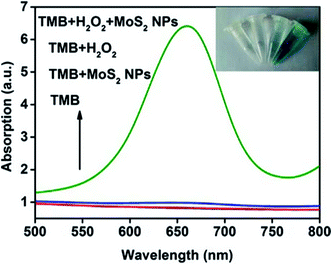
A series of absorption spectra for comparison is shown in Fig. 3. The absorption spectra of TMB and TMB–MoS2 NPs systems have no significant difference in the range between 500 nm and 800 nm. Although TMB–H2O2 system exhibits a little absorption in 656 nm, it could be ignored compared to spectrum collected from TMB–H2O2–MoS2 NPs system, which displays a strong absorption peak at 656 nm. This is a typical peak of the oxidation products of TMB. The inset images represent different systems after 10 min of reaction. TMB and TMB–MoS2 NPs systems remained colorless, which means no oxidation reaction occurred. TMB–H2O2 system shows a pale green color, which is due to the slow oxidation of TMB by the presence of H2O2. However, the TMB solution turns to be blue after the addition of MoS2 NPs and H2O2. These results suggest that the prepared MoS2 NPs have the peroxidase-like catalysis capability and could effectively catalyze the oxidation of TMB by H2O2. MoS2 NPs could facilitate the electron transfer between TMB and H2O2 in the oxidation of TMB catalyzed by MoS2 NPs. In this process, TMB molecules are absorbed on the surface of MoS2 NPs, and donate lone-pair electrons from the amino groups to MoS2 NPs, resulting in the increase in electron density and mobility in the MoS2 NPs. This will accelerate the electron transfer from the MoS2 NPs to H2O2, and promote the decomposition of H2O2 in acidic media into ·OH, which oxidizes TMB to form a blue product.18,20
Optimization of experimental conditions
The optimization of detection parameters, including the concentration of MoS2 NPs, temperature, pH and reaction time were investigated to enhance the catalysis capability of MoS2 NPs. Fig. S1(a)† shows the time-dependent absorption changes against the different concentrations of MoS2 NPs. The change in absorption approached the maximum when 10 μg mL−1 MoS2 NPs was utilized. The effect of temperatures on the catalytic activity was shown in Fig. S1(b).† It can be found that MoS2 NPs show good activity over a wide temperature range from 25 °C to 45 °C, and 30 °C was chosen as the optimal temperature for the subsequent experiments. In addition, the catalytic activity was also pH-dependent (Fig. S1(c)†) and the buffer solution with pH of 3.5 was employed for the further experiments. Finally, the effect of reaction time on the catalytic activity of MoS2 NPs was investigated, which was shown in Fig. S1(d).† Obviously, 20 min was the optimal reaction time because the maximum change in the spectra could be achieved and the absorption had little change from this time. Therefore, the optimal MoS2 NPs concentration, temperature, pH, and reaction time were selected which are 10 μg mL−1, 30 °C, 3.5, and 20 min, respectively, and they are used in the following experiments.Kinetic assay of peroxidase-like activity of MoS2 NPs
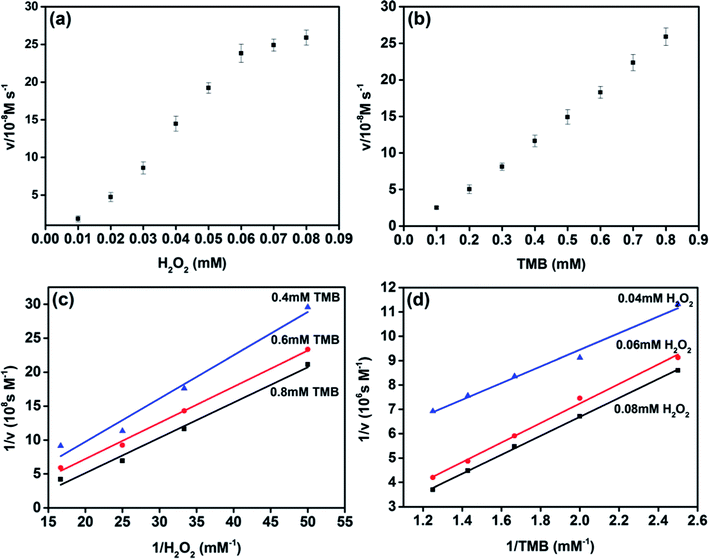
The peroxidase-like catalytic behavior of MoS2 NPs was studied by employing the steady-state kinetics. The kinetic parameters were harvested through altering the concentration of H2O2 or TMB while holding the other one invariable. The adsorption of MoS2 NPs monitored at the wavelength of 656 nm was recorded from 0–60 s during the peroxidase-like activity. The transient of adsorption can be fitted to the equation, I = vt + b, where v is the velocity characterized the adsorption change rate between the initial and final stage, b is the initial value of adsorption and t is the time. The maximum initial velocity (Vmax) and Michaelis–Menten constant (Km) shown in Table S1† were calculated using the Lineweaver–Burk plot, 1/v = (Km/Vmax) × (1/S) + 1/Vmax, where v is the initial velocity and S is the concentration of the substrate.27–30 Km usually indicates the affinity of the enzyme to the substrate. The smaller the Km is, the stronger the affinity will be.20,31,32The apparent Km value of MoS2 NPs with H2O2 as substrate was obviously lower than that of HRP, which indicates that MoS2 NPs exhibits higher affinity to H2O2 than that of HRP. The apparent Km value of MoS2 NPs with TMB as substrate was higher than that of HRP, is in accordance with the observation that a higher concentration of TMB was required to utilize to achieve the optimal activity of MoS2 NPs. To further investigate the catalysis mechanism of MoS2 NPs, the activity employing a wide range of TMB and H2O2 concentrations was carried out. The double-reciprocal plots of initial velocity versus different concentrations of one substrate were acquired while the concentration of this counterpart is set to be invariable (Fig. 4). The parallel slope of the lines reveals a ping-pong mechanism.18,20,33 In other words, the MoS2 NPs bond and react with the first substrate, and then release the first product before reacting with the second substrate.
Detection of H2O2 and glucose
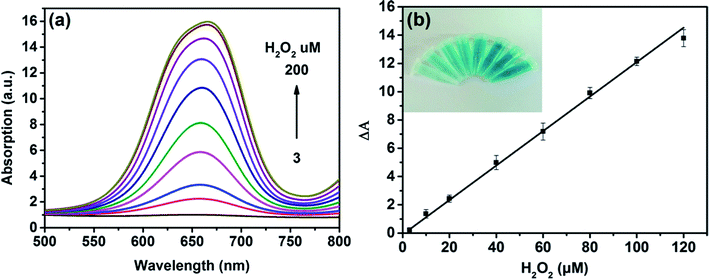
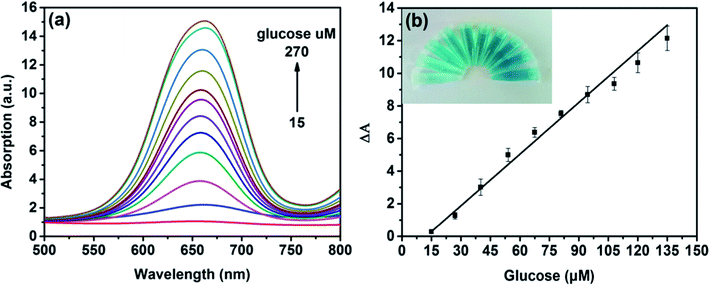
On the basis of the intrinsic peroxidase-like property of MoS2 NPs, a colorimetric approach for the detection of H2O2 and glucose was utilized under the optimal experimental conditions. Fig. 5(a) shows the absorption spectra of MoS2 NPs with different H2O2 concentration. It can be seen that the catalytic activity of MoS2 NPs is dependent on the concentration of H2O2. Fig. 5(b) exhibits the calibration plot for H2O2. It shows that the absorption of the mixture has a good linear relationship with the concentration of H2O2 ranging from 3 to 120 μM (R2 = 0.9983) and the detection limit is calculated to be 1.25 μM. A comparison of this approach with other colorimetric methods for H2O2 detection is listed in Table S2.† It is shown that by employing MoS2 NPs produced by this method, this sensing ability to H2O2 is comparable to other colorimetric methods.Since H2O2 is the main product during the GOx-catalyzed reaction, this sensing strategy can be employed for the determination of glucose when MoS2 NPs is combined with glucose oxidase (GOx). GOx can catalyze glucose oxidation to generate H2O2 and the following produced H2O2 can further oxidize TMB to bring a blue-color product through the catalysis of MoS2 NPs. Fig. 6(a) illustrates the absorption spectra of different concentrations of glucose from 15 to 270 μM. Fig. 6(b) shows the calibration curve for detecting glucose with a linear range from 15 to 135 μM. Moreover, the detection limit was found to be as low as 7 μM. Also, the LOD obtained by this method was much lower than the level of about 1 mM for glucose in human blood serum, indicating that the probe was suitable for detection of glucose in clinic.
The recovery was performed and the results are shown in Tables S4 and S5.† It can be seen that the recoveries are range from 95% to 99%. These results showed that this proposed method has a promising reliability for the detection of H2O2 and glucose.
Selectivity analysis for glucose detection
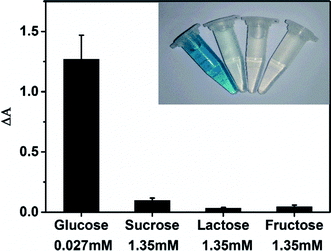
NaCl, KCl, NH4Cl etc. are utilized to test the selectivity of the MoS2 NPs-based sensing method towards H2O2. Mixture of 40 μM H2O2 and various concentrations of these impurities were measured and the results are shown in Table S6.† The negligible change in absorption of these mixtures compared to that collected from pure H2O2 indicates the presence of these impurities has little effect on the detection of H2O2.The selectivity of MoS2 NPs-based sensing method towards glucose was investigated in a series of control experiments. Some alternative sugars including sucrose, lactose and fructose were examined using the same detection procedure as that of glucose. Fig. 7 shows that no obvious interference is observed even the concentrations of other sugars are 50 times higher than that of glucose. Therefore, this MoS2 NPs produced here has a high selectivity for glucose detection because of the high specificity of GOx towards glucose.
Serum analysis
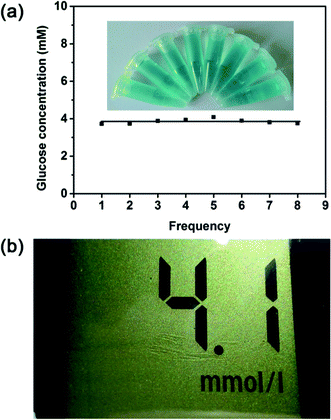
The glucose detection strategy reported here was then employed to detect the real samples to prove its practicability. Serums were selected as the detecting targets. On the basis of this method described in previous section, the glucose level in the human serum sample was tested. As shown in Fig. 8a, the glucose concentration of the serum sample is calculated to be 3.85 mM, which is not much different from the result obtained from the medical electronic glucometer (4.1 mM) (Fig. 8b). To confirm its suitability for real assay, another three different serum samples were used for comparison. As illustrated in Table S7,† the detection results of proposed method agree well with the detection results of glucometer. Therefore, this proves that this method initiated here is applicable for the glucose detection in human serum.The serum used in this paper was kindly donated from the Department of Clinical Laboratory at the University Town Hospital of Chongqing Medical University (Shapingba District, Chongqing Municipality) with the permission of the patients.
This donation and experiment conducted in study was in compliance with the regulations issued by the Chongqing Medical University.
Conclusions
MoS2 NPs in ultra-small size was synthesized by a simple facile liquid exfoliation method in the presence of ethanol/water mixture. This environmental friendly and economic fabrication method benefits from the utilization of ethanol/water mixture. A custom-made thermal controlled spectrometer has been established to analyze the intrinsic peroxidase-like activity of the prepared MoS2 NPs. The detection of H2O2 and glucose with high sensitivity and good selectivity by using this MoS2 NPs has been demonstrated and the MoS2 NPs exhibits a good performance to sense the serum. The potential of MoS2 NPs used as catalyst for other oxidation reactions could be carried out in the future research and this could create a new opportunity for this enzyme-mimicking MoS2 NPs in various significant fields.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is sponsored by the National Natural Science Foundation of China (Grant no. 21707137), the Key Application and Development Program of Chongqing (No. cstc2017shmsA0159), Research Funding Project of Yangtze Normal University (No. 2017KYQD23), Young Scientist Research Fund of Yangtze Normal University, Open Research Fund Program of Key Laboratory of Reservoir Aquatic Environment of Chinese Academy of Sciences.References
- V. Sanz, M. S. De, J. R. Castillo and J. Galbán, J. Am. Chem. Soc., 2005, 127, 1038 CrossRef CAS PubMed.
- C. Xu, J. Ren, L. Feng and X. Qu, Chem. Commun., 2012, 48, 3739 RSC.
- T. Wang, H. Zhu, J. Zhuo, Z. Zhu, P. Papakonstantinou, G. Lubarsky, J. Lin and M. Li, Anal. Chem., 2013, 85, 10289 CrossRef CAS PubMed.
- Z. G. Wang, O. I. Wilner and I. Willner, Nano Lett., 2009, 9, 4098 CrossRef CAS PubMed.
- J. Chen, Q. Chen, J. Chen and H. Qiu, Microchim. Acta, 2016, 183, 3191 CrossRef CAS.
- Y. Tao, E. Ju, J. Ren and X. Qu, Chem. Commun., 2014, 50, 3030 RSC.
- Q. Chen, J. Chen, C. Gao, M. Zhang, J. Chen and H. Qiu, Analyst, 2015, 140, 2857 RSC.
- C. Gao, H. Zhu, J. Chen and H. Qiu, Chin. Chem. Lett., 2017, 28, 1006 CrossRef CAS.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577 CrossRef CAS PubMed.
- J. Mu, Y. Wang, M. Zhao and L. Zhang, Chem. Commun., 2012, 48, 2540 RSC.
- Y. Jv, B. Li and R. Cao, Chem. Commun., 2010, 46, 8017 RSC.
- W. Zhang, D. Ma and J. Du, Talanta, 2014, 120, 362 CrossRef CAS PubMed.
- W. Shi, Q. Wang, Y. Long, Z. Cheng, S. Chen, H. Zheng and Y. Huang, Chem. Commun., 2011, 47, 6695 RSC.
- J. Xie, H. Zhang, S. Li, R. Wang, X. Sun, M. Zhou, J. Zhou, X. W. Lou and Y. Xie, Adv. Mater., 2013, 25, 5807 CrossRef CAS PubMed.
- M. Chhowalla, Z. Liu and H. Zhang, Chem. Soc. Rev., 2015, 44, 2584 RSC.
- Q. H. Wang, K. Kalantar-Zadeh, A. Kis, J. N. Coleman and M. S. Strano, Nat. Nanotechnol., 2012, 7, 699 CrossRef CAS PubMed.
- J. N. Coleman, M. Lotya, A. O'Neill, S. D. Bergin, P. J. King, U. Khan, K. Young, A. Gaucher, S. De and R. J. Smith, Science, 2011, 42, 568 CrossRef PubMed.
- T. Lin, L. Zhong, L. Guo, F. Fu and G. Chen, Nanoscale, 2014, 6, 11856 RSC.
- X. Guo, Y. Wang, F. Wu, Y. Ni and S. Kokot, Analyst, 2015, 140, 1119 RSC.
- K. Zhao, W. Gu, S. Zheng, C. Zhang and Y. Xian, Talanta, 2015, 141, 47 CrossRef CAS PubMed.
- J. Yu, X. Ma, W. Yin and Z. Gu, RSC Adv., 2016, 6, 81174 RSC.
- B. L. Li, H. Q. Luo, J. L. Lei and N. B. Li, RSC Adv., 2014, 4, 24256 RSC.
- Z. Sun, Q. Zhao, G. Zhang, Y. Li, G. Zhang, F. Zhang and X. Fan, RSC Adv., 2015, 5, 10352 RSC.
- K. G. Zhou, N. N. Mao, H. X. Wang, Y. Peng and H. L. Zhang, Angew. Chem., 2011, 50, 10839 CrossRef CAS PubMed.
- L. Jia, L. Ding, J. Tian, L. Bao, Y. Hu, H. Ju and J. S. Yu, Nanoscale, 2015, 7, 15953 RSC.
- J. Lei, X. Lu, G. Nie, Z. Jiang and C. Wang, Part. Part. Syst. Charact., 2015, 32, 886 CrossRef CAS.
- L. Ai, L. Li, C. Zhang, J. Fu and J. Jiang, Chem.–Eur. J., 2013, 19, 15105 CrossRef CAS PubMed.
- L. Su, J. Feng, X. Zhou, C. Ren, H. Li and X. Chen, Anal. Chem., 2012, 84, 5753 CrossRef CAS PubMed.
- X. Sun, S. Guo, C. S. Chung, W. Zhu and S. Sun, Adv. Mater., 2013, 25, 132 CrossRef CAS PubMed.
- L. Zhang, L. Han, P. Hu, L. Wang and S. Dong, Chem. Commun., 2013, 49, 10480 RSC.
- M. A. Lukowski, A. S. Daniel, F. Meng, A. Forticaux, L. Li and S. Jin, J. Am. Chem. Soc., 2013, 135, 10274 CrossRef CAS PubMed.
- Q. Wang, J. Lei, S. Deng, L. Zhang and H. Ju, Chem. Commun., 2013, 49, 916 RSC.
- D. J. Porter and H. J. Bright, J. Biol. Chem., 1983, 258, 9913 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra12584d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |